Despite recent studies, the organization of coagulation factor VIII (FVIII) on a phospholipid (PL) membrane is not known in detail. Thus, 2-dimensional (2D) crystals of human FVIII lacking the B domain were prepared for electron microscopy onto negatively charged PL monolayers. The 3-dimensional (3D) density map of the PL-bound FVIII protein was calculated at 1.5 nm. Existing atomic data and models for FVIII domains were fitted unambiguously within the 3D density map of the molecule. FVIII domains arrangement followed a compact spiral organization with the A3 domains in close association with the C1 and C2 domains near the PL surface. Viewed toward the membrane the A domains' heterotrimer is oriented side-on with the pseudo–3-fold axis almost parallel to the PL surface and A1 fully covering C1. The C2 domain is partially overlapped by the A2 domain of an adjacent molecule in the 2D crystal, favoring close packing. Viewed parallel to the membrane, C2 is slightly inclined to the PL surface covering an area of 12 nm2. Four C2 loops are embedded within the lipid monolayer at about 0.7 to 1.0 nm depth. C1 forms almost a right angle with C2, its long axis nearly parallel to the membrane. The proposed structure for membrane-bound FVIII results from modeling of the FVIII domains within a 3D density map obtained from electron crystallography and accords with the main biochemical and structural information known to date. A model is proposed for FVIIIa and factor IXa assembly within the membrane-bound factor X-activating complex.
Introduction
Factor VIII (FVIII) is a plasma protein essential for blood coagulation and is deficient or defective in individuals with hemophilia A. FVIII is synthesized as a 300-kd precursor protein with domain structure A1-A2-B-A3-C1-C2.1,2 In plasma it circulates as a series of noncovalently bound heterodimers, produced by proteolytic cleavage at the B-A3 junction or within the B domain.3 Thus, FVIII in vivo consists of a heavy chain (HC) of variable molecular mass (from 90 kd, when only the A1-A2 domains are present, to 220 kd when the full-length B domain is present) and a light chain (A3-C1-C2; LC) of 76 kd molecular mass (for a review, see Lenting et al4).
In the process of blood coagulation, FVIII acts as a cofactor to the serine protease factor IXa (FIXa). Before the initiation of coagulation, FVIII circulates in a noncovalent complex with von Willebrand factor (VWF). Limited proteolysis near the N-terminus of LC by traces of thrombin removes a peptide containing a VWF-binding site, resulting in release of free FVIII. Further proteolysis between the A1 and A2 domains results in generation of FVIIIa, the active heterotrimeric cofactor. Binding of FVIIIa to negatively charged phospholipid (PL) and to factor IXa (FIXa) form the factor X-ase (FX-ase) complex. The product of the reaction, activated factor X (FXa), forms with activated factor V (cofactor FVa) a similar membrane-bound system, the prothrombinase complex that converts prothrombin to thrombin.6 The serine proteases FIXa and FXa are capable of cleaving their substrates without the presence of protein cofactors. However, localizing the proteolytic reaction onto negatively charged PL surface and locking the protein conformation in complex with FVIIIa and FVa, respectively, amplifies the reaction rates by up to 5 orders of magnitude.6
Malfunctioning at the level of FVIII causes a mild to severe bleeding disorder called hemophilia A.7 Failure to form an active FX-ase complex results in a dramatic reduction of FXa production, which is required for the explosive formation of thrombin. There is therefore great interest in elucidating the exact structure and conformation of these proteins and the FX-ase complex.
To date, the structure of human FVIII and FV C2 domains9,10 and of porcine FIXa has been solved by x-ray crystallography9 (although the membrane-binding Gla domain of FIXa was poorly resolved). Attempts have been made to purify FVIII in a quantity and form suitable for 3-dimensional (3D) crystallization; however, the low plasma concentration (about 0.5 nM), its heterogeneity, and sensitivity to proteolysis have made direct crystallographic studies of the whole protein impossible thus far.
We have therefore used the method of 2-dimensional (2D) crystallization onto coagulant-active PL monolayers followed by electron microscopy and crystallography to solve the FVIII structure at 1.5 nm resolution in its membrane-bound state.11-14 Within the 2D crystals, FVIII was present as a noncovalently linked heterodimer of HCs (domains A1-A2) and LCs (domains A3-C1-C2).14 Very little material (of the order of micrograms) is needed to define a low-resolution 3D density map of a protein organized in 2D crystals by electron crystallography. The crystallization/incubation times can be kept as short as 3 hours allowing us to preserve the integrity of the protein and the following steps must be achieved successfully: (1) generation of good-quality 2D crystals, (2) estimation of the exact height of the molecule above the membrane, and (3) collection of data from tilted 2D crystals and generation of a 3D reconstruction, combining different projections.
By fitting and modeling of the atomic structures of the 5 major domains (A1, A2, A3, C1, and C2)10,15,16 within the 3D envelope of membrane-bound FVIII, it was possible to define their orientations within the molecule and relative to the membrane surface. Further biochemical information about the FIXa binding sites, the B domain and activated protein C (APC) binding site positions within the FVIII molecule were also taken into account during the modeling process. Data for key point mutations,17 as well as for amino acid residues potentially located at interdomain contact sites were also considered. A model for the FVIII/FIXa complex was constructed based on the crystal structure of FIXa,9 the membrane-bound FVIII structure, and the FIXa/FVIIIa binding sites.
Materials and methods
Electron microscopy
Purified human FVIII was crystallized in 2D and negatively stained as described.14 FVIII was also attached to liposomes containing dioleoylphosphatidylserine (DOPS) and dioleoylphosphatidylcholine (DOPC); solvent from 0.5 mg/mL DOPS/DOPC at 1:10 ratio solution in chloroform was evaporated under argon in a rotary evaporator and the lipids resuspended in μL buffer (10 mM MES, 2 mM Ca++, 100 mM NaCl, at pH 6.0). Unilamellar liposomes were obtained by extrusion18 and incubated with 100 μL purified FVIII at 0.04 mg/mL in the same buffer. The FVIII-liposomes were filtered through a Sephadex 25G gel filtration column to remove the nonattached protein; 5 μL was deposited onto a hydrophilic carbon-coated electron microscopy grid and negatively stained as for the 2D crystals.14
The samples were observed in a Phillips CM10 transmission electron microscope operated at 100 kV. The 2D crystals were tilted from −60° to +60° in a rotational holder,19 at 4 positions: 0°, 90°, 180°, and 270°. Electron micrographs were recorded at a calibrated magnification of × 44 600 for the 2D crystals and at both × 50 300 and × 70 800 for the liposomes with attached FVIII. The same 2D crystals from the same electron microscopy grids described previously14 were tilted. Three of the 7 tilt series were considered for further analysis.
Data analysis
The 3D crystallographic analysis followed essentially the procedure described previously.19 Briefly, the selected best tilt series images were digitized at 25 μm/pixel and analyzed by the Medical Research Council software for 2D crystal analysis.20 21 Thirty-three files, of which 28 tilted from −54° to +60° (by 10° to +/−40° and then by 5°) at 2 orthogonal tilt angle to astar (TAXA) values (67°/80° and 164°/169°) were included in the final 3D reconstruction.
The 3D structure was calculated by merging to a common origin all of the 1083 structure factors or reflections, in reciprocal space. The IQ value of the included reflections was restricted to 7 and the sampling in the direction perpendicular to the 2D crystal plane set to zstar = 0.006. The overall average phase residual was 21.8°. Smooth curves were fitted manually along the lattice lines sampled at 1/20 (0.05) to obtain the correct amplitudes and phases for the 243 unique reflections. The final 3D map was calculated using the CCP4 crystallographic suite.22
The height of the FVIII was estimated from side-on views of liposome-attached molecules. The FVIII 3D structure was manually fitted within the ‘Ono 6.2.2’ software package (TA Jones, Department of Molecular Biology, BMC, Uppsala University, Sweden, and M. Kjeldgaard, Department of Chemistry, Aarhus, University of Denmark).
Fitting of individual FVIII domains within the 3D envelope
The atomic coordinates of C2,10 the homology models for C1,16 and the 3 A domains15 were fitted within the 3D density of the membrane-bound FVIII using the ‘Ono’ software package. This procedure was performed interactively, because the low symmetry, limited resolution, and tight packing of the FVIII molecules within the 2D crystals rendered automatic fitting ineffective.23
The resultant atomic model for membrane-bound FVIII was geometrically optimized and refined in Insight II with the simulation program Discover (MSI, San Diego, CA). A3-C1 and C1-C2 domain interfaces were slightly modified to reduce steric clashes. Linker segments at A3-C1 and C1-C2 were built interactively using loop search protocols24 and secondary structure prediction25 followed by short energy refinement and 5 to 10 ps molecular dynamics (only the linker segments were allowed to move while the remaining structure was held fixed). Several structural models were imported back into ‘Ono’ and superimposed over the 3D density map. The models best fitting the electron crystallography data were selected for further assessment and evaluated against known biochemical data.
A preliminary model of the FVIII-FIXa-PL complex was assembled using Insight II. The x-ray structure of FIXa8 was docked onto the FVIII-PL model guided by experimental information including synthetic peptide studies and site-directed mutagenesis.
The overall FVIII protein data bank file can be found on the following sites: http://europium.csc.mrc.ac.uk andhttp://klkemi.mas.lu.se/dahlback.
Results
FVIII binding to liposomes
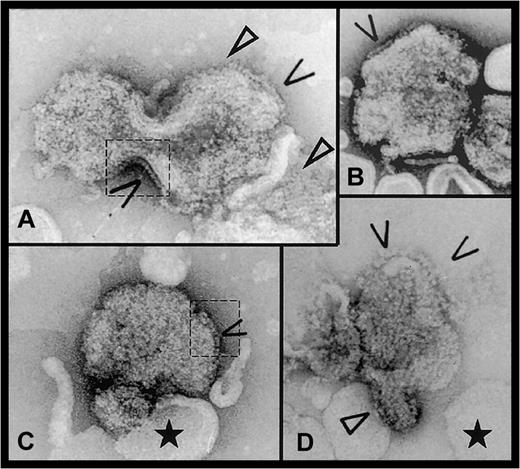
Factor VIII was attached to liposomes and the height above the membrane estimated as described.12 Only a proportion of the liposomes was covered with protein; however, on Figure1 it is easy to differentiate between naked and protein-coated liposomes. On larger liposomes, some FVIII molecules formed 2D arrays, also observed where the liposome surface was invaginated or curved. The above observations suggest a cooperative binding of the protein. From the edge-on view of 125 FVIII molecules picked from 9 different micrographs, the average height of the membrane-bound protein was 9.5 ± 0.5 nm.
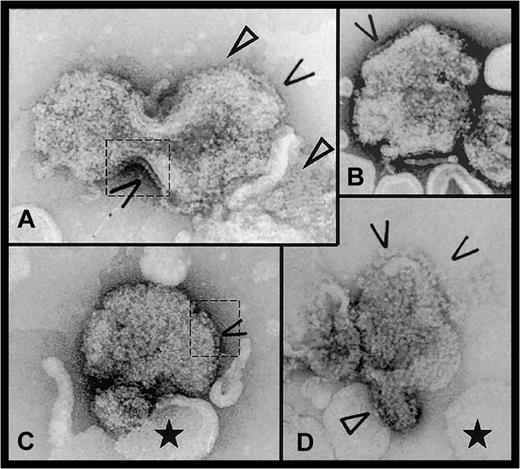
Electron micrographs of FVIII bound to DOPS/DOPC liposomes.
Open arrows indicate side-on views of membrane-attached FVIII molecules. In panels A and C, dashed line areas where the protein molecules are clearly visible on the liposome edges are contoured. Closed arrows show 2D arrays of FVIII molecules. The black star indicates the ‘naked’ liposomes, without attached FVIII, showing the cooperative effect of FVIII binding. Scale bar is 100 nm.
Electron micrographs of FVIII bound to DOPS/DOPC liposomes.
Open arrows indicate side-on views of membrane-attached FVIII molecules. In panels A and C, dashed line areas where the protein molecules are clearly visible on the liposome edges are contoured. Closed arrows show 2D arrays of FVIII molecules. The black star indicates the ‘naked’ liposomes, without attached FVIII, showing the cooperative effect of FVIII binding. Scale bar is 100 nm.
3D density map of membrane-bound FVIII
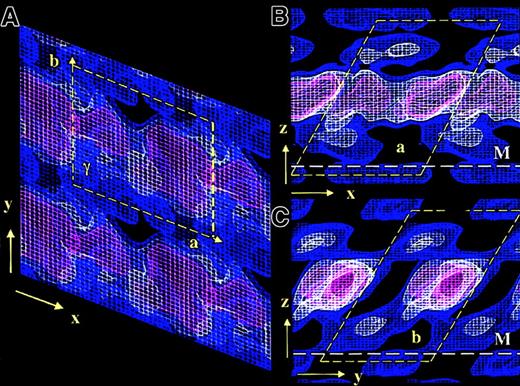
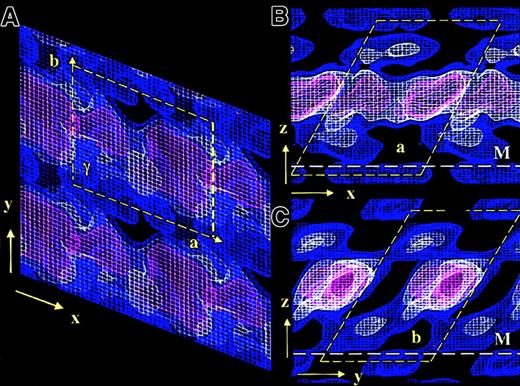
The overall 3D unit cell dimensions of membrane-bound FVIII were: a = 8.1 nm, b = 7.0 nm, c = 10 nm, and γ = 67°. The symmetry of the plane group was P1.14 The 3D density contours were displayed at a spacing of 0.7 ς, the lowest density being 0.4 ς comparable to the previously published 2D contour map14 (Figure 2). Sigma values equal zero when the protein density signal is comparable to the background noise of the electron micrograph. Because the obtained FVIII 2D crystals were small (∼1300 molecules) and imperfectly ordered, a low signal-noise ratio and consequently low sigma density contours were observed in the final 3D map.
3D electron density map of FVIII bound to PL monolayers.
The 3 density contours are blue at 0.4ς, white at 1.1ς , and red at 1.8ς. The density corresponding to one FVIII molecule is delineated by the yellow dashed box. The height of the unit cell above the monolayer is 10 nm. Panel A shows the top view, looking toward the monolayer along the z-axis. The unit cell dimensions are as for the 2D map: a = 8.1 nm, b = 7.0 nm, and γ = 113°. Panels B and C show side-on views along the y-axis and the x-axis, respectively. The white dashed line denotes the membrane surface (M).
3D electron density map of FVIII bound to PL monolayers.
The 3 density contours are blue at 0.4ς, white at 1.1ς , and red at 1.8ς. The density corresponding to one FVIII molecule is delineated by the yellow dashed box. The height of the unit cell above the monolayer is 10 nm. Panel A shows the top view, looking toward the monolayer along the z-axis. The unit cell dimensions are as for the 2D map: a = 8.1 nm, b = 7.0 nm, and γ = 113°. Panels B and C show side-on views along the y-axis and the x-axis, respectively. The white dashed line denotes the membrane surface (M).
Viewed toward the membrane plane (along the z-axis) the protein density corresponding to each molecule was well defined (Figure 2A). In the 2 orthogonal views (along the x- and y-axes), an overlapping head-to-tail arrangement between molecules was observed, FVIII being inclined 60° to 65° to the membrane plane, in both the X and Y directions (Figure 2B,C).
The main protein mass was centered approximately 5 nm above the membrane and was not well differentiated at contour levels below 1.8 ς (red contour in Figure 2). Viewed along the y-axis, FVIII molecules were not clearly separated in the main protein density (Figure 2B). The best-defined molecular contour was seen along the x-axis (Figure 2C). In this view the A domains, constituting 70% of the crystallized FVIII molecular mass, were well localized (red contoured area), as were the less dense C domains underneath. The contact area of C2 with the PL membrane was not well differentiated at a higher than 0.4 ς (blue contour), due to similar signals from the lipid molecules and the low protein mass of this part of FVIII. We expect part of C2, up to 1 nm in depth, to be buried within the lipid layer.
The larger mass of the A domains, situated above the C domains and unconnected to the membrane surface, prevents close interactions between C domains of adjacent molecules. This results in the C2 domains being less well ordered than the A domains, giving a less pronounced density in the 3D map and also favoring tight packing in the main protein density, with significant gaps close to the phospholipid surface, when membrane-bound FVIII is organized in 2D crystals (Figure2).
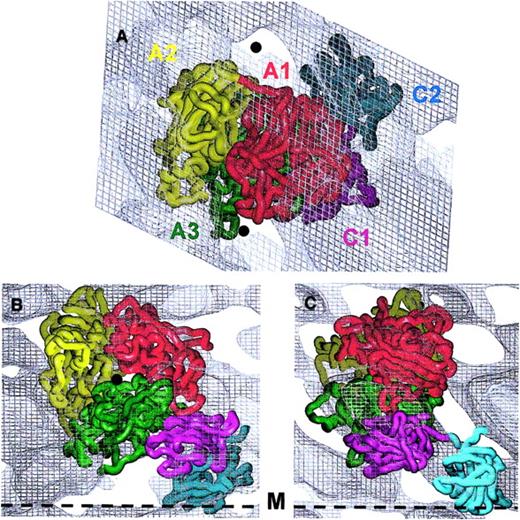
Fitting of the FVIII domains with the 3D electron density map
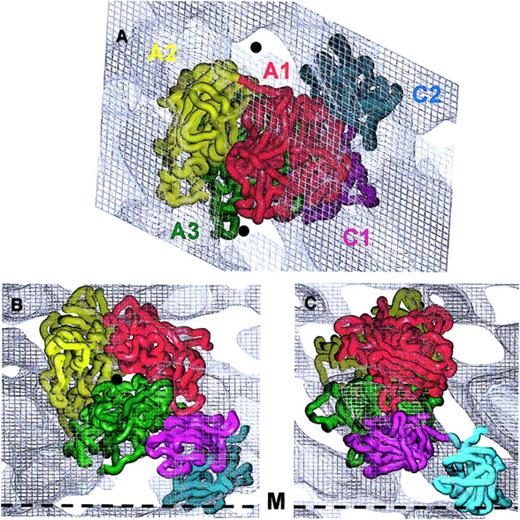
The existing model for the A domains,15 the atomic structure of C210 and the corresponding atomic model of C116 were oriented to match as closely as possible the 3D protein density distribution. The FVIII domain structures fitted the 3D map well when viewed toward the membrane (Figure3A). However, part of A2 and a few loops from A3 and A1 deriving from the homology model15 were found outside the main contour when viewed parallel to the membrane (Figure 3B,C). We therefore continued manual fitting using the experimentally determined height of the molecule and the low-density areas (Figure 3) as guides. The placing of the A domain heterotrimer to align the A3 C-terminus to the C1 N-terminus oriented A2 in a position allowing most of the defined FIXa binding sites to be accessible to a macromolecular ligand.
Fitting of the FVIII domains within the 3D electron density map.
The alpha-carbon (Cα) chains of the 5 FVIII domains are presented as worms: A1 domain residues colored in red, A2 in yellow, A3 in green, C1 in magenta, and C2 in cyan. Domains were fitted within the 3D density corresponding to one membrane-bound molecule contoured at 0.4ς. Panel A indicates the top view, toward the membrane surface. The 5 protein domains fit well within the electron density. The matching minimum protein density areas are marked with black spheres. Panels B and C show views along the y- and x-axes, respectively, parallel to the membrane surface (black dashed line marked M). The Cα chains are fitted to the 3D density of one molecule following the strongest density distribution, as the contour at 0.4ς could not accommodate all loops of the protein model.
Fitting of the FVIII domains within the 3D electron density map.
The alpha-carbon (Cα) chains of the 5 FVIII domains are presented as worms: A1 domain residues colored in red, A2 in yellow, A3 in green, C1 in magenta, and C2 in cyan. Domains were fitted within the 3D density corresponding to one membrane-bound molecule contoured at 0.4ς. Panel A indicates the top view, toward the membrane surface. The 5 protein domains fit well within the electron density. The matching minimum protein density areas are marked with black spheres. Panels B and C show views along the y- and x-axes, respectively, parallel to the membrane surface (black dashed line marked M). The Cα chains are fitted to the 3D density of one molecule following the strongest density distribution, as the contour at 0.4ς could not accommodate all loops of the protein model.
Placement of the A domain model15 within the 3D map confirmed the previously suggested orientation of the heterotrimer.14 Viewed from the side, that is, along the x- and y-axes (Figure 3B,C), the A domains are oriented with the pseudo–3-fold axis forming an angle of approximately 15° with the membrane surface.
The C domains were fitted manually within the electron density using the following additional information: (a) FVIII C2 has a cylindrical shape 4.5 nm by 3 nm10; (2) the fold of C domains is very well conserved,9,10 predicting C1 to have a very similar structure and shape to C2, as defined from homology modeling16,26; and (3) C1 and C2 were connected head-to-head as required by the positions of their C- and N-termini respectively.10 27
The C2 structure was fitted first, inclined at 60° toward the membrane surface (Figure 3B). The C2-PL contact area was estimated at 3 nm in diameter, and 4 of the loops from the lower part of the C2 domain insert approximately 0.7 nm into the lipid layer (Figure 3B,C). C1 was fitted between the A3 and C2 density with its long axis almost perpendicular to that of C2, such that the A3, C1, and C2 domains are associated in close proximity with each other and the PL surface. Viewed from the top C1 is largely covered by A3 and A1, whereas C2 lies outside the contour of the A domains (Figure 3A).
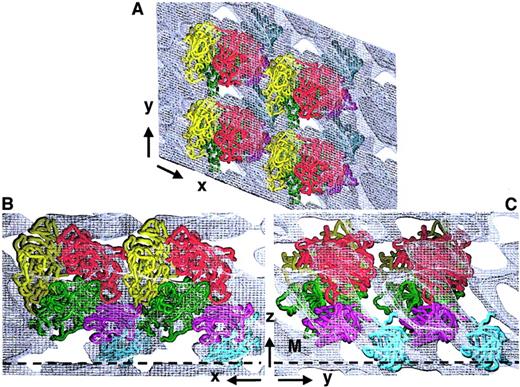
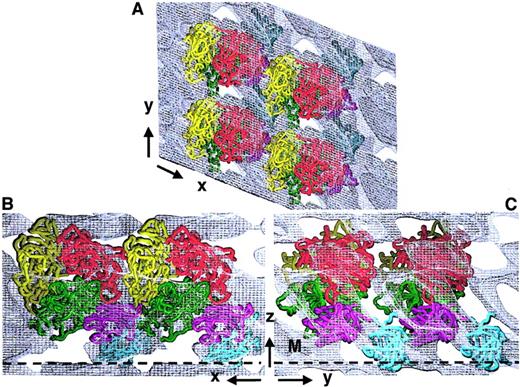
Although the overall FVIII domain organization was consistent with many available experimental data, we evaluated further our 3D structure by considering also the interactions between adjacent FVIII molecules within the membrane-bound 2D crystal (Figure4). Four identical FVIII molecules were fitted within the 3D map. The top view shows FVIII molecules arranged in rows, tightly packed along the x-axis and a little further spaced apart along the y-axis (Figure 4A). The C2 domains from one molecule are partially overlapped by the A2 and A3 domains of an FVIII molecule from the adjacent row (Figure 4A,B). Viewed parallel to the membrane (Figure 4B,C), FVIII molecules are tightly packed in the 2D crystal, with C1 and C2 domains in close proximity to the A3 and A2 domains of the next molecule in the same row (Figure 4B,C). Although some FVIII loops protrude out of the main molecular contour, there were no clashes between neighboring molecules or within one FVIII molecule. The tight packing of the membrane-bound molecule into the 2D crystal allows only one overall organization and orientation of the FVIII domains, when fitted within the 3D map. No density within the 3D unit cell could be ascribed to any extension of A2 into the B domain and the tight packing did not leave any free volume or indication for a possible fitting of a portion of B domain within the proposed 3D structure.
Fitting of FVIII molecule arrays within the 3D map of the 2D crystal.
Four membrane-bound FVIII molecules were fitted within the calculated 3D electron density of the FVIII 2D crystal. Cα chains are shown in worm presentation: A1, red; A2, yellow; A3, green; C1, magenta; and C2, cyan. Panel A shows the top view, along the z-axis. Panel B shows the side-on view along the y-axis. Panel C shows the side-on view along the x-axis. The membrane surface is denoted by a black dashed line marked M.
Fitting of FVIII molecule arrays within the 3D map of the 2D crystal.
Four membrane-bound FVIII molecules were fitted within the calculated 3D electron density of the FVIII 2D crystal. Cα chains are shown in worm presentation: A1, red; A2, yellow; A3, green; C1, magenta; and C2, cyan. Panel A shows the top view, along the z-axis. Panel B shows the side-on view along the y-axis. Panel C shows the side-on view along the x-axis. The membrane surface is denoted by a black dashed line marked M.
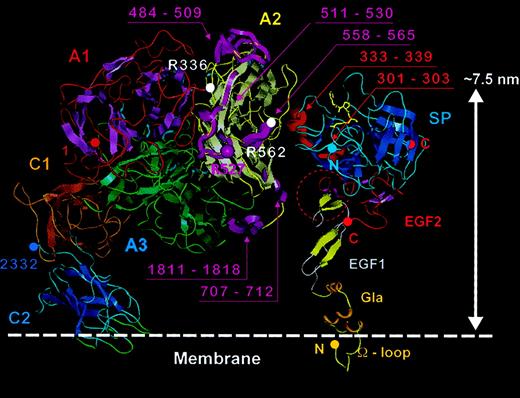
The best model structure for FVIII bound to the membrane obtained from our analysis is presented in Figure 5. To complete this model in detail, the N- and C-termini at the A3-C1 and C1-C2 boundaries were linked, optimizing the resultant orientation of the domains within the 3D map and the known secondary structures including disulfide links.
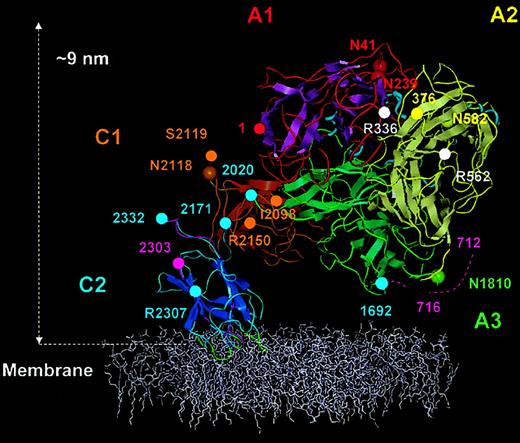
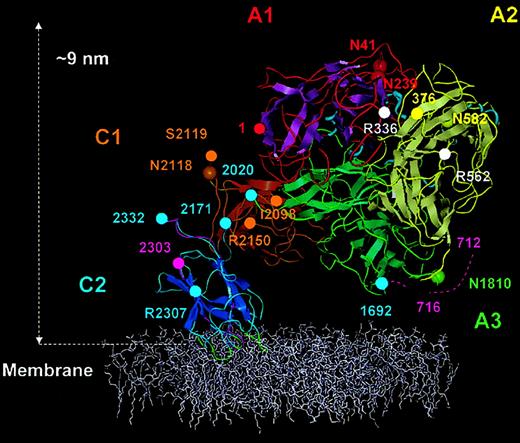
Membrane-bound FVIII model.
The FVIII model is shown anchored into the top of a phospholipid membrane (colored white). The β strands are shown as flattened ribbons. A1 domain (residues 1-336) is in red with magenta β strands. A2 domain (residues 376-716) is in yellow with silver β strands and A3 domain (residues 1692-2020) is in green with β strands also in green. C1 and C2 domains and β strands are in gold-brown and blue, respectively. Short α helices within the A domains are presented as light-blue spirals. The 4 predicted membrane-binding loops of the C2 domain are shown in green. Domain boundaries were defined thus: A1, from amino acid residues 1 (red sphere) to 336 (white sphere); A2, 376 (yellow sphere) to 716; A3, 1692 to 2020 (green spheres); C1, 2021 to 2170; and C2 2171 to 2332 (blue spheres). Asparagine residues known to be glycosylated (Asn41, Asn239, Asn582, Asn1810, and Asn 2118), are represented with large spheres colored as for the Cα traces of each domain. APC cleavage sites Arg336 and Arg562 are marked by white spheres. The beginning of the A2-B connecting acidic peptide (a2) is shown as residues 712-716 (dashed magenta trace). In C2, peptide 2303 (magenta sphere) to 2332 is also shown in magenta, which is reported to accommodate VWF and membrane binding sites. The side chains of residues, Arg2307 (blue sphere) Ile2098, Ser2119, and Arg2150 (golden-brown sphere) are solvent exposed and implicated in VWf binding.
Membrane-bound FVIII model.
The FVIII model is shown anchored into the top of a phospholipid membrane (colored white). The β strands are shown as flattened ribbons. A1 domain (residues 1-336) is in red with magenta β strands. A2 domain (residues 376-716) is in yellow with silver β strands and A3 domain (residues 1692-2020) is in green with β strands also in green. C1 and C2 domains and β strands are in gold-brown and blue, respectively. Short α helices within the A domains are presented as light-blue spirals. The 4 predicted membrane-binding loops of the C2 domain are shown in green. Domain boundaries were defined thus: A1, from amino acid residues 1 (red sphere) to 336 (white sphere); A2, 376 (yellow sphere) to 716; A3, 1692 to 2020 (green spheres); C1, 2021 to 2170; and C2 2171 to 2332 (blue spheres). Asparagine residues known to be glycosylated (Asn41, Asn239, Asn582, Asn1810, and Asn 2118), are represented with large spheres colored as for the Cα traces of each domain. APC cleavage sites Arg336 and Arg562 are marked by white spheres. The beginning of the A2-B connecting acidic peptide (a2) is shown as residues 712-716 (dashed magenta trace). In C2, peptide 2303 (magenta sphere) to 2332 is also shown in magenta, which is reported to accommodate VWF and membrane binding sites. The side chains of residues, Arg2307 (blue sphere) Ile2098, Ser2119, and Arg2150 (golden-brown sphere) are solvent exposed and implicated in VWf binding.
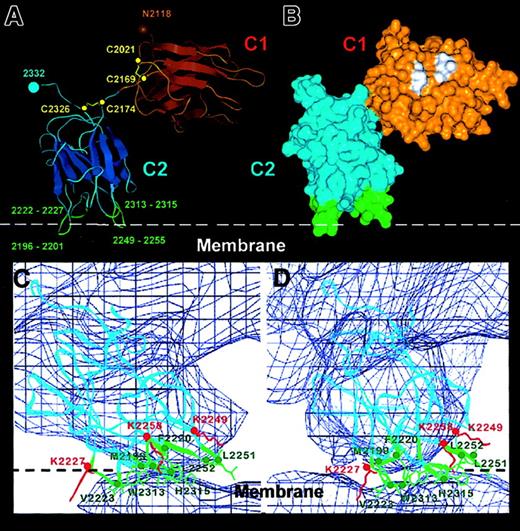
For connection of the C1 and C2 domains, we built a relatively extended and bent linker segment (Figure 6A) taking into consideration 3 factors: (1) the positioning of the C1 and C2 domains and their orientation relative to the membrane; (2) the position of C1 residue Asp2170, following Cys2169, is constrained in space due to the presence of the 2021-2069 disulfide link; and (3) location of Ser2173 at the N-terminus of the C2 domain, whose spatial position is well defined because it abuts the Cys2174-Cys2326 disulfide link in the FVIII C2 crystal structure.10 These constructions, based on the 3D map, led to our proposal that only limited contacts are present between the 2 C domains (Figure 6A).
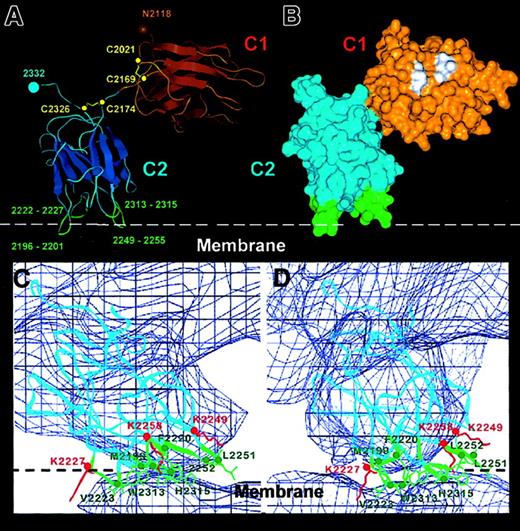
Details of the C1-C2 domain orientation and C2–membrane-binding region derived from the FVIII model.
Panel A shows the Cα trace of the C1-C2 domains as fitted within the FVIII 3D map. The main β strands are depicted as flattened ribbons. Intradomain disulfide bonds are shown in yellow and the cysteine Cα atoms as yellow spheres. The glycosylated Asn2118 residue is shown as a brown sphere. The 4 predicted membrane-binding loops of the C2 domain are shown in green with the loop residues numbered. Panel B shows the surface representation of the C1-C2 domains in the same orientation and domain coloring as in panel A. Some hydrophobic residues (see text) predicted in our model to have direct contact with the A1 and A3 domains are shown in white. Panels C and D show the membrane-binding region of the C2 domain viewed along the x-axis (panel C) and rotated clockwise 10° around the z-axis (panel D). The Cα chain (blue ribbon) is fitted within the 3D map (0.4ς blue contour). The hydrophobic side chains and residue numbers of the 4 loops inserting into the membrane are shown in green. The side chains of 3 lysine residues predicted to interact electrostatically with phosphatidyl serine head groups from the phospholipid surface (white dashed line) are shown in red.
Details of the C1-C2 domain orientation and C2–membrane-binding region derived from the FVIII model.
Panel A shows the Cα trace of the C1-C2 domains as fitted within the FVIII 3D map. The main β strands are depicted as flattened ribbons. Intradomain disulfide bonds are shown in yellow and the cysteine Cα atoms as yellow spheres. The glycosylated Asn2118 residue is shown as a brown sphere. The 4 predicted membrane-binding loops of the C2 domain are shown in green with the loop residues numbered. Panel B shows the surface representation of the C1-C2 domains in the same orientation and domain coloring as in panel A. Some hydrophobic residues (see text) predicted in our model to have direct contact with the A1 and A3 domains are shown in white. Panels C and D show the membrane-binding region of the C2 domain viewed along the x-axis (panel C) and rotated clockwise 10° around the z-axis (panel D). The Cα chain (blue ribbon) is fitted within the 3D map (0.4ς blue contour). The hydrophobic side chains and residue numbers of the 4 loops inserting into the membrane are shown in green. The side chains of 3 lysine residues predicted to interact electrostatically with phosphatidyl serine head groups from the phospholipid surface (white dashed line) are shown in red.
Secondary structure prediction suggested that the C-terminal A3 residues (up to residue Tyr2017 approximately) would form a β strand, continued by a loop segment. We therefore built a loop between residues Ser2018 and Cys2021 (the C1 N-terminus). The location of Tyr2017 in the A domain model is well defined and its predicted position within the A3-C1 interface is determined by an appropriate distance to the C1 N-terminal region and absence of steric clashes at domain interfaces. As Cys2021 in the C1 domain forms a disulfide bond with Cys2169, its position is locked in space, allowing for the construction of the short linker peptide (Figure 5). With this orientation, approximately 1200 Å2 of the C1 domain surface including hydrophobic amino acids Leu2053, Ile2135, Met2167, and Pro2024 (Figure 6B) are buried by the A1 and A3 domains (including residues Pro1715, Val1933, Ala1935, Ile1995, Leu2013, Leu2015, and Tyr2017). Although the existence of hydrogen bonds and salt bridges at the domain interfaces was predicted, the present medium resolution of the 3D model renders detailed listing of such interactions too speculative.
Interface of C2 domain with PL membrane
The domain orientation described above should also be consistent with the positions of residues predicted or expected to be surface exposed in the FVIII structure; in particular those involved in PL binding. The depth of C2 insertion within the PL can only be determined with an accuracy reflecting the resolution along the z-axis in the 3D density map, which is slightly better than 2. However, fitting the x-ray structure of C2 within the low-resolution FVIII 3D envelope allows the membrane-binding loops to be unambiguously defined at atomic resolution. Four loops inserting into the membrane were characterized as the C2–membrane-binding area (Figure 6C,D). From left to right (Figure 6C) they are: loop 1 (2222-2227) containing Val2223; loop 2 (2196-2201) with Met2199 and Phe2200; loop3 (2313-2315) with Trp2313 and Val2314; and loop 4 (2249-2255) with Leu2251 and Leu2252 (3 of which contain 2 adjacent hydrophobic residues). Above loops 1 and 4 were situated 3 lysine residues (Lys2227, Lys2249, and Lys2258), which appeared close enough to the membrane surface for electrostatic interaction with negatively charged DOPS head groups. The C2-membrane binding area totalled 7 to 10 nm,2 or approximately 16 to 25 lipid molecules. Figure 6D is rotated 10° clockwise (viewed from above the membrane) to better illustrate the 4 membrane-binding loops and charged lysine residues. Loops 1, 2, and 4 correspond to the membrane-binding loops proposed from the x-ray structure.10
Model of the FVIII/FIXa complex
Although the active FX-ase complex is formed from PL, FIXa and proteolytically activated FVIIIa, the binding characteristics of FVIII and FVIIIa for FIXa and PL are similar28-30 and the FVIII/FIXa complex should share considerable structural similarity with that of FVIIIa/FIXa. Therefore, we built a model of the FVIII/FIXa complex to aid interpretation of FVIIIa/FIXa function at the molecular level, based on FVIII surface regions known to be implicated in FIXa binding,31 areas of FIXa known to play a role in FVIII interactions, and the overall orientations of the 2 molecules with regard to the membrane plane. Several key areas important for FVIIIa/FIXa interactions are appropriately distributed on each molecule (Figure 7), including (1) FIXa protease domain residues 301-303 and 333-33932,33 with FVIII A2 domain residues 558-56534 and 698-71235,36and (2) the FIXa EGF1-EGF2 interface with FVIII A3 domain residues 1811-1818.37 Particularly good agreement was obtained between our model and that recently proposed for a region of the FVIIIa A2-FIXa protease domain interface.38 After positioning the cofactor and protease with appropriate membrane insertion of the FVIII C2 and FIXa Gla domains, the height of the FIXa active site corresponded to that reported by fluorescence measurements.39 In addition, the arrangement suggests availability of a face of the complex for potential docking of the substrate zymogen FX in the FX-ase complex, placing it in close proximity to the origin of the FVIII interdomain sequence a1(approximately residues 337-372), which spans between A1 and A2. Although this highly acidic peptide cannot be modeled by homology, it has been implicated in FX interaction40 and our FVIII-FIXa model predicts it would lie in proximity to FX zymogen. The model also appears to allow the involvement of the FVIII C2 domain in FX interaction as suggested by recent studies.41
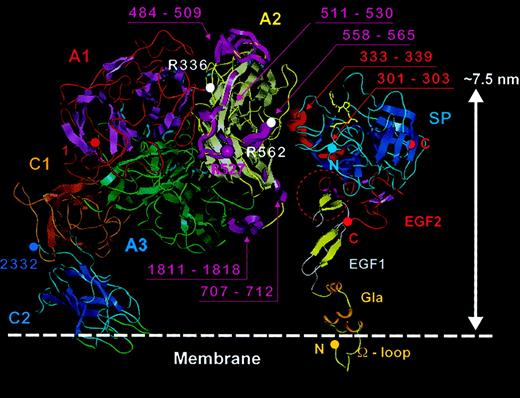
Model for membrane-bound FVIII/FIXa complex.
To the left the FVIII model (domains as described in Figure 5) is anchored into a phospholipid membrane (denoted with white dashed line). The FVIII domains, helices, PL-binding loops, and chain termini are colored as in Figure 5, and β sheets are shown as flat ribbon arrows. APC cleavage sites Arg336 and Arg562 are marked with white spheres. FVIII regions implicated in FIXa binding (484-509, 511-530, 558-565, 707-712, and 1811-1818) are highlighted as magenta ribbons and labeled in the same color, as is residue Arg527. To the right, the crystal structure for porcine FIXa9 is placed with the Gla domain (modeled based on that of prothrombin fragment I) inserted into the membrane surface, and with the Gla, EGF1, EGF2, and serine protease (SP) domains marked. The β sheets are shown as flat ribbon arrows, N- and C-termini of the FIXa light and heavy chains are shown as solid spheres, and the covalent active site inhibitor FPR-CK9 as yellow sticks. Sequences implicated in FVIII binding (301-303 and 333-339) are highlighted as red ribbons and labeled in the same color. A red dashed circle also highlights a possible FVIII interaction site at the EGF1-EGF2 interface. The FVIII and FIXa structures were docked manually such that most of the interactive regions were placed in reasonable proximity, then separated along the horizontal axis by approximately 1.0 to 1.5 nm for clarity of display.
Model for membrane-bound FVIII/FIXa complex.
To the left the FVIII model (domains as described in Figure 5) is anchored into a phospholipid membrane (denoted with white dashed line). The FVIII domains, helices, PL-binding loops, and chain termini are colored as in Figure 5, and β sheets are shown as flat ribbon arrows. APC cleavage sites Arg336 and Arg562 are marked with white spheres. FVIII regions implicated in FIXa binding (484-509, 511-530, 558-565, 707-712, and 1811-1818) are highlighted as magenta ribbons and labeled in the same color, as is residue Arg527. To the right, the crystal structure for porcine FIXa9 is placed with the Gla domain (modeled based on that of prothrombin fragment I) inserted into the membrane surface, and with the Gla, EGF1, EGF2, and serine protease (SP) domains marked. The β sheets are shown as flat ribbon arrows, N- and C-termini of the FIXa light and heavy chains are shown as solid spheres, and the covalent active site inhibitor FPR-CK9 as yellow sticks. Sequences implicated in FVIII binding (301-303 and 333-339) are highlighted as red ribbons and labeled in the same color. A red dashed circle also highlights a possible FVIII interaction site at the EGF1-EGF2 interface. The FVIII and FIXa structures were docked manually such that most of the interactive regions were placed in reasonable proximity, then separated along the horizontal axis by approximately 1.0 to 1.5 nm for clarity of display.
Discussion
The purpose of this study was to define membrane-bound FVIII structure and conformation, fitting the available structural, biochemical, biophysical, and mutation information within an experimentally derived 3D envelope of the molecule. Solving the structure of FVIII will improve our understanding of its structure-function relationship in both health and disease.
The FVIII 3D density map revealed the main protein density distribution within the 2D crystals (Figure 2). Direct visualization of secondary structures is not possible at 1.5 to 2.0 nm resolution; however, for a 170-kd molecule, a 3D structure solved at 1.5 nm provides excellent guidance for the correct arrangement of all 5 FVIII domains in membrane-bound conformation.
Because data were collected from small, negatively stained 2D crystals, some areas within the molecule may be less well resolved than others due to preferential staining of charged and hydrophilic surfaces and low signal-noise ratio of the 3D map. However, the 3D structure of the complete FVIII 2D crystal (Figure 2) provided a unique template for the organization of membrane-bound FVIII molecules in conditions close to those found in vivo.
The 4-step manual fitting of the domain structures to the density map was performed iteratively: (1) matching the main 3D map with the main protein density derived from the A and C domains; (2) checking domain interfaces to optimize the orientation; (3) verifying that no clashes occurred between adjacent molecules within the 2D crystal; and (4) considering the resultant model with respect to published biochemical and structural data such as hemophilic mutations, surface-exposed residues, and binding sites for other proteins, enabling us to produce a reliable 5-domain structure for PL-bound FVIII (Figure 5).
The highest protein density area within the 3D map (Figure 2, red contour) was well defined and located unambiguously the predominant mass of the FVIII molecule, the A domain heterotrimer (∼130 kd). The C domains (each ∼ 20 kd) were oriented with careful attention to placements of domain interfaces. C1 orientation was constrained by the shortness of the A3-C1 linker and the C1 intradomain disulfide bridge. The A3-C1 angle could be varied only slightly without affecting the quality of fitting to the electron density. For example, rotation of the C1 domain along its long axis displaced its N-terminus away from the A3 C-terminus (Figure 5).
C1 orientation further determines the length and orientation of the C1-C2 link (Figure 6A) suggesting that a “vertical”orientation of C1 on top of C2 for FVIII, as proposed for both FVIIIa and FVa42,43 may be unlikely. The experimentally determined height and dimensions of membrane-bound FVIII impose that C1 is sandwiched between the A domains and C2, assuming a rather horizontal orientation in respect to the membrane plane (Figures 5 and 6). It is possible that FVa assumes a membrane-bound conformation allowing the C domains to form a cylinder out of the main contour of the A domains.12 14 Without direct structural data, however, even at low resolution, comparison of our experimentally based FVIII model with FVa models is difficult, and further structural data on the C1-C2 domain pair will be needed to define the organization of the FVa domains.
The tight packing of the 5 FVIII domains within the 3D unit cell (Figure 2) resulted in a compact “spiral” organization of the FVIII domains within the unit cell volume (8 × 7 × 10 nm), with the A3, C1, and C2 domains in close association near the PL membrane. As described previously,14 no electron density could be assigned for extension of the A2 domain into the B domain. Because the purified human FVIII used here bore the expected heterogeneous heavy chains indicating the presence of variable lengths of B domain following the acidic a2 sequence, this tight domain packing suggests either that only FVIII molecules with no B domain extension are capable of forming regular arrays on the surface, or that fragments of the highly glycosylated and hydrophilic B domain extend past and above the A domain heterotrimer into solvent, poorly structured and therefore not resolved in the 3D density map. No density, however, corresponding to the B domain mass (up to 100 kd) could be seen in the side-on view of FVIII attached to liposomes.
Increased confidence in this A3-C1-C2 organization and orientation derives from the fact that the whole structure is in agreement with the overall dimensions of membrane-bound FVIII and places particular FVIII features in accordance with known biochemical and mutational data as given below.
First, the 5 glycosylated asparagine residues (Asn41 and Asn239 in A1, Asn582 in A2, Asn1810 in A3, Asn2118 in C1)44 45 are surface exposed in the FVIII structure (Figure 5).
Second, FIXa binding sites on FVIII (511-530, 558-565 and 707-712 in A2, and 1811-1818 in A3), determined by synthetic peptide studies,34-37 are surface exposed on one face of A2 at the appropriate height to interact with the FIXa protease domain (sequences 130-132 and 172-171)38 and epidermal growth factor–like domains (Figure 7). In addition, mutation at Arg527 (Figure 7) reduced apparent FIXa binding affinity.46 Recently, a further A2 sequence (482-493) was suggested to be a FIXa recognition site47,48; however, we have been unable to modify the FVIII-FIXa proposed model to include this region, in agreement with another recent report.38This apparent discrepancy may indicate more than one productive conformation of FVIII bound to FIXa; alternatively, the domain arrangements of FVIII and FVIIIa may differ slightly in exposure of FIXa-binding regions as suggested for the 558-565 sequence.49
Third, APC cleavage sites at Arg336 and Arg562 (for a review, see Lenting et al4) are fully surface exposed (Figure 5). Residue Arg562 is also referred as being part of a FIXa binding site and is correctly positioned within our FVIII-FIXa model for this role (Figure 7).
Fourth, there are 3 acidic peptides (a1, a2, anda3) at the interdomain interfaces (A1-A2, A2-B, and B-A3), each bearing proteolytic cleavage sites and containing sulfated tyrosine residues, important for protein-protein interactions such as A1-A2 interdomain contacts or membrane-assembled FVIII binding to factor X (a1),40 association with thrombin prior to FVIII activation (a2) or VWF (a3) (for a review, see Lenting et al4). These peptides cannot be modeled by homology (being absent in ceruloplasmin) and our structure resolution is insufficient to define their orientation; however, they can be accommodated as exposed structures accessible for protein-protein interaction and proteolytic degradation.
Fifth, we considered interactions of the C domains. There are 2 notable solvent-exposed clusters of hydrophobic residues on the flank of C1 opposite to the A domains, including residues Ile2080, Leu2171, Ile2145, and Leu2107 in the first patch and residues Leu2123, Val2125, Met2124, Ile2102, Phe2068, and Phe2127 in the second (not shown). Such clusters frequently form part of protein recognition sites.
The first cluster is close to Arg2116, Ser2119, and Tyr2105; mutation of these 3 residues results in hemophilia A and may be involved directly or indirectly in the binding of VWF.26,50 The second cluster may also be directly involved with VWF binding, because it is located close to Arg2150 (Figure 5), a residue closely associated with VWF interaction.51-53 Our structural model is in accord with these clinical data and structure-function studies.
The C2 domain possesses functionally important binding sites, which may be evaluated in our model. These involve interaction with other proteins such as VWF, factor Xa, and thrombin as well as PL membranes (for a review, see Pratt54). Binding site(s) on C2 for PL and VWF are either identical or closely adjacent55,56 and direct binding of the isolated C2 domain to both VWF and negatively charged PL membranes has been demonstrated.55,57 The C-terminal 2303-2332 sequence was suggested to form an interactive site for both VWF and PL58 and this peptide has been shown to inhibit binding of FVIII and isolated C2 domain to both VWF and PL. Although the sequence 2303-2324 was shown by nuclear magnetic resonance studies to form an amphipathic helix, which bound to synthetic PL vesicles,59 resolution of the C2 structure by x-ray crystallography10 demonstrated that 2303-2332 forms a part of the domain core, requiring major rearrangement to expose the sequence as a helix. This suggests that the 2303-2332 peptide, in solution, mimics in a nonspecific way the binding characteristics of the complete domain, although recent studies support the concept of conformational changes within the C2 domain on membrane binding or FVIIIa/FIXa complex formation.60,61 The C2 crystal structure suggested explicitly a different molecular mechanism for PL binding involving insertion of hydrophobic side chains from 3 C2 loops while several positively charged side chains made contact with negatively charged PL head groups. Although none of these 3 hydrophobic loops or basic residues are contained within the sequence 2303-2332, several are within the sequence 2181-2243, which is also involved in PL binding,62 and alanine scanning mutagenesis recently demonstrated involvement of Met2199, Phe2200, Leu2251, and Leu2252 in forming a PL-binding site on C2.63,64 In our FVIII-PL model we position these 3 hydrophobic loops at the C2-PL interface, together with a fourth loop bearing Trp2313, Val2314, and His2315, and a similar grouping of basic residues (Figure 6). This fourth loop is within the 2303-2332 sequence closely associated with PL binding in previous studies; Figure 5 shows that the termini of the 2303-2332 sequence are exposed distal to the membrane while the mid portion, bearing the hydrophobic residues 2313-2315, is buried within the PL bilayer. Similar arguments as to the nature of the protein-lipid interface have resulted from solution of the structure of the homologous factor V C2 domain,9 and mutational analysis65 66 supporting a general model of FV/FVIII-PL binding.
There is considerable evidence for closely related or even identical binding sites on the C2 domain for VWF and PL with the 2303-2332 sequence implicated, and the substitution Arg2307Gln is found in a patient with mild hemophilia A with a defect in VWF binding.26 However, for VWF there is also strong evidence that the N-terminal acidic region of the A3 domain (the a3sequence) contains a key binding site67-69; although this sequence cannot be modeled by homology, we can place the termini speculatively in the region of the A2-A3 interface (residues 716 and 1692, Figure 5). C1 domain residues Ile2098, Ser2119, and Arg2150 have also been implicated in VWF interaction,50-53 and a recombinant C1-C2 molecule bound to immobilized VWF at least 2 orders of magnitude better than isolated C2,70 confirming a role for both C domains in addition to the a3 sequence. Figure 5shows that in our FVIII-PL model the hypothetical a3 region and residues on the C domains associated with VWF binding are found surface exposed and spread in a patch across the membrane-proximal region of the FVIII molecule. We propose that binding of the FVIII molecule via a3, C1, and C2 to the carrier VWF multimer interferes grossly with binding of FVIII to a membrane via the C2 domain, until released by cleavage at Arg1689 removing thea3 peptide.
This novel FVIII domain arrangement, with the A3 domain anda3 peptide position close to the membrane and compactly associated with the C domains, shares one feature (the membrane-proximal orientation of A3 in the A domain heterotrimer) with that predicted in a recent modeling study of membrane-bound FVa,65 although an alternative arrangement for modeling FVa with the A2 domain closest to C2-PL has also been proposed.71 In the present FVIII-PL study, however, the domain geometry is based directly on the experimental 3D density map and extensive biochemical data, supporting the presented domain arrangement.
C2 has also been reported to interact with FXa and thrombin, both of which selectively cleave and activate FVIII. Binding of intact FVIII light chain and of the isolated C2 domain to FXa (inactivated at the active site) could be competed with the C2-derived synthetic peptide 2253-2270.41 This peptide binding was also shown to be specific for FXa as against FVIII binding to VWF, PL, FIXa, or FX, and inhibited FXa-dependent FVIII activation.72 Residues in the 2253-2270 sequence are exposed in our model, forming well-distinguished solvent-exposed clusters near the membrane binding loop containing Ile2251 and Ile2252 and in proximity to the C1-C2 binding interface. Interaction of C2 with active site-inhibited thrombin has also been described, and isolated C2 domain inhibited thrombin activation cleavage of FVIII at the N-terminus of the A3 domain (Arg1689).73 This site has not yet been localized; however, it appears distinct from the FXa-binding site.72
The major goals of this study were first to define the domain arrangements of FVIII bound to PL, and second to propose a docking structure for the FVIII/FIXa complex, which could stand as a model for the tenase complex of activated FVIII with FIXa on a PL surface. To build and evaluate the tenase model, we mapped proposed FVIIIa/FIXa binding sites as well as functionally significant point mutations. The model satisfies the various criteria described above and we suggest that it may prove useful in discussion of the structural and functional aspects of the tenase complex. Refinement of the present FVIII-FIXa model structure using both modeling and further experimental studies is in progress and should shed light on a clinically and structurally important macromolecular system.
This work was accomplished in the School of Biochemistry and Molecular Biology, University of Leeds. S.S.-M. would like to thank Dorit Hanein (The Burnham Institute) for her support and Jo Jaeger and Simon Phillips from the Astbury Centre of Structural Biology (The University of Leeds) for their support and use of the computer.
Supported by a 3-year research grant from the Wellcome Trust (United Kingdom) (S.S.-M. and A.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Svetla Stoilova McPhie, Wadsworth Center, NY State Department of Health, Empire State Plaza, PO Box 509, Albany, NY 12201-0509; e-mail: svetla@burnham.org.