Abstract
Iron is essential for cell proliferation, heme synthesis, and a variety of cellular metabolic processes. In most cells, transferrin receptor–mediated endocytosis is a major pathway for cellular iron uptake. Recently, transferrin receptor 2 (TfR2), another receptor for transferrin, was cloned. High levels of expression of TfR2 messenger RNA (mRNA) occur in the liver, as well as in HepG2 (a hepatoma cell line) and K562 (an erythroid leukemia cell line). In this study, TfR2 mRNA expression was analyzed in hematological cell lines, normal erythroid cells at various stages of differentiation, and leukemia and preleukemia cells. High levels ofTfR2 expression occurred in all of the erythroid cell lines that were examined. Erythroid-specific expression of TfR2 protein in bone marrow cells was confirmed by immunohistochemical staining. Expression of TfR2 mRNA was high in normal CD34+ erythroid precursor cells, and levels decreased during erythroid differentiation in vitro. Levels of expression of TfR2-α mRNA were significantly higher in erythroleukemia (M6) marrow samples than in nonmalignant control marrow samples. In addition, relatively higher levels of TfR2-αmRNA expression occurred in some samples of myelodysplastic syndrome that had erythroid hyperplasia in bone marrow, acute myelogenous leukemia M1, M2, and chronic myelogenous leukemia. Expression profiles of normal members of the erythroid lineage suggest that TfR2-α may be a useful marker of early erythroid precursor cells. The clinical significance of TfR2-α expression in leukemia cells remains to be determined.
Introduction
Iron is essential for a variety of physiological activities of cells, such as electron transport and DNA synthesis, and it is used as a cofactor of cytochromes, aconitases, ribonucleotide reductase, and heme proteins.1,2 Transferrin receptor 1 (TfR1) is a type II membrane protein that mediates cellular iron uptake. In the serum, most iron exists in a transferrin-bound form.3 On the cell surface, iron-bound transferrin binds to TfR1; this is followed by internalization of this complex.
Recently, another receptor for transferrin, transferrin receptor 2(TfR2), was cloned in our laboratory.4 At least 2 alternatively spliced forms of transcripts, α andβ, are transcribed from the TfR2 gene. TfR2-α protein is a type II membrane–integrated glycoprotein similar to TfR1 that can mediate iron uptake through transferrin. TfR2-β lacks both intracellular and transmembrane domains and may be an intracellular protein. Although TfR2-β transcripts were detectable by a very sensitive reverse-transcriptase polymerase chain reaction (RT-PCR), it was not detectable by Northern blot analysis. In contrast,TfR2-α was easily detectable by Northern blotting in the liver and in K562 cells, indicating that the α-form transcript was far more prevalent than the β-form in these cells. Among various tissues and cell lines that we tested, high levels of expression of TfR2 messenger RNA (mRNA) occurred only in the liver, and in HepG2 hepatoma and K562 erythroid cell lines, suggesting that its expression was tissue specific.4 5
Considering our limited knowledge of the expression profile ofTfR2, we asked the following questions: (1) IsTfR2 expression, as a previous report6suggested, specific to erythroid cells within the hematopoietic population? (2) Iron is essential for cell growth, and we previously showed that expression of TfR2-α supported cell growth in TfR-deficient Chinese hamster ovary (CHO) cells.7Therefore, is expression of TfR2-α particularly predominant in hematological diseases such as leukemia? (3) In murine erythroleukemia (MEL) cells, TfR1 was upregulated and TfR2 was downregulated as the cells underwent erythroid differentiation.6 Do these changes occur during normal erythroid differentiation? In this study, we address these questions by analyzing TfR2 expression in normal erythroid precursors as well as in preleukemia and leukemia cells.
Materials and methods
Cell lines
TfR1-deficient CHO-TRVb cells were kindly provided by Dr T. McGraw.8 MOLT-16 (T-lymphoid),9 NB4 (a myeloid cell line derived from a patient with acute myelogenous leukemia [AML]–M3),10 and KCL22 (a myeloid cell line from a patient with chronic myelogenous leukemia [CML])11 were kind gifts from Drs J. Minowada, M. Lanotte, and I. Miyoshi, respectively. Erythroid cell lines HEL-R (from a patient with erythroleukemia),12 DCI-M1 (from a patient with erythroleukemia),12 and KU-812-F (derived from a patient with CML and spontaneously differentiating toward erythroid lineage in vitro)13,14 were kindly provided by Dr T. Papayannopoulou. KG-1 (a myeloid cell line) was established by our group.15 TfR2 stable transfectants of CHO-TRVb cells were established as previously described.4 Raji (B-lymphoid), U937 (monoblastoid), HL-60 (myeloid), and K562 (erythromegakaryocytoid) cell lines were obtained from American Type Culture Collection (Manassas, VA).
Immunohistochemistry
Cytospin specimens of CHO-TRVb cells (stably transfected with either TfR1, TfR2, or neomycin-resistant control plasmid) and normal bone marrow (BM) mononuclear cells were fixed with acetone and then incubated with either diluted rabbit anti-TfR2 antiserum7 or normal rabbit serum for 30 minutes, followed by immunohistochemical staining with Envision/AP (Dako, Carpinteria, CA). The cells were counterstained with Mayer hematoxylin (Wako Pure Chemicals, Osaka, Japan).
Preparation of human erythroid cells at various stages of differentiation
Human erythroid cells from buffy coats were cultured as previously described.16 Briefly, the mononuclear cells were obtained by density centrifugation in lymphocyte separation media (Organon Teknika, Durham, NC). The cells were cultured in the Phase I α–Eagle minimum essential medium (α-MEM) (Sigma, St Louis, MO) containing 10% fetal bovine serum (FBS) (Intergen, Purchase, NY), 10% conditioned media prepared from supernatant of 5637 carcinoma bladder cells, and 1 μg/mL cyclosporin A (Sigma). After 7 days' incubation, the cells were washed with phosphate-buffered saline and transferred to Phase II α-MEM media containing 30% FBS; 10% bovine serum albumin; 10−5 M 2-mercaptoethanol; 10−6M dexamethasone; 0.033 g/L (33 μg/mL) holotransferrin; 10 ng/mL stem cell factor (Sigma); and 1000 U/L (1 U/mL) erythropoietin (Amgen, Thousand Oaks, CA). After 4 days in Phase II culture, the cells were collected and incubated for 30 minutes with a cocktail of antibodies against the following cell surface markers: CD2, CD19, CD33, and CD66b. After incubation for an additional 30 minutes with Stemsep Magnetic colloid, the cells of interest were isolated by negative selection with a 0.6 Tesla magnet (StemCell Technologies, Vancouver, BC, Canada). Stage-specific cell populations (CD34+/d4, and glycophorin A [GPA]/d4) have been isolated from the erythroid population by positive selection with anti-CD34 and anti-GPA antibodies (Stem Cell Technologies), respectively. To obtain erythroid cells from a later stage of maturation, both cell depletion and positive selection by means of GPA have been applied to erythroid cultures that have been incubated for 10 days in Phase II (GPA/d10). Alternatively, the CD34+/d4 progenitors were cultured in the same media for up to 14 days. The purity and homogeneity of these populations were confirmed by flow-cytometry analysis.
Quantitative RT-PCR for human erythroid cells at various stages of differentiation
The mRNA was extracted from human erythroid cells prepared as above, and complementary DNA (cDNA) was synthesized. Real-time quantitative PCR (Q-PCR) was performed with primers specific for TfR1 (5′-AAA ATCCGGTGTAGGCACAG-3′ and 5′-CCTTTAAATGCAGGGACG AA-3′); TfR2 (5′-TACCCATTCCTGCACACA AA-3′ and 5′-AGTACACCCACTGCAGGG TC-3′); and TfR2-α (5′-ACCTGGAGGAGGAAGAGGAA-3′ and 5′-CGACGTAGCCCAGTAGGAAG-3′). Primer specificity was confirmed by restriction endonuclease and agarose gel analysis. Sybr green I dye (Molecular Probes, Eugene, OR) was used as the reporter dye for Q-PCR, which was performed in a PE Biosystems SDS 7700 thermal cycler (PerkinElmer, Boston, MA). Results are presented as attomoles per microgram mRNA.
RT-PCR for clinical samples
From a collection at Showa University School of Medicine (Tokyo, Japan), 107 leukemia and preleukemia samples from 90 patients were analyzed for TfR1, TfR2-α, and TfR2-βexpression by semiquantitative RT-PCR. Either BM or peripheral blood mononuclear cells (PBMNCs) were used. These samples were from patients with AML (38 BMs and 25 PBMNCs); CML (chronic or accelerated phase, 5 BMs and 4 PBMNCs); acute lymphocytic leukemia (ALL) (3 BMs and 5 PBMNCs); myelodysplastic syndromes (MDSs) (20 BMs and 6 PBMNCs); and aplastic anemia (1 BM). The French-American-British classification of AML and MDS is presented here, along with an enumeration of the numbers and types of samples we obtained for each: M1, undifferentiated (4 BMs and 3 PBMNCs); M2, myeloblastic (6 BMs and 3 PBMNCs); M3, promyelocytic (6 BMs); M4, myelomonocytic (6 BMs and 6 PBMNCs); M5a, poorly differentiated monoblastic (3 BMs and 1 PBMNC); M5b, well-differentiated monocytic (4 BMs and 5 PBMNCs); M6, erythroleukemia (7 BMs); refractory anemia (RA) (3 BMs); RA with ring sideroblasts (RARS) (4 BMs); RA with excess blasts (RAEB) (8 BMs and 4 PBMNCs); RAEB in transformation (RAEBT) (5 BMs and 2 PBMNCs). As controls, 8 nonmalignant BM (idiopathic thrombocytopenic purpura, iron deficiency anemia, and normal BM) and 6 normal PBMNC samples were also analyzed. The semiquantitative RT-PCR was performed essentially as previously described.17 Primers and cycle numbers were as follows: for TfR1, primers 5′-AGGAACCGAGTCTCCAGTGA-3′ and 5′-ATCAACTATGATCACCGAGT-3′, 22 cycles; for TfR2-α, primers 5′-GTGGTCAGTGAGGATGTCAA-3′ and 5′-CCACACGTGGTCCAGCTTCTGGCGGGAG-3′, 22 cycles; for TfR2-β, primers 5′-ACGTCTCTGGCATCCTTCC-3′ and 5′-TGTAGGGGCAGTAGACGTCA-3′, 25 cycles; for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), primers 5′-TACATGGCTGGGGTGTTGAA-3′ and 5′-AAGAGAGGCATCCTCACCCT-3′, 13 cycles. The PCR products were transferred to nylon membranes after agarose gel electrophoresis and hybridized with32P-labeled cDNAs, and the ratio of band intensities of TfR1, TfR2-α, and TfR2-β versus GAPDH were calculated with the use of a densitometer.
Other methods
Results
Expression of TfR2 mRNA in hematopoietic cell lines
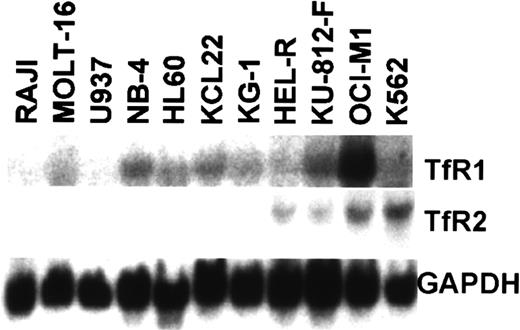
Our previous study showed that high levels ofTfR2 expression occurred in K562, an erythroid leukemia cell line derived from a patient with CML, whereas expression ofTfR2 mRNA was not detectable in the myeloid cell lines KG-1, U937, and HL-60 by a standard Northern analysis.4 To examine whether TfR2 expression is specific for erythroid cells in the hematopoietic system, we performed Northern analysis using a variety of hematopoietic cell lines, including 4 erythroid cell lines: HEL-R, KU-812-F, OCI-M1, and K562. Among the cell lines that we tested, all of the erythroid cell lines expressed high levels ofTfR2 mRNA, while all the lymphoid (Raji and MOLT-16) and myeloid (U937, NB4, HL-60, KCL22, and KG-1) cell lines expressed either low or undetectable levels of TfR2 mRNA (Figure1). In contrast, TfR1expression was detectable in all the cell lines that we tested, though levels of TfR1 mRNA expression were very high in OCI-M1 cells and were relatively low in Raji, U937, HEL-R, and K562 cells (Figure 1).
Northern blot analysis of TfR expression in hematopoietic cell lines.
Total RNA (15 μg) from Raji (B lymphoid), MOLT-16 (T lymphoid), U937, NB4, HL-60, KG-1, KCL22 (myeloid leukemia), HEL-R, OCI-M1, KU-812-F, and K562 (erythroid leukemia) was used to analyze TfR1, TfR2, and GAPDH expression. Levels of TfR1expression in Raji and U937 were relatively low, but their expression was confirmed by a longer autoradiography as well as by a prior study.4
Northern blot analysis of TfR expression in hematopoietic cell lines.
Total RNA (15 μg) from Raji (B lymphoid), MOLT-16 (T lymphoid), U937, NB4, HL-60, KG-1, KCL22 (myeloid leukemia), HEL-R, OCI-M1, KU-812-F, and K562 (erythroid leukemia) was used to analyze TfR1, TfR2, and GAPDH expression. Levels of TfR1expression in Raji and U937 were relatively low, but their expression was confirmed by a longer autoradiography as well as by a prior study.4
Immunohistochemical staining of TfR2-transfected CHO-TRVb and normal BM cells
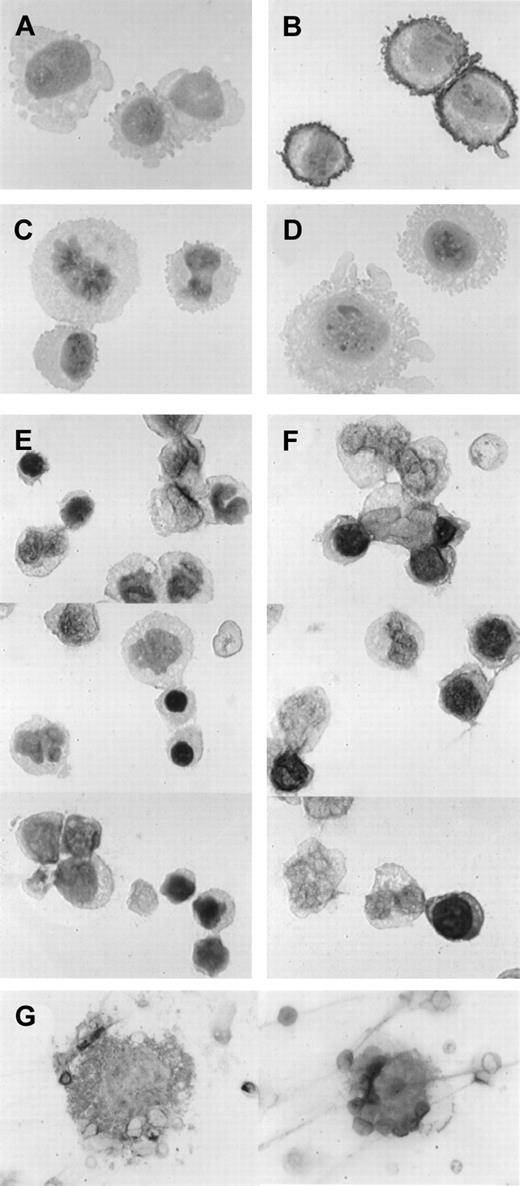
To examine if expression of TfR2 is lineage specific among hematopoietic cells, we used the immunohistochemical technique. First we tested our anti-TfR2 antibody for this technique using CHO-TRVb cells that had been stably transfected with eitherTfR1 or TfR2-α. Cytospin specimens of neomycin-resistant control cells as well as TfR1-transfected and TfR2-α–transfected cells were incubated with 50-fold diluted anti-TfR2 antiserum or the same concentration of normal rabbit serum and were immunohistochemically stained (Figure 2A-D). TfR2-transfected cells stained strongly, especially on their cell surfaces (panel B as compared with panel A), whereas neomycin-resistant control cells and TfR1-transfected cells stained only at background levels (Figure 2C and 2D, respectively). These results prompted us to examine normal BM mononuclear cells using this technique. Low levels of staining occurred in both erythroid and nonerythroid cells with nonimmune rabbit serum (Figure 2E), but the majority of erythroblasts clearly stained with the anti-TfR2 antibody at more than the background levels (Figure 2F). Some of the erythroid cells at the late stages of differentiation as well as mature erythrocytes did not stain clearly with the anti-TfR2 antibody. Most of the myeloid cells that we examined were negative for TfR2, although we could not distinguish eosinophils and basophils from neutrophils with this staining. A few large cells on the cytospin specimens were probably megakaryocytes, and some of them were positive for TfR2 (Figure 2G).
Immunohistochemical staining for TfR2.
Cells were incubated with either 50-fold diluted normal rabbit serum (panels A and E) or 50-fold diluted rabbit anti-TfR2 antiserum (panels B-D, F-G) followed by immunohistochemical staining. Magnifications: panels A-F, × 1000; panel G, × 300. (A-B) TfR2-α stably transfected CHO-TRVb cells. (C) Neomycin-resistant control CHO-TRVb cells. (D) TfR1 stably transfected CHO-TRVb cells. (E-F) Normal human BM cells. (G) Representatives of TfR2-positive megakaryocytes.
Immunohistochemical staining for TfR2.
Cells were incubated with either 50-fold diluted normal rabbit serum (panels A and E) or 50-fold diluted rabbit anti-TfR2 antiserum (panels B-D, F-G) followed by immunohistochemical staining. Magnifications: panels A-F, × 1000; panel G, × 300. (A-B) TfR2-α stably transfected CHO-TRVb cells. (C) Neomycin-resistant control CHO-TRVb cells. (D) TfR1 stably transfected CHO-TRVb cells. (E-F) Normal human BM cells. (G) Representatives of TfR2-positive megakaryocytes.
Expression of TfR2 mRNA in cultured normal erythroid cells
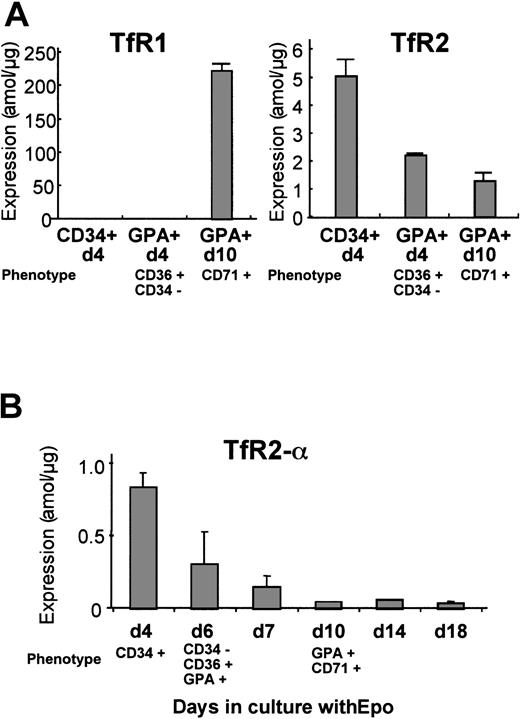
Human erythroid cells at various stages of differentiation were analyzed for expression of TfR1 and TfR2 mRNA by real-time Q-PCR. We prepared 3 erythroid cell populations from normal peripheral blood: CD34+/d4, GPA+/d4, and GPA+/d10. The CD34+/d4 cells represent mostly immature erythroid progenitors. The GPA+/d4 cells were CD36+ and CD34− by flow-cytometric analysis, indicating that these cells were at either the erythroid colony-forming unit or the erythroblast stage. The results from Q-PCR are shown in Figure 3A. Levels ofTfR1 mRNA dramatically increased during erythrocytic maturation between GPA+/d4 and GPA+/d10, whileTfR2 mRNA gradually decreased during erythroid differentiation. The primers that we used for TfR2 could amplify both α and β forms, although our previous study indicated that the majority of TfR2transcripts in erythroid cell lines were the α form. To confirm the expression profile of the α transcripts during normal erythroid differentiation, we designed another set of primers that can amplify only the α form. For this analysis, CD34+/d4 cells were cultured in the presence of erythropoietin, and the cells were harvested at days 4, 6, 7, 10, 14, and 18. The cells harvested at day 6 expressed CD36 and GPA but not CD34, so this population was the equivalent of the population of GPA+/d4 in Figure 3A. The cells harvested at day 10 expressed both GPA and CD71, and these cells were the equivalent of the GPA+/d10 cells in Figure 3A. Results of quantitative RT-PCR shown in Figure 3B demonstrated that, in agreement with our first analysis, the levels of TfR2-αtranscript declined as the erythroid progenitors matured.
Levels of expression of TfR1 andTfR2 mRNA during erythroid cell differentiation.
Human erythroid cell populations at different stages of differentiation were analyzed by real-time Q-PCR. Bars show mean ± SD. (A) Levels of TfR1 and TfR2 mRNA were analyzed. CD34+/d4 represents human erythroid cells positively selected for CD34 expression after 4 days in culture. GPA+/d4 and GPA+/d10 represent human erythroid cells positively selected for glycophorin A expression at day 4 and day 10 of culture, respectively. In all these experiments, the cells were cultured in the presence of erythropoietin. (B) Levels ofTfR2-α transcripts were analyzed by means of α-form–specific primers. The CD34+/d4 erythroid progenitors were cultured in the presence of erythropoietin and harvested on different days of culture. The purity and homogeneity of these populations were confirmed by flow-cytometric analysis, and the phenotypes of each population are shown in each panel. The absolute values (attomoles per microgram) of expression shown in panels A and B are not directly comparable, since the cells used in these experiments were from different donors.
Levels of expression of TfR1 andTfR2 mRNA during erythroid cell differentiation.
Human erythroid cell populations at different stages of differentiation were analyzed by real-time Q-PCR. Bars show mean ± SD. (A) Levels of TfR1 and TfR2 mRNA were analyzed. CD34+/d4 represents human erythroid cells positively selected for CD34 expression after 4 days in culture. GPA+/d4 and GPA+/d10 represent human erythroid cells positively selected for glycophorin A expression at day 4 and day 10 of culture, respectively. In all these experiments, the cells were cultured in the presence of erythropoietin. (B) Levels ofTfR2-α transcripts were analyzed by means of α-form–specific primers. The CD34+/d4 erythroid progenitors were cultured in the presence of erythropoietin and harvested on different days of culture. The purity and homogeneity of these populations were confirmed by flow-cytometric analysis, and the phenotypes of each population are shown in each panel. The absolute values (attomoles per microgram) of expression shown in panels A and B are not directly comparable, since the cells used in these experiments were from different donors.
Expression of TfR2 mRNA in BM and PBMNCs from patients with hematological disorders
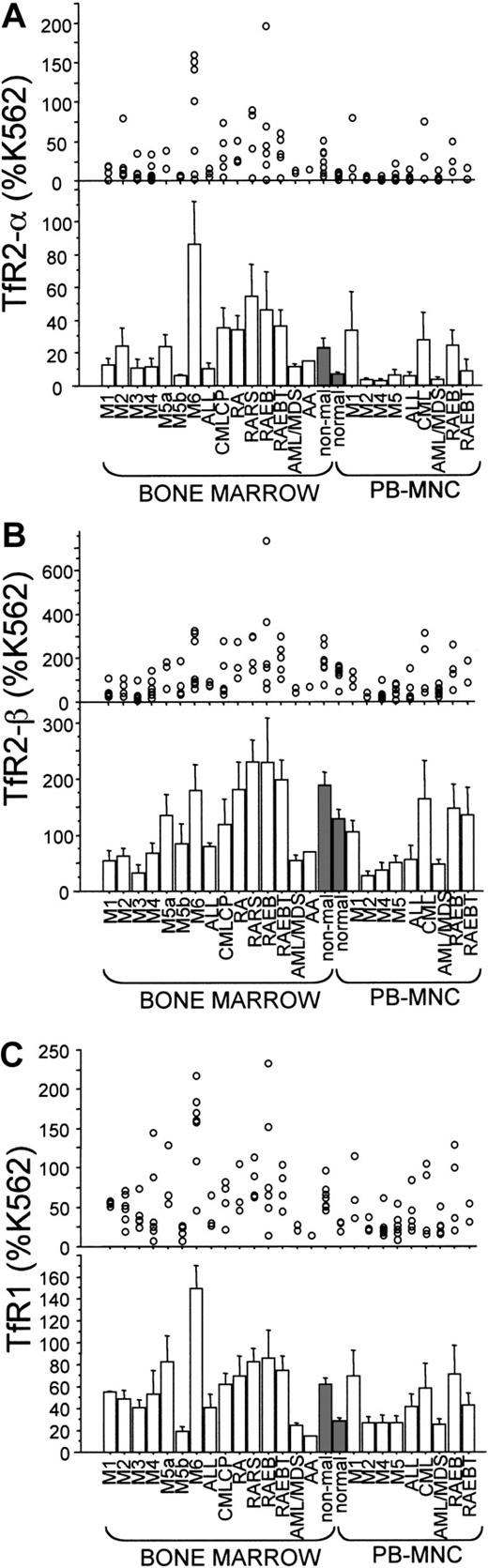
We analyzed 107 samples from 90 individuals with leukemia and preleukemia together with 8 nonmalignant BM and 6 normal PBMNC samples for expression of TfR1, TfR2-α, and TfR2-βmRNAs. Some of the samples were taken serially from the same patients during disease progression. Expression levels of TfR2-αwere higher in nonmalignant BM samples than in normal PBMNC samples (P = .038; Figure 4A, shaded bars). Among the BM samples, levels of TfR2-α expression in M6 were clearly higher than those of nonmalignant BM samples (P = .247; Figure 4A). High levels of expression ofTfR2-α (greater than 60% of K562) occurred in 13 samples: 1 M2-BM, 4 M6-BM (erythroleukemias), 1 CML-BM, 2 RARS-BM, 2 RAEB-BM, 1 RAEBT-BM, 1 M1-PB, and 1 CML-PB.
Semiquantitative RT-PCR for expression ofTfR2-α, TfR2-β, and TfR1 mRNAs.
Expression levels of TfR2-α, TfR2-β, and TfR1mRNA are shown in panels A, B, and C, respectively. Each value was calculated as a percentage of the level detected in K562 cells (% K562). In every panel, each circle represents one sample, and every bar shows a mean ± SD in its clinical category. Nonmalignant BMs and normal PBMNCs were used as controls, and the bars corresponding to them were shaded. AML/MDS indicates AML evolved from MDS.
Semiquantitative RT-PCR for expression ofTfR2-α, TfR2-β, and TfR1 mRNAs.
Expression levels of TfR2-α, TfR2-β, and TfR1mRNA are shown in panels A, B, and C, respectively. Each value was calculated as a percentage of the level detected in K562 cells (% K562). In every panel, each circle represents one sample, and every bar shows a mean ± SD in its clinical category. Nonmalignant BMs and normal PBMNCs were used as controls, and the bars corresponding to them were shaded. AML/MDS indicates AML evolved from MDS.
Differential cell counts of the BM were available from 9 of the 11 BM samples that showed high levels of TfR2-α expression (Table 1; all the M6 cases are also shown). Samples 28 and 198 were obtained from the same patient at different times: when initial diagnosis was made (sample 28) and 4 months later when blast cells became nearly 70% in the BM (sample 198). Similarly, samples M6-3 and M6-4 were from another patient at different times: at the initial diagnosis (M6-3) and at relapse (M6-4). The M6-6 sample was obtained after the individual had received repeated units of red blood cell transfusion, which may have suppressed the marrow erythroid number. Most cases with high levels of expression ofTfR2-α mRNA showed BM erythroid hyperplasia, except for sample 219, a CML case that had myeloid hyperplasia. In samples 28 and 198 (the same M6 patient), TfR2-α expression decreased (from 141% to 3% of K562) during disease progression (myeloblasts increased and erythroid cells decreased during this period). Among PB samples, one M1 and one CML (accelerated phase) sample showed high levels of TfR2-α expression; the M1 sample contained 98% blast cells, and the CML sample contained immature myeloid cells that included blast cells.
The profile of TfR2-β expression largely paralleled levels of the α form, being significantly lower in AML (except M5a and M6) and ALL as compared with nonmalignant BM samples (Figure 4B). UnlikeTfR2-α, expression of the β-form was not increased in M6 samples. Also, the expression profile ofTfR1 mRNA was largely, but not always, similar to that ofTfR2-α (Figure 4C). For example, in sample M6-6 (Table 1), expression of TfR1 was very elevated (158% of K562), and levels TfR2-α were low (only 9% of K562).
Discussion
Among the hematological cell lines that we tested, high levels ofTfR2 occurred only in erythroid cell lines (Figure 1). This suggests that expression of this gene may be selective to the erythroid lineage of hematopoietic cells. This idea was supported by our immunohistochemical staining of normal BM cells using anti-TfR2 antibody, in which a majority of erythroblasts, but not myeloid cells, were clearly stained. These results are consistent with our previous study that showed enhancement of murine TfR2 promoter activity by GATA-1.6 GATA-1 is highly expressed in erythroid cells as well as in megakaryocytes, eosinophils, and mast cells, and the putative GATA-1–binding sites of TfR2 are well conserved between human and mouse. We observed a few large cells, probably megakaryocytes, that were positive for TfR2 staining (Figure2G). However, expression of TfR2 in megakaryocytes, eosinophils, and mast cells still remains to be studied, because the numbers of these cells in our materials were very small and we could not distinguish eosinophils from neutrophils in our immunohistochemical staining method.
In the leukemic and preleukemic BM samples, levels of TfR2mRNA roughly correlated with the proportion of erythroid cells in the marrow. High levels of TfR2-α mRNA expression occurred in 4 of 7 M6 erythroleukemias, 1 of 6 M2, 1 of 5 CML, 2 of 4 RARS, 2 of 8 RAEB, and 1 of 5 RAEBT samples. Among them, marrow cell classification was available in 9 cases; all but 1 CML case, had an increased percentage of erythroid cells in the BM (Table 1) (erythroid cells greater than 30%), which is consistent with our hypothesis that the levels of TfR2 expression may be related to mass of immature erythroid cells. But this was not always the case. Two CML cases with myeloid hyperplasia (one BM and one PBMNC) and one individual with AML-M1 (PBMNCs with 98% blast cells) also highly expressedTfR2-α mRNA (Table 1 and data not shown). These results suggest that, in addition to immature erythroid cells, some myeloid, nonerythroid leukemia cells also expressed high levels ofTfR2-α. On the other hand, sample M6-5 had low levels ofTfR2 mRNA but had remarkable erythroid hyperplasia (greater than 80% erythroid cells) (Table 1).
In MEL cells, we have shown that expression of TfR2decreased and TfR1 increased during dimethyl sulfoxide–induced erythrocytic differentiation.6 Similar expression profiles were observed in normal human erythroid cells as they differentiated in vitro in the presence of erythropoietin (Figure3). The CD34+ erythroid precursors expressed high levels ofTfR2 and very low levels of TfR1 mRNAs. During their differentiation, levels of TfR2 mRNA decreased gradually and expression of TfR1 mRNA increased dramatically (Figure 3B). TfR2 has at least 2 transcripts, αand β, and our previous observations indicated that the majority of TfR2 transcripts were the αform.4 The expression profile of TfR2-αduring erythrocytic differentiation was almost the same as that ofTfR2 (Figure 3B).
We have shown that TfR2-α, similarly to TfR1, can mediate cellular iron uptake and support cell growth in TfR-deficient CHO cells. CHO cells stably expressing TfR2-α were grown in nude mice; the resulting tumors developed much faster and became much larger than those of neomycin-resistant control cells. This reflects the growth-supporting effects of TfR2-α.7 However, the physiological function of TfR2-α is still unclear. Prior studies have demonstrated that expression of TfR1 was downregulated by iron loading and upregulated by iron deficiency in the liver and K562 cells.4,5 The main mechanism of this regulation is through binding of iron-regulatory proteins (IRPs) to the iron-responsive elements (IREs) of the 3′-untranslated region of the TfR1mRNA. In a low-iron environment, IRPs bind to the IREs of theTfR1 mRNA and stabilize it, whereas in the presence of excess iron IRPs are released from IREs, resulting in degradation of these transcripts.2,19 In contrast, TfR2 is not known to be regulated by cellular iron status, has no IREs in the region of the gene, and is expressed constitutively in the liver and K562 cells.4 5 These TfR2 expression profiles raise the question of whether the only function of TfR2-α is to facilitate intracellular iron uptake.
Recently, Camaschella et al20 and Roetto et al21 reported on patients with hereditary hemochromatosis from 4 families, who had homozygous nonsense mutations (Tyr250Xaa, Glu60Xaa, or Met172Lys) of the TfR2 gene. Both Tyr250Xaa and Met172Lys mutations affect both the α andβ forms of TfR2, and the Glu60Xaa mutation affects only the α form. These reports are paradoxical in terms of our previous studies, which suggested that a function of TfR2 was to enhance cellular uptake of iron4,7; thus, the authors proposed that the major function of TfR2 may be involved in iron regulation rather than iron uptake. Intriguingly, according to the report by Camaschella et al,20 none of the hemochromatosis patients with homozygous TfR2 mutations showed erythroid abnormalities,21 although our current study showed high levels of expression of TfR2 in erythroid precursors. TfR1 or other molecules may be able to substitute for the function of TfR2 in erythroid precursors of these patients.
From the current study, we believe that TfR2 may be a useful marker for early erythroid cells. We also identified several myeloid, nonerythroid leukemia cases in which levels of expression of TfR2 were relatively high. Expression of TfR2 may have some relevance to clinical features of these cases. Functional significance of theTfR2 gene in hematopoietic cells remains to be delineated by study of TfR2-deletional mice.
We thank Drs T. McGraw, J. Minowada, M. Lanotte, I. Miyoshi, and T. Papayannopoulou for providing us with valuable cell lines and K. Yoshida (Kanazawa Medical University) for his technical assistance.
Supported in part by grants from National Institutes of Health, C. and H. Koeffler Foundation, Horn Foundation, Parker Hughes Trust, Ko-So Foundation and Kanazawa Medical University (S00-2); H.P.K. holds the Mark Goodson endowed chair of Oncology at Cedars-Sinai Medical Center and is a member of the Jonsson Cancer Center of University of California–Los Angeles.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
H. Phillip Koeffler, The Division of Hematology/Oncology, Department of Medicine, Cedars-Sinai Medical Center, Burns and Allen Research Institute, UCLA School of Medicine, Los Angeles, CA 90048.