Abstract
The derivation of follicular lymphomas (FLs) from germinal centers is not only supported by their morphologic appearance with a nodular growth pattern and a germinal center–like cellular composition, but also by the presence of ongoing somatic hypermutation (a germinal center B cell–specific process) during their clonal expansion. The intraclonal sequence diversity of the tumor cells and their follicular growth pattern allows one to analyze lymphoma cell dissemination and the way the tumor “metastasizes” to distinct follicles. In the present study, we analyzed individual follicles of 3 FLs by micromanipulation of single cells from individual lymphoma follicles and amplification of immunoglobulin V region genes. Genealogical trees for the VH and the VL gene rearrangements were constructed to analyze the clonal relationship among individual cells of 3 distinct follicles of each case. In all 3 cases there is evidence that distinct tumor follicles are founded by many tumor cells, suggesting that there is extensive migration of the tumor cells among follicles. The observation that the tumor cells of FLs retain their follicular growth patterns despite this cellular migration supports the idea that they depend on the follicular microenvironment for their clonal expansion.
Introduction
Follicular lymphoma (FL) is one of the most common B-cell lymphomas in Europe and the United States, accounting for up to 40% of all non-Hodgkin lymphomas. It is a low-grade lymphoma with a median survival of 8 to 10 years after diagnosis.1-4 A majority of the cases (80% to 90%) are characterized by the t(14;18)(q32;q21) chromosomal translocation, resulting in overexpression of the proto-oncogene bcl-2 and protecting the cell against apoptosis.5-8 FLs show a characteristic nodular growth pattern resembling the architecture of reactive nonmalignant germinal centers (GCs) in secondary lymphoid organs. The cellular composition of FLs is also similar to that of reactive GCs, consisting of a mixture of centrocytelike and centroblastlike cells, reactive T cells, follicular dendritic cells, and a few macrophages.9 10
The follicular growth pattern of the lymphoma and the morphologic similarity of the tumor cells in FL with normal GC centroblasts and centrocytes indicates a derivation of FLs from GC B cells. This is also supported by molecular analysis of rearranged immunoglobulin (Ig) genes in FL cells. In the GC reaction, Ig genes of GC B cells undergo the process of somatic hypermutation, which introduces mutations at a high rate into the Ig variable (V) region genes in the course of clonal expansion of the cells.11 On this background, the observation that FLs carry somatically mutated Ig genes and show intraclonal sequence diversity suggests that the hypermutation machinery is active in the FL cells, and hence indicates a derivation of the lymphoma cells from transformed GC B cells.12 13
As the clonal tumor cell population in FL is the descendent of a single B cell, the establishment of the malignant clone presumably started with a single transformed GC B cell. This founder cell of the tumor clone likely first proliferated in a GC where it underwent malignant transformation, before descendents of the clone either migrated to neighboring B cell follicles and/or established novel follicles in the involved lymph node. In most tumors, the dissemination of the malignant clone in the tissue cannot be studied because of lack of appropriate markers, so that details of tumor cell dissemination are largely unknown in most malignant diseases. However, the follicular growth pattern of FLs defines distinct histologic tumor cell subsets, and the intraclonal sequence diversity of the lymphoma cells represents a suitable and unique marker to analyze the way in which the tumor clone “metastasizes” to distinct follicles.
In the present analysis, we studied lymphoma cell dissemination in FL by isolating single tumor cells from 3 distinct follicles from each of 3 cases. By amplification and sequence analysis of rearranged Ig heavy and light chain V genes, the somatic mutation pattern was used to delineate the distribution of distinct members of the tumor clone among separate follicles and hence the migration of the FL cells among follicles.
Patients, materials, and methods
Patients and tissues
Patient 1 was a 45-year-old male, patient 2 a 47-year-old female, and patient 3 a 50-year-old male. In all patients, lymph node biopsies were performed for diagnostic purposes and FL grade I was diagnosed (see “Results”).
Approval was obtained from the Instiutional Review Board for these studies. Informed consent was provided according to the Declaration of Helsinki.
Immunostaining and micromanipulation
For diagnostic purposes, 3-μm-thick sections of paraffin-embedded tissue of the 3 cases were stained with mouse anti–bcl-2 antibody (monoclonal; Dako, Hamburg, Germany). Frozen tissue sections, 5-μm- to 10-μm-thick, were stained with mouse anti-CD20 (monoclonal; Dako) or rabbit anti-CD3 (polyclonal; Dako) antibodies for the isolation of FL B cells and reactive T cells, respectively. Anti-CD3 staining was followed by an incubation step with mouse anti–rabbit immunoglobulin (Dako). Monoclonal biotinylated rabbit-anti–mouse antibody (Dako) was used for both CD3- and CD20-staining as secondary antibody, followed by an incubation step with Streptavidin-bound alkaline phosphatase (StreptABcomplex/AP; Dako). Bound alkaline phosphatase was visualized with Fast Red. Stained single cells were dissected from surrounding tissue with a UV-Laser beam (Palm, Wolfratshausen, Germany) to isolate the cells. The sections were then overlaid with Tris-buffered saline and cells were aspirated with micropipettes and the help of a hydraulic micromanipulator under a microscope as described.11 Aspirated cells were transferred to polymerase chain reaction (PCR) tubes containing 20 μL Expand high-fidelity PCR buffer (Boehringer Mannheim, Germany) and stored at −20°C. From each section used for micromanipulation, aliquots of the buffer covering the section were aspirated and analyzed in parallel to the B cells as negative controls. Furthermore, CD3+ T cells were micromanipulated from adjacent tissue sections of the same cases as additional negative controls.
DNA isolation from tissue sections
For DNA isolation, QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) was used. Tissue sections, 20-μm- to 30-μm-thick, were digested overnight with 40 μL proteinase K solution (20 mg/mL) and 360 μL ATL (QIAmp Tissue Lysis) buffer at 55°C. From this solution, DNA was extracted and dissolved in H2O at a concentration of 100 μg/mL. A quantity of 10 μL of this solution was used in each amplification reaction.
Whole-tissue and single-cell PCR
The clonal VH and VL gene rearrangements of the FL clones were first identified by whole-tissue DNA PCR using either family–specific VH leader or framework region (FR) I primers and Vκ or Vλ FR I family-specific primers together with the respective J gene segment primers.11 14-16 In case 2, the clonal Vκrearrangement was only identified in the single-cell analysis, perhaps due to somatic mutations at the primer binding sites that reduced the amplification efficiency in the whole-tissue PCR below detection level.
Rearranged Ig genes from single cells were amplified in a seminested PCR using primers for VH and Vκ or Vλ genes together with the respective J segment primers. In the second round of amplification, 1.5 μL aliquots of the first round were further amplified with the same V gene primers in separate reactions and internal J segment primer mixes.11,15 16 B cells and control T cells were analyzed in parallel, together with buffer controls.
In the single-cell analyses, the first 15 cells of each case were analyzed with all VH and VL FR I primers, with the exception of 2 VH rearrangements where the whole-tissue PCR with VH leader primers identified mutations at the VH FR I primer binding sites. For those VHrearrangements, rearrangement-specific primers binding to sequences in the respective VH leader introns were used (case 1: 5′AGTGAATATGTGTG(AG)CAGTTTCTG3′ [2 mutations different from germline], case 3: 5′GTTGTTGCGCTTGCAGGTGTCC3′ [4 mutations different from germline]). The VH leader primers were not used for single-cell PCR, because longer PCR products are often amplified with lower efficiency. All V genes amplified from these cells showed the clonal rearrangements identified with the whole-tissue DNA PCR. The remaining cells were analyzed only with the family-specific VH and VL primers recognizing the tumor-specific rearrangements and the corresponding J primer mixes.
Sequence analysis and construction of genealogical trees
PCR products were gel-purified and directly sequenced on automatic sequencers (ABI377 and 3100; Applied Biosystems, Weiterstadt, Germany). Sequences were analyzed using the EMBL IMGT (international imMunoGenetics database) (www.genetik.uni-koeln.de/dnaplot/) and Genbank databases. Based on multiple sequence alignment obtained with the clustal W program of MacVector software 7.0 (Accelrys, München, Germany), genealogical trees for the VH and the VL gene rearrangements were constructed to analyze the clonal relationship among the individual cells of the 3 different follicles of each case. In some of the trees, modifications were made (ie, either by introducing backmutations or by assuming that some identical nucleotide exchanges happened independently in distinct cells) to optimize the congruence between the VH and the VL trees or to correct obvious mistakes in the trees generated by the software (eg, not discriminating 2 distinct mutations at the same position in separate cells; see figure legends for additional explanations).
Results
Histologic analysis of the cases
The lymph nodes of the 3 patients showed a histologic picture typical of FL with only a few centroblastlike cells (FL grade I). In case 3, the tumor showed both a follicular and diffuse growth pattern. In all cases, immunohistochemistry revealed positivity of the tumor cells for CD20 and Bcl- 2.
Amplification and sequence analysis of the tumor-derived V gene rearrangements
For the identification of the VH and VLgene rearrangements carried by the FL cells, whole-tissue DNA PCR was carried out with V gene family–specific primers and the respective J gene primers, and the resulting PCR products were directly sequenced. In case 1 and case 3, potentially functional VH and VL gene rearrangements were identified in the whole-tissue PCR. In case 2, a productive VH gene was amplified in the whole-tissue PCR, whereas the clonal Vκ gene was amplified only in the single-cell analysis (Table1; the data for the clonal Vκ rearrangement of case 2 is also presented here). All rearrangements were somatically mutated (Table 1). From the FL of patient 1, a VH3-23 rearrangement with 13.6% mutation frequency and a VκB3 rearrangement with 6.8% mutation frequency were obtained. Case 2 carries a VH3-23 and a VκL2 rearrangement, with 6.7% and 3.1% mutation frequencies, respectively. The VH gene of case 3 uses the VH3-49 gene with 10.5% mutation, the light chain gene rearrangement uses the Vλ3r gene and shows 11.2% mutation frequency.
Analysis of mutation pattern
The ratio of replacement (R) to silent (S) mutations in the FRs of functional V gene rearrangements is an indicator whether cells have been under selective pressure for expression of an antigen receptor. In that case, R mutations are counterselected in the FRs to preserve the evolutionary optimized structure of the antibody V domain. In the absence of selection, like in out-of-frame rearrangements, R mutations in the FRs are not counterselected. The R/S ratios of the mutations within the FRs in productive rearrangements of various B-cell subsets with mutated V gene rearrangements have been shown to be in a range between 1.0 and 1.6, whereas for a collection of nonfunctional out-of-frame rearrangements, the ratio is 3.0, the value expected assuming random mutagenesis.17 18 The 79 shared mutations in the FRs of the 6 potentially functional VH and VL rearrangements amplified from the 3 FLs showed an R/S ratio of 1.1, which is in the range typical for antigen-selected cells.
PCR analysis of single micromanipulated cells for VH, Vκ, and Vλ gene rearrangements
For each patient, a total of 150 FL B cells from 3 different follicles were micromanipulated (50 cells per follicle, Figure1) and analyzed for VH and VL gene rearrangements. The first 15 cells were analyzed with primers for all VH and VL families with the exception of 2 VH rearrangements where the whole-tissue PCR with VH leader primers identified mutations at the VH FR I primer binding sites, so that gene-specific primers were designed for these rearrangements. All V genes amplified from these cells corresponded to the clonal rearrangements identified in the whole-tissue PCR, confirming that the follicles chosen for cell isolation represent tumor follicles. Therefore, the remaining cells were analyzed only with the primers recognizing the tumor-specific rearrangements.
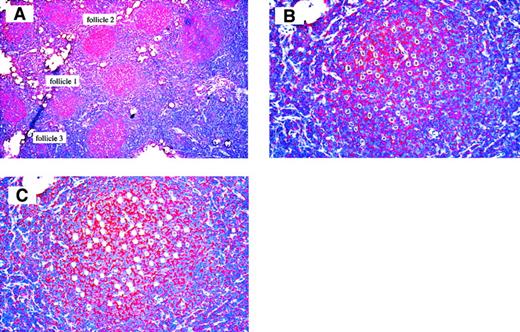
Frozen section of case 3 used for micromanipulation.
Anti-CD20 antibody staining, hematoxylin counterstain. (A) 150 FL B cells from 3 different follicles were micromanipulated (50 cells per follicle), 50 × magnification. (B) Follicle 1 after dissection from surrounding tissue with UV laser beam, 200 × magnification, and (C) after aspirating the cells with micropipettes, 200 × magnification.
Frozen section of case 3 used for micromanipulation.
Anti-CD20 antibody staining, hematoxylin counterstain. (A) 150 FL B cells from 3 different follicles were micromanipulated (50 cells per follicle), 50 × magnification. (B) Follicle 1 after dissection from surrounding tissue with UV laser beam, 200 × magnification, and (C) after aspirating the cells with micropipettes, 200 × magnification.
For patient 1, the clonal VH and VLrearrangements were obtained from 30 and 25 of the cells, respectively (Table 2). One cell isolated from follicle 3 harbored a unique VH3-11 rearrangement, and 2 cells with an identical unmutated VH3-23 and VκB3 rearrangement were distinct from the tumor clone. These cells likely represent members of a normal B cell clone present in that follicle. For patient 2, the clonal VH gene rearrangement was amplified from 71 and the clonal Vκ rearrangement from 55 of the cells (Table 2). For patient 3, the clonal VH gene rearrangement was amplified from 37 and the clonal Vλ rearrangement from 30 of the cells (Table 2).
For each of the 3 cases, 60 T cells were micromanipulated and 60 aliquots of buffer covering the sections during micromanipulation were aspirated. These samples were analyzed as negative controls in parallel to the FL B cells (Table 2). All these controls were negative in case 1. In case 2, the clonal VH rearrangement was amplified from 3 T cell samples and one buffer control, and the clonal Vκ rearrangement was obtained from 4 T cell samples. In case 3, the clonal VH and Vλ genes were each amplified once from a T cell sample and a buffer control. Since the frequency of these contaminations is much lower than the frequency of rearrangements amplified from the isolated FL cells (Table 2), their rare occurrence does not hamper the reliability and interpretation of the results.
Intraclonal V gene diversity
For each of the 6 clonal rearrangements amplified from the 3 FLs, intraclonal sequence diversity was detected in the single-cell analysis (Table 1; Figures2-4). In case 1, 19 sequence variants among 30 sequences of the VH rearrangement and 12 sequence variants among 25 sequences of the Vκ rearrangement were found. In case 2, 44 sequence variants were observed among 71 sequences of the VH rearrangement and 17 sequence variants among 55 sequences of the Vκ rearrangement. In case 3, 11 sequence variants were observed among 37 VH sequences and 13 sequence variants among 30 Vλ sequences.
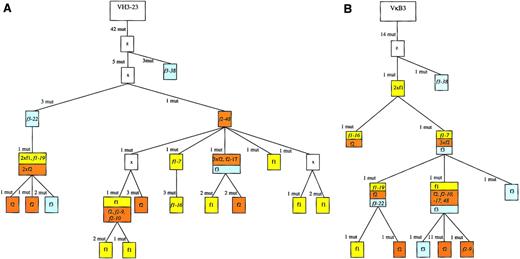
Genealogical trees for the VH and VL rearrangements of case 1 to analyze the clonal relationship among the individual cells of the 3 different follicles.
Genealogical trees are based on multiple sequence alignments obtained with the MacVector software 7.0 (Accelrys). “z” denotes the presumed precursers. Assumed intermediates are included in the genealogical tree and marked as “x.” When a branch in the tree is defined by more than one mutation, it may well be that they occurred in separate steps. However, this would not change the basic structure of the tree and the relationship of the distinct cells identified. Cells from the same follicle are marked with a follicle-specific color: follicle 1, yellow; follicle 2, orange; and follicle 3, blue. There were 9 cells with a VH as well as a Vκ gene amplificate (the names of these cells are given in the trees). We made some corrections of the VH tree generated by the MacVector software 7.0: at 2 positions the software regarded different point mutations habored by all cells at the same position of the sequences as a shared mutation (97 C to A or 97 C to G, 277 C to A or 277 C to G; see sequences available from the EMBL data library). As these were clearly distinct mutation events, the tree was accordingly corrected, including introduction of one backmutation into 2 VHsequences at position 97 (the most right branch with 2 f1 cells). There were 2 point mutations shared by 2 sequences that were regarded as independent mutation events; importantly, the number of mutation events remained the same with this modification. The branch with f2-9 and f2-10 could also originate from f3-22, but to aquire congruence with the Vκ tree origination from f2-48 with the same number of mutations was assumed. Additional details are available from the authors upon request. f1 indicates follicle 1; f2, follicle 2; f3, follicle 3; mut, mutation.
Genealogical trees for the VH and VL rearrangements of case 1 to analyze the clonal relationship among the individual cells of the 3 different follicles.
Genealogical trees are based on multiple sequence alignments obtained with the MacVector software 7.0 (Accelrys). “z” denotes the presumed precursers. Assumed intermediates are included in the genealogical tree and marked as “x.” When a branch in the tree is defined by more than one mutation, it may well be that they occurred in separate steps. However, this would not change the basic structure of the tree and the relationship of the distinct cells identified. Cells from the same follicle are marked with a follicle-specific color: follicle 1, yellow; follicle 2, orange; and follicle 3, blue. There were 9 cells with a VH as well as a Vκ gene amplificate (the names of these cells are given in the trees). We made some corrections of the VH tree generated by the MacVector software 7.0: at 2 positions the software regarded different point mutations habored by all cells at the same position of the sequences as a shared mutation (97 C to A or 97 C to G, 277 C to A or 277 C to G; see sequences available from the EMBL data library). As these were clearly distinct mutation events, the tree was accordingly corrected, including introduction of one backmutation into 2 VHsequences at position 97 (the most right branch with 2 f1 cells). There were 2 point mutations shared by 2 sequences that were regarded as independent mutation events; importantly, the number of mutation events remained the same with this modification. The branch with f2-9 and f2-10 could also originate from f3-22, but to aquire congruence with the Vκ tree origination from f2-48 with the same number of mutations was assumed. Additional details are available from the authors upon request. f1 indicates follicle 1; f2, follicle 2; f3, follicle 3; mut, mutation.
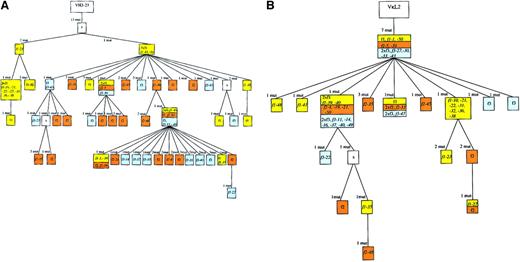
Genealogical trees for the VH and the Vκ rearrangements of case 2.
There were 38 cells with a VH as well as a Vκgene amplificate (the names of the cells are given in the tree). For the VH tree we made one correction of the tree generated by the software: one point mutation (71 C; see sequences available from the EMBL data library) was regarded as a shared mutation, although a group of cells had no mutation at this position. Consequently, we included 71 C as a mutation event in the construction of the genealogical tree. For the Vκ tree we made some modifications of the MacVector trees to aquire congruence of heavy and light chain genes: we observed a point mutation shared by 21 sequences incompatible regarding the construction of the VH- and Vκ-based trees (see the 3 branches with f3-47, f1-21, and the branch with the assumed intermediate originating from f1-39). Therefore, it was assumed that this mutation happened 3 times independently. F3-22 was assumed to originate from f1-39, although the software-generated tree suggested another position. Importantly, the number of mutation events remained the same for this modification. For f1-27, f1-35, and f2-48, one backmutation was introduced in each of the Vκ sequences. See Figure 2 for additional explanations.
Genealogical trees for the VH and the Vκ rearrangements of case 2.
There were 38 cells with a VH as well as a Vκgene amplificate (the names of the cells are given in the tree). For the VH tree we made one correction of the tree generated by the software: one point mutation (71 C; see sequences available from the EMBL data library) was regarded as a shared mutation, although a group of cells had no mutation at this position. Consequently, we included 71 C as a mutation event in the construction of the genealogical tree. For the Vκ tree we made some modifications of the MacVector trees to aquire congruence of heavy and light chain genes: we observed a point mutation shared by 21 sequences incompatible regarding the construction of the VH- and Vκ-based trees (see the 3 branches with f3-47, f1-21, and the branch with the assumed intermediate originating from f1-39). Therefore, it was assumed that this mutation happened 3 times independently. F3-22 was assumed to originate from f1-39, although the software-generated tree suggested another position. Importantly, the number of mutation events remained the same for this modification. For f1-27, f1-35, and f2-48, one backmutation was introduced in each of the Vκ sequences. See Figure 2 for additional explanations.
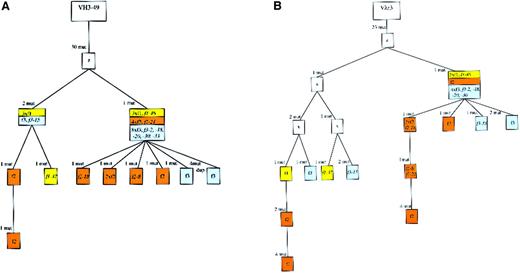
Genealogical trees for the VH and the Vλ rearrangements of case 3.
There were 11 cells that carried a VH as well as a Vλ gene amplificate (the names of the cells are given in the trees). In the Vλ tree generated by the software, one mutation (either 200 G to A or 200 G to C in all sequences; see sequences available from the EMBL data library) was erroneously regarded as a shared mutation. This was corrected, defining the mutations as distinct events. Furthermore, we positioned f1-37 into the left main branch of the Vλ tree to get compatibility in the heavy and light chain genealogical trees. Dup indicates duplication. See Figure 2 for additional explanations.
Genealogical trees for the VH and the Vλ rearrangements of case 3.
There were 11 cells that carried a VH as well as a Vλ gene amplificate (the names of the cells are given in the trees). In the Vλ tree generated by the software, one mutation (either 200 G to A or 200 G to C in all sequences; see sequences available from the EMBL data library) was erroneously regarded as a shared mutation. This was corrected, defining the mutations as distinct events. Furthermore, we positioned f1-37 into the left main branch of the Vλ tree to get compatibility in the heavy and light chain genealogical trees. Dup indicates duplication. See Figure 2 for additional explanations.
The sequence variants differed from each other by few to many distinct mutations (between 1 to 17). Consequently, there was considerable variation in the mutation load of individual cells (Table 1). For example, in case 1 the mutation load of the Vκ genes varied between 7.3% to 13.7%. Most of the sequence variants of the 3 cases are due to single nucleotide exchanges. Only in case 3 a duplication of 3 base pairs in the VH gene of one cell was observed. None of these diversifying point mutations and the duplication in the potentially functional rearrangements rendered a gene nonfunctional, indicating that the tumor clone was still under selection to preserve antibody expression. In line with this view, the overall R/S value in the framework regions for the nonshared mutations is below 1.2 (43 R/36 S).
Construction of genealogical trees
For each case, a genealogical tree for the VH and the VL gene rearrangements was constructed to demonstrate the clonal relationship among the individual cells of the 3 different follicles (Figures 2-4). The genealogical trees are based on the clustal W alignment obtained with MacVector software 7.0 (see “Patients, materials, and methods” and the figure legends for additional explanations).
All 6 genealogical trees demonstrate that the cells of the 3 individual follicles (marked with the same “follicle-specific” color in each case, Figures 2-4) were distributed diffusely among the branches of the trees. Members of distinct subclones in the tree were usually found in 2 or 3 of the follicles. No indication was found that one of the follicles was founded by a single or few FL cells which then expanded and mutated restricted to that follicle. There was only in case 2 one subclone of 13 cells in the VH tree that was restricted to follicle 1 and in the Vλ tree of case 3 one subclone of 6 cells restricted to follicle 2. However, follicle 1 in case 2 and follicle 2 in case 3 also harbored cells belonging to several other branches of the genealogical tree. Thus, taking the results of the 3 cases together, the V gene mutation patterns reveal considerable cell migration among follicles in the course of clonal expansion of the FL clones.
Discussion
The classification of FL as a GC B cell tumor is not only supported by the follicular growth pattern and the morphologic appearance of the lymphoma cells, but also by the regular detection of ongoing somatic hypermutation,12,19 which represents a GC B cell–specific process.11 Intraclonal V gene diversity was indeed observed for each of the 6 clonal rearrangements amplified from the 3 cases. As also observed by others,20 there was variation of the frequency of sequence variants, ranging from 11 in 37 (30%) sequences for the VH gene of case 3, to 19 in 30 (63%) for the VH gene of case 1. Nevertheless, due to the large number of cells analyzed, informative genealogical trees could also be generated for the rearrangements with less variability.
In the construction of the trees it has to be considered that identical mutations found in separate follicles may have happened independently. Thus, there may have been a parallel development of the same mutations in separate follicles instead of cell migration among follicles, perhaps because of intrinsic mutation hot spots and/or selection of the tumor cells for recognition of a particular antigen. Indeed, to optimize the congruence of the VH and VL trees, independent occurence of some identical mutations in distinct cells was assumed (see the legends to Figures 2-4). However, there are several arguments why it is unlikely that there was significant parallel development of identical mutations in separate follicles. First, nearly half (75 R, 57 S) of the mutations are silent, and hence are not selectable. Second, only 37.7% of the nonclonal mutations shared by at least 2 cells are found at the main hypermutation hot spot in human V region genes, namely the RGYW/WRCY motif, a value similar to what is reported in the literature.21 Moreover, even typical hot spot mutations found at the same position in distinct cells may often derive from a single mutation event. Finally, some of the main branches are defined by more than one mutation, and/or are identified in the VH as well as in the respective VL trees. A parallel development of multiple mutations, however, appears highly unprobable. Taken together, although we cannot exclude that some identical mutations that occurred independently may have remained undetected, it is likely that the frequency of such events is low and that the trees therefore reliably reflect the evolution of the intraclonal V gene diversification.
FLs grow in a follicular pattern, thereby defining spatially separated histologic subsets of the tumor clone, and the intraclonal V gene diversity allows one to define the genealogical relationship of members of the tumor clone. On this basis, a single-cell analysis of FL cells isolated from distinct follicles in the tissue represents an attractive model to study tumor cell dissemination. More specifically, we wanted to address the questions of whether distinct tumor follicles are founded by single, few, or many FL cells, and of whether there is evidence for considerable migration of tumor cells among follicles. As the present analysis revealed, there is indeed indication for extensive migration of FL cells among follicles. Cells belonging to branches in the genealogical trees with multiple members were usually found in each of the 3 follicles analyzed. In several instances members of branches in the genealogical trees were found in a single follicle (eg, the most left branch in Figure 3A). This is not unexpected, as the FL B cells continue to mutate while proliferating within the follicles. However, all 9 follicles harbored members of several separate branches in the genealogical trees (Figures 2-4). Hence, it appears that FLs are characterized by extensive tumor cell traffic among follicles in the course of the clonal expansion of the lymphoma cells and that each follicle is composed of various members of the tumor clone stemming from several distinct branches in the genealogical trees. Based on these features, it is likely that the “metastatic” dissemination of FL cells to new follicles is a process involving many tumor cells and not only rare members of the tumor clone that perhaps possess particular migration properties.
While the present analysis is focused on tumor cell dissemination among follicles in a given lymph node, in some cases of FL mutation patterns among distinct lymph nodes showing FL infiltration were compared.22,23 These studies did not reveal any distinct mutation patterns in the separate lymph nodes, indicating that many cells of the FL clone are involved in tumor cell metastasis—in line with the findings of the present investigation. Whether disseminating FL cells normally establish novel follicles or migrate to established reactive follicles was recently analyzed by Su et al.24Their work supports the latter idea, as evidence for the regular presence of reactive B cells in FL follicles was found. In our investigation, the presence of reactive tissue was not systematically analyzed, because 90% of the cells were analyzed only with the tumor clone specific primers. It is noteworthy that FL cells do not only migrate among follicles, since members of the tumor clone can occasionally also be found in interfollicular areas, where they acquire a phenotype resembling memory B cells.22
The migratory features of the FL cells appear to distinguish them from normal GC B cells. Although there are not many data published on this issue, studies in the mouse indicate that each GC represents a distinct entity and that there is little if any exchange of cells among these structures.25 This may be advantagous in a normal immune response to allow for the selection of many different and clonally unrelated memory B cells expressing Ig receptors with increased affinity to the immunizing antigen, thereby establishing a high-affinity yet diverse memory B cell population.
While the extensive cell traffic among FL follicles shows that the follicular growth pattern is not due to migratory inabilities of the lymphoma cells, the growth of the tumor clone in distinct follicles underscores the idea that FL cells depend on the follicular microenvironment for their survival and clonal expansion.26-28 Therapeutical strategies targeting the interaction of FL cells with their follicular microenvironment might therefore represent attractive approaches to fight tumor progression.
We thank Ekaterini Hadzoglou for excellent technical assistance.
Supported by the Deutsche Krebshilfe, Dr Mildred Scheel Stiftung Bonn, Germany to M.-L.H., and a Heisenberg Award, Deutsche Forschungsgemeinschaft, Bonn, Germany to R.K.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sabine Oeschger, University of Frankfurt, Senckenberg Department of Pathology, Theodor-Stern-Kai 7, D-60590 Frankfurt am Main, Germany; e-mail: oeschger@em.uni-frankfurt.de.