Abstract
To identify new markers of minimal residual disease (MRD) in B-lineage acute lymphoblastic leukemia (ALL), gene expression of leukemic cells obtained from 4 patients with newly diagnosed ALL was compared with that of normal CD19+CD10+ B-cell progenitors obtained from 2 healthy donors. By cDNA array analysis, 334 of 4132 genes studied were expressed 1.5- to 5.8-fold higher in leukemic cells relative to both normal samples; 238 of these genes were also overexpressed in the leukemic cell line RS4;11. Nine genes were selected among the 274 overexpressed in at least 2 leukemic samples, and expression of the encoded proteins was measured by flow cytometry. Two proteins (caldesmon and myeloid nuclear differentiation antigen) were only weakly expressed in leukemic cells despite strong hybridization signals in the array. By contrast, 7 proteins (CD58, creatine kinase B, ninjurin1, Ref1, calpastatin, HDJ-2, and annexin VI) were expressed in B-lineage ALL cells at higher levels than in normal CD19+CD10+ B-cell progenitors (P < .05 in all comparisons). CD58 was chosen for further analysis because of its abundant and prevalent overexpression. An anti-CD58 antibody identified residual leukemic cells (0.01% to 1.13%; median, 0.03%) in 9 of 104 bone marrow samples from children with ALL in clinical remission. MRD estimates by CD58 staining correlated well with those of polymerase chain reaction amplification of immunoglobulin genes. These results indicate that studies of gene expression with cDNA arrays can aid the discovery of leukemia markers.
Introduction
In B-lineage acute lymphoblastic leukemia (ALL), the most common form of leukemia in children, the level of minimal (ie, submicroscopic) residual disease (MRD) during clinical remission is one of the most powerful prognostic indicators.1 Correlative studies have demonstrated that detection of MRD by flow cytometric or polymerase chain reaction (PCR) analysis of leukemia-specific markers is strongly associated with subsequent relapse.2-10Therefore, MRD assays are being introduced into treatment protocols as a tool to gauge treatment response and aid in the selection of therapeutic strategies.
The greatest remaining obstacle to the routine use of MRD studies in ALL therapy protocols is that none of the techniques currently available for MRD detection can be applied to all patients. PCR amplification of chromosomal breakpoints can be applied to fewer than half of all children with ALL, that is, those whose leukemic cells express nonrandom genetic abnormalities.11 The success rate of PCR analysis of antigen-receptor genes ranges from 80% to 90% because of lack of sufficiently specific leukemia sequences, oligoclonality, and clonal evolution.12-15 This method is also laborious and can be performed only in a few specialized centers. Flow cytometry is widely used for the diagnosis and classification of leukemia but can monitor MRD in only 80% to 85% of cases of B-lineage ALL because of lack of leukemia-specific immunophenotypes.16 Moreover, MRD studies by this technique require extensive panels of complex antibody combinations to distinguish leukemic lymphoblasts from their normal counterparts, the B-lymphoid progenitors of the bone marrow, and to prevent false-negative findings due to immunophenotypic changes during the course of the disease.16 Thus, the identification of new leukemia markers that are easily detectable and are stably expressed in a large proportion of B-lineage cases would greatly simplify the application of MRD studies and help to extend their benefit to all patients.
For productive detection of MRD in ALL, it is necessary to distinguish leukemic lymphoblasts from their normal counterparts, B-lymphoid progenitors that normally reside in the bone marrow. By comparing the gene expression of these 2 cell populations, it should be possible to identify differentially expressed molecules that could be used as new markers for MRD studies by flow cytometry. Leukemic and normal B-lymphoid progenitors are ideally amenable to comparative studies of gene expression because they are immunophenotypically well defined and can be isolated to a high degree of purity.
Oligonucleotide or cDNA arrays allow simultaneous testing of the expression levels of thousands of genes.17,18 This approach has been applied to define gene profiles potentially useful for the subclassification of acute leukemia19 and B-cell lymphoma20 and to identify genes upregulated in epithelial cancer cells.21-23 In this study, we first used cDNA arrays to screen for differences in gene expression between leukemic lymphoblasts and their normal counterparts, CD19+CD10+ B-lymphoid progenitors purified from bone marrow. Genes that were overexpressed in leukemic cells by cDNA array analysis were tested by flow cytometry to assess levels of protein expression and to validate their usefulness as leukemia cell markers.
Materials and methods
Cells
Bone marrow samples were obtained from children aged 1 month to 18 years (median, 5 years) at the time of diagnosis of ALL and during clinical remission, and from healthy donors aged 1.5 to 33 years (median, 13 years) during the harvest of bone marrow for stem cell transplantation. These studies were approved by the Institutional Review Board, with informed consent obtained from donors, patients, and their parents or their guardians. Immunophenotyping of leukemic cells at diagnosis was performed by standard methods.24 Leukemic and normal mononuclear cells were collected after centrifugation on a Lymphoprep density gradient (Nycomed, Oslo, Norway) and were washed 3 times in phosphate-buffered saline. For the microarray experiments, CD19+ leukemic cells were enriched using a magnetic cell separation system (Miltenyi Biotec, Bergisch Gladbach, Germany), yielding more than 98% CD19+ cell purity. Normal CD19+ cells were enriched using the same system; this step was followed by staining with anti-CD19 conjugated to phycoerythrin (PE) and anti-CD10 conjugated to fluorescein isothiocyanate (FITC) (both from Becton Dickinson, San Jose, CA) and sorting of CD19+CD10+ cells using a MoFlo high-speed fluorescence-activated cell sorter (Cytomation, Fort Collins, CO). All samples were processed or cryopreserved within 5 hours of collection.
The cell line RS4;11 was available in our laboratory. It was established from the relapse sample of a patient with acute leukemia.25 RS4;11 cells carry the t(4;11)(q21;q23) chromosomal abnormality and the corresponding MLL-AF4 gene fusion and have rearrangements of immunoglobulin (Ig) heavy chains and kappa light chains.25,26 The immunophenotypic, karyotypic, and molecular features of this cell line are characteristic of B-lineage ALL with MLL gene rearrangements.25 27
cDNA array assay
To screen cells for gene expression, we used high-density filter-based cDNA arrays purchased from Research Genetics (Huntsville, AL).28,29 The GF211 “Known genes” Genefilter array contains 4132 named human genes. Each spot on the 5-cm × 7-cm positively charged nylon membrane contains approximately 0.5 ng cDNA. The cDNAs are approximately 1 kb in length, contain the entire 3′UTR, and are all sequence-verified I.M.A.G.E./LLNL clones.30 Samples were prepared according to instructions provided by the array's manufacturer. Briefly, we used the RNeasy Mini Kit (Qiagen, Valencia, CA) to extract total RNA from CD19+cells purified from 4 ALL samples obtained at diagnosis, from CD19+CD10+ cells purified from 2 normal bone marrow samples, and from the cell line RS4;11. Total RNA (0.5-1.0 μg) was converted to cDNA by using 1.5 μL of reverse transcriptase (Superscript II; Gibco, Rockville, MD) and 2 μL of oligo dT primer in the presence of 10 μL 33P-dCTP. Labeled cDNA was purified with a Bio-Spin 6 chromatography column (BioRad, Hercules, CA), denatured, and hybridized to the gene filter for 18 hours in a hybridization roller oven at 42°C. The filter was washed and exposed to a phosphorimager storage screen that was then scanned with a Storm 860 phosphorimager (Molecular Dynamics, Sunnyvale, CA). After scanning, the filter was stripped by immersion in boiling 0.5% sodium dodecyl sulfate solution and agitation for 1 hour. To avoid inaccuracies due to inter-filter variability, we compared the gene expression of primary leukemic cells and normal lymphoid progenitors by performing sequential hybridizations on the same filter. Multiple hybridizations were performed under identical conditions; after each stripping, the filter was scanned to ensure that no residual radioactivity was detectable.
cDNA array data analysis
The phosphorimager files were imported as tagged image files into the Pathways 2.01 software (Research Genetics).29,31-33 Individual elements were mapped to an internal reference database by aligning the images in a software-based matrix using spots containing total genomic control DNA. The intensity of these spots was used to normalize the intensity of all spots in inter-hybridization comparisons. Changes in levels of expression were calculated using normalized intensity readings that were given as ratios and visualized by using green-red false-color images. An intensity mask of 5000 arbitrary units was used to exclude background and very weak hybridization signals. In all pairwise comparisons, we used 1.5-fold increase as the minimum expression change to define overexpression. This arbitrary cutoff was previously applied to the results of GF211 Genefilter to identify differentially expressed genes in resting and activated human lymphocytes.29 In preliminary experiments comparing gene expression in RS4;11 cells harvested at 2 successive days during culture, we found that in 4075 of the 4132 genes (98.6%) examined, the level of expression varied less than 1.5-fold. Among the remaining 57 genes, expression varied by 1.5-fold to less than 2-fold in 50 (1.2%) and by more than 2-fold (range, 2.1-3.5; median, 2.2) in 7 (0.17%).
Flow cytometric analysis
To verify the overexpression of gene products, we used flow cytometry with the following antibodies: an anti-CD58 monoclonal antibody conjugated to FITC (Beckman-Coulter, Miami, FL), unconjugated IgG monoclonal antibodies to creatine kinase B (a gift of Dr B. Wieringa, University of Nijmegen, The Netherlands),34ninjurin1, annexin VI (both from Transduction Laboratories, Lexington, KY), Ref1 (Novus Biologicals, Littleton, CO), calpastatin (Alexis Biochemicals, San Diego, CA), HDJ-2 (NeoMarkers, Freemont, CA), caldesmon (Novocastra, Newcastle, United Kingdom), and rabbit antiserum to myeloid nuclear differentiation antigen (Chemicon, Temecula, CA). These antibodies were used in combination with IgM monoclonal antibodies to CD10 (Boehringer Mannheim, Indianapolis, IN) or CD19 (Research Diagnostics, Flanders, NJ). Antibody binding was detected with the use of fluorochrome-conjugated species- and isotype-specific goat antisera from Jackson Immunodiagnostics (West Grove, PA) and Southern Biotechnology Associates (Birmingham, AL). Species- and isotype-matched nonreactive Igs were used as controls. Because all markers except CD58, CD10, and CD19 were expressed intracellularly, cell membranes were permeabilized with ORTHO Permeafix (ORTHO, Raritan, NJ) during the cell-labeling procedure, as described previously.16 Measurements of antibody labeling were performed by multiparameter flow cytometry using a FACSCalibur flow cytometer equipped with the CellQuest software (Becton Dickinson). In some experiments, antibody labeling of cytocentrifuge preparations was examined with a fluorescence microscope (Zeiss Axioskop, Oberkochen, Germany).
MRD assays
Studies of MRD by flow cytometry were performed with various combinations of monoclonal antibodies conjugated to FITC, PE, peridinin chlorophyll protein, and allophycocyanin, as described previously.2,5,16 The method used allows the identification of one leukemic cell among 10 000 normal bone marrow cells or greater.2,5,16 Determination of MRD by PCR amplification of Ig genes was performed as described previously.35 MRD studies by the 2 methods were done independently in separate laboratories.
Results
cDNA array screening
To identify genes overexpressed in leukemic cells, we compared the expression of genes in leukemic lymphoblasts purified from bone marrow samples taken from 4 children with B-lineage ALL at diagnosis with that of CD19+CD10+ lymphoid progenitors from bone marrow samples taken from 2 healthy individuals (Figure 1). Of the 4132 genes examined, 495 were expressed at 1.5-fold or greater in leukemic cells in at least one pairwise comparison. Each of the 495 spots of different intensities contained visually detectable hybridization signals; none of the differentially expressed genes were expressed exclusively in leukemic cells. The level of overexpression ranged from 1.5-fold to 5.8-fold; 478 of the 495 genes were overexpressed 2-fold or more in at least one comparison.
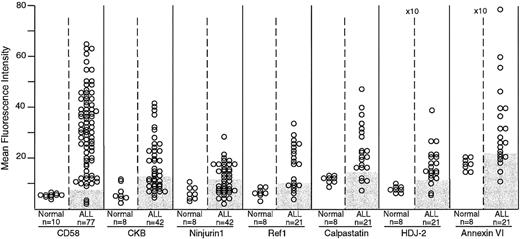
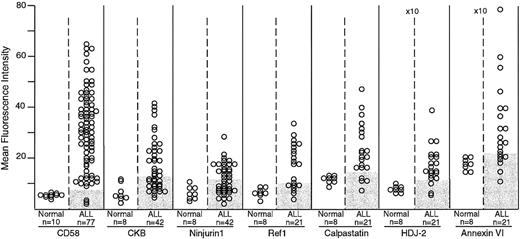
Expression of CD58, creatine kinase B (CKB), ninjurin1, Ref1, calpastatin, HDJ-2, and annexin VI in normal and leukemic immature B cells by flow cytometry.
The number of samples studied is indicated. Shaded areas in the ALL columns indicate the corresponding level of expression in normal samples for each marker. For all 7 markers, differences in mean fluorescence intensity between normal and leukemic cells were significant by t test (CD58, P < .0001; creatine kinase B, P = .007; ninjurin1,P = .013; Ref1, P = .002; calpastatin,P = .006; HDJ-2, P = .003; and annexin VI,P = .023). The mean fluorescence intensity scale for HDJ-2 and annexin VI is higher (×10) than that of the other molecules.
Expression of CD58, creatine kinase B (CKB), ninjurin1, Ref1, calpastatin, HDJ-2, and annexin VI in normal and leukemic immature B cells by flow cytometry.
The number of samples studied is indicated. Shaded areas in the ALL columns indicate the corresponding level of expression in normal samples for each marker. For all 7 markers, differences in mean fluorescence intensity between normal and leukemic cells were significant by t test (CD58, P < .0001; creatine kinase B, P = .007; ninjurin1,P = .013; Ref1, P = .002; calpastatin,P = .006; HDJ-2, P = .003; and annexin VI,P = .023). The mean fluorescence intensity scale for HDJ-2 and annexin VI is higher (×10) than that of the other molecules.
When the gene profiles of the 2 normal samples were compared, 25 of the 4132 genes were overexpressed (at least 1.5-fold) in one sample and 43 were overexpressed in the other. Because of this variability in gene expression among normal cells, we restricted our subsequent analysis to genes that were overexpressed in leukemic cells relative to both normal samples. We found that 334 genes fulfilled this criterion. Notably, 238 were also overexpressed in the cell line RS4;11 when compared with both samples of normal B-cell progenitors. In total, 274 of the 334 genes were overexpressed in more than one leukemic sample: 210 were overexpressed in 2 samples and 64 were overexpressed in 3 samples. No gene was expressed at least 1.5-fold in more than 3 samples.
Although several genes were underexpressed in ALL cells in pairwise comparisons with either normal sample, only one gene, Gal β(1-3/1-4) GlcNAc α 2,3-sialyltransferase,36 was underexpressed by 1.5-fold or greater in one case of ALL when compared with both normal samples.
Flow cytometric studies of protein expression
Because we wished to identify markers of leukemia that are suitable for MRD detection by flow cytometric analysis, we tested the expression of proteins encoded by genes found to be overexpressed in the cDNA array screening with the use of specific antibodies. For these studies, we selected 9 molecules encoded by genes overexpressed in at least 2 leukemic samples. The selected molecules had an overexpression of 2-fold or greater in at least one comparison between leukemic and normal cells. A further criterion for selecting these molecules was that specific antibodies, proven to work in immunofluorescence, were available.
Two of the 9 proteins (caldesmon and myeloid nuclear differentiation antigen) were very weakly expressed in leukemic cells by flow cytometry and fluorescence microscopy, despite strong hybridization signals in the cDNA array testing. These findings are likely to reflect low protein expression rather than poor antibody binding because cells known to express these proteins, including tonsillar follicular dendritic cells (caldesmon)37 and normal and leukemic myeloid cells (myeloid nuclear differentiation antigen),38were strongly labeled (data not shown).
The remaining 7 proteins were clearly detectable in lymphoid leukemic cells and were selected for further testing. These represented a wide variety of putative cellular functions. CD58, also known as lymphocyte function–associated antigen-3 (LFA-3), is an adhesion molecule that binds to CD2 on T cells and natural killer cells39; creatine kinase B is a cytoplasmic molecule involved in adenosine triphosphate/adenosine diphosphate metabolism40; ninjurin1 is an adhesion molecule primarily expressed in the central nervous system and epithelium41; Ref1 (also known as apurinic/apyrimidinic endonuclease [APE], APEX, HAP1) is a multifunctional enzyme that functions as an endonuclease and as a redox-modifying factor for a variety of transcription factors42; calpastatin is an inhibitor of calpain, a Ca++-dependent cysteine protease that promotes apoptosis and necrosis43,44; HDJ-2 (HSDJ, DNAJ) is a human heat-shock protein-40 homologue involved in protein transport and folding45,46; and annexin VI is a Ca++- and phospholipid-binding protein that is required for budding of clathrin-coated pits.47 In the cDNA array analysis, 5 of these genes (creatine kinase B, calpastatin, Ref1, HDJ-2, and annexin VI) were overexpressed in 2 leukemic samples, whereas 2 (CD58 and ninjurin1) were overexpressed in 3 leukemic samples. None of these 7 genes had fluctuations of 1.5-fold or greater in the preliminary studies with RS4;11 cells harvested at 2 different points during culture.
The flow cytometric studies demonstrated that the expression of these proteins was higher in ALL cells than in their normal counterparts, CD19+CD10+ B-cell progenitors (P < .05 by t test in all comparisons; Figure1). In a substantial proportion of B-lineage ALL cases, the expression of these molecules was abnormally high (Figures 1 and2). CD58, creatine kinase B, ninjurin1, and Ref1 were expressed at the lowest detectable level or were virtually undetectable in normal CD19+CD10+lymphoid progenitors, but were expressed at levels 2 to 6 times greater in multiple samples from diagnostic B-lineage ALL cases. Although the remaining 3 proteins (calpastatin, HDJ-2, and annexin VI) were expressed by normal B-lymphoid cells, cases of B-lineage ALL overexpressed these molecules. As expected from the cDNA array observations and from the known immunophenotypic and genetic heterogeneity of ALL,48 none of the molecules were overexpressed in all leukemic cases studied.
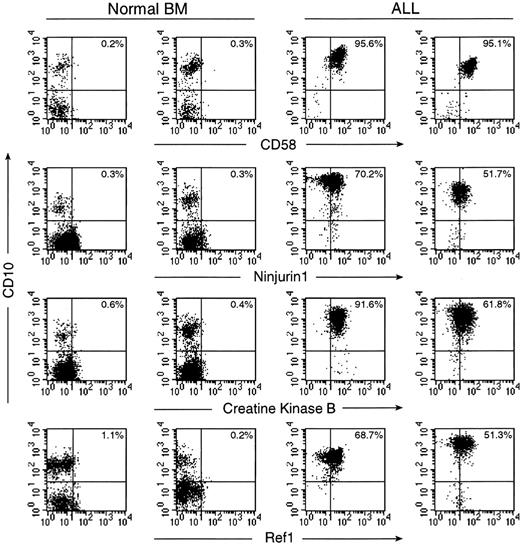
Expression of CD58, ninjurin1, creatine kinase B, and Ref1 in normal and leukemic immature B cells by flow cytometry.
For each protein, results obtained with 2 normal bone marrow samples and 2 cases of B-lineage ALL are shown. Flow cytometric dot plots illustrate labeling of CD19+ lymphoid cells with antibodies against the tested protein (FITC; x axes) and with anti-CD10 (PE; y axes). Drawing of quadrants was based on staining with isotype-matched fluorochrome–conjugated nonreactive Igs. The percentage of cells in the upper right quadrant is indicated in each plot.
Expression of CD58, ninjurin1, creatine kinase B, and Ref1 in normal and leukemic immature B cells by flow cytometry.
For each protein, results obtained with 2 normal bone marrow samples and 2 cases of B-lineage ALL are shown. Flow cytometric dot plots illustrate labeling of CD19+ lymphoid cells with antibodies against the tested protein (FITC; x axes) and with anti-CD10 (PE; y axes). Drawing of quadrants was based on staining with isotype-matched fluorochrome–conjugated nonreactive Igs. The percentage of cells in the upper right quadrant is indicated in each plot.
Validation of the CD58 marker for MRD detection
In further studies, we determined the usefulness of CD58 as a marker to monitor MRD. CD58 was selected because (1) it is highly overexpressed in a large proportion of ALL cases and (2) anti-CD58 antibodies directly conjugated to fluorochromes are commercially available; such antibodies are required for multiparametric studies of MRD by flow cytometry.5,16 We tested CD58 as a marker of MRD in 104 bone marrow samples obtained from 33 children with ALL in clinical remission. Because studies of MRD by flow cytometry require multiparameter analysis,2,5 16 we used an anti-CD58 antibody in combination with antibodies to CD19, CD10, and CD34. In 95 samples from 28 patients, CD19+ cells expressing CD10 or CD34 had a CD58 expression level that was consistently identical to that of normal samples. The remaining 9 samples from 5 patients contained B-lymphoid progenitor cells overexpressing CD58, indicative of MRD (Figure 3). The level of MRD in these samples ranged from 0.01% (the limit of detection by flow cytometry) to 1.13% (median, 0.03%). Figure4 illustrates the results of MRD monitoring in 4 patients with multiple sequential samples.
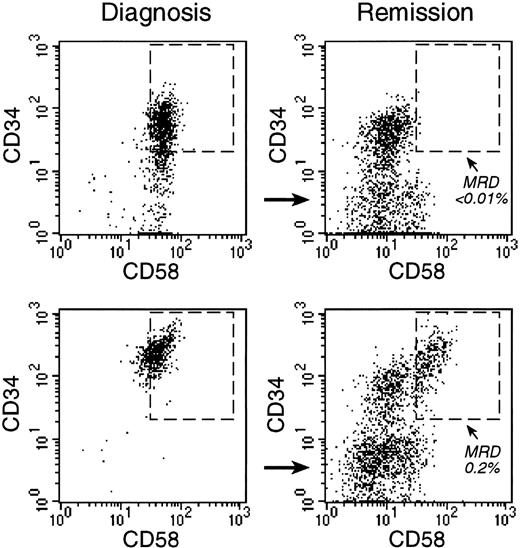
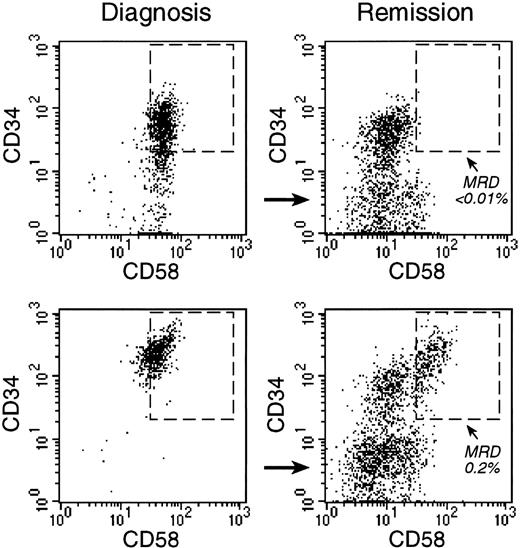
Monitoring of MRD with anti-CD58 labeling.
Bone marrow cells from 2 children with B-lineage ALL were studied at diagnosis (left panels) and at the end of remission-induction therapy (right panels), when both patients were in complete morphologic remission. Flow cytometric dot plots illustrate the expression of CD34 and CD58 on CD19+ cells. In both patients, most CD19+ cells at diagnosis were CD34+ and CD58+. At the end of remission induction, CD19+CD34+CD58+ cells represented less than 0.01% of bone marrow mononuclear cells in one patient (top). In the other patient (bottom), 0.2% of cells expressed this phenotype, indicative of MRD. PCR amplification of Ig genes confirmed these findings (less than 0.01% and 0.4% MRD, respectively).
Monitoring of MRD with anti-CD58 labeling.
Bone marrow cells from 2 children with B-lineage ALL were studied at diagnosis (left panels) and at the end of remission-induction therapy (right panels), when both patients were in complete morphologic remission. Flow cytometric dot plots illustrate the expression of CD34 and CD58 on CD19+ cells. In both patients, most CD19+ cells at diagnosis were CD34+ and CD58+. At the end of remission induction, CD19+CD34+CD58+ cells represented less than 0.01% of bone marrow mononuclear cells in one patient (top). In the other patient (bottom), 0.2% of cells expressed this phenotype, indicative of MRD. PCR amplification of Ig genes confirmed these findings (less than 0.01% and 0.4% MRD, respectively).
Sequential monitoring of MRD in 4 children with ALL.
All samples were studied during morphologic remission with the use of anti-CD58 in combination with antibodies to CD19, CD10, and CD34.
Sequential monitoring of MRD in 4 children with ALL.
All samples were studied during morphologic remission with the use of anti-CD58 in combination with antibodies to CD19, CD10, and CD34.
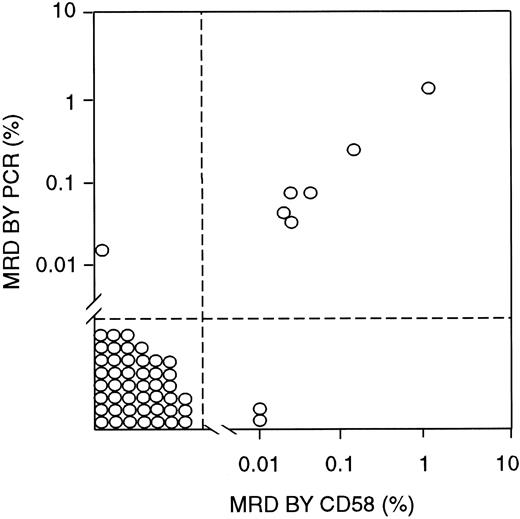
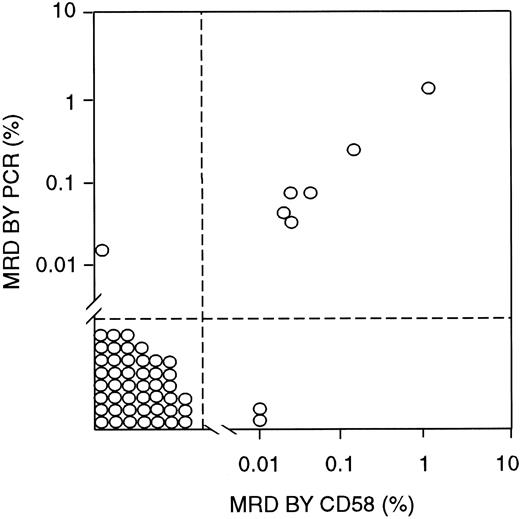
To determine the validity of MRD findings obtained by using CD58 overexpression as a marker, we compared the flow cytometry results with those of PCR amplification of Ig μ heavy-chain genes, which were available for 55 samples obtained from 29 patients (Figure5). In 46 samples from 23 patients, MRD was less than 0.01% by PCR analysis and negative by CD58 flow cytometric studies. By contrast, greater than 0.01% leukemic cells were detected by both methods in 6 samples from 3 patients, with a good correlation in the MRD estimates by the 2 techniques (r2 = 0.9993 by regression analysis; Figure5). In only 3 of the 55 samples, the 2 methods yielded discrepant results: One showed 0.02% leukemic cells by PCR but no detectable cells by CD58 analysis, and 2 others showed 0.01% MRD by CD58 analysis but less than 0.01% (0.0014% and 0.0022%) by PCR.
Correlation between MRD estimates by labeling with an anti-CD58 antibody (in combination with anti-CD19, anti-CD34, and anti-CD10 antibodies) and PCR amplification of Ig genes.
All bone marrow samples were from patients in morphologic remission.
Correlation between MRD estimates by labeling with an anti-CD58 antibody (in combination with anti-CD19, anti-CD34, and anti-CD10 antibodies) and PCR amplification of Ig genes.
All bone marrow samples were from patients in morphologic remission.
Discussion
Large-scale gene expression in cancer cells and in their normal counterparts may facilitate the identification of molecular profiles associated with neoplasia. These profiles should include markers that can be used to track residual tumor cells, measure response to therapy, and predict disease recurrence with a sensitivity and accuracy significantly superior to those afforded by current approaches. In this study, we used cDNA array analysis to identify differences between normal and leukemic immature B cells and to screen for candidate markers of leukemia to be used for monitoring residual disease. Because normal bone marrow contains cells from at least 8 lineages at multiple stages of differentiation, it was important to isolate cells with immunophenotypic features similar to those of leukemic lymphoblasts, ie, bone marrow CD19+CD10+ cells, which cannot be easily distinguished from leukemic cells in studies of MRD.16 By comparing the expression of more than 4000 genes in this relatively homogeneous cell population of B-cell progenitors to that of B-lineage leukemic lymphoblasts, we identified several genes that appeared to be frequently overexpressed in leukemic cells. Seven of the 9 genes whose expression was tested with specific antibodies to the encoded proteins were indeed overexpressed in leukemia. Therefore, these molecules represent novel markers of ALL that could be used for monitoring residual disease.
Measurement of treatment response and detection of impending relapse in patients with acute leukemia are traditionally done by microscopic examination of bone marrow smears.49 However, this procedure lacks sensitivity and accuracy, and patients who are in remission by this technique may still harbor as many as 1010 leukemic cells.50 To date, progress in the identification of new leukemia-specific markers has relied on testing the expression of markers commonly used for leukemia classification in normal bone marrow cells.16 This approach, largely based on trial and error, is slow and is hampered by the relatively limited number of markers used for routine leukemia immunophenotyping. The cDNA array screening identified molecules whose differential expression in normal and leukemic immature B-lymphoid cells had not been characterized previously, such as creatine kinase B, ninjurin1, Ref1, calpastatin, HDJ-2, and annexin VI. It also identified CD58 overexpression, in agreement with a previous report indicating high expression of this molecule in leukemic cells.51 The study of CD58 expression in combination with CD19, CD34, and CD10 identified residual leukemic cells in bone marrow samples from patients with ALL in clinical remission. Although only 9 of the 334 overexpressed molecules highlighted by the array screening were investigated at the protein level in this study, the analysis resulted in 7 markers of leukemia potentially useful for MRD monitoring. It seems likely that several additional markers will emerge from the investigation of the remaining genes, particularly those found to be overexpressed in more than one case.
Because only a limited number of cells can be obtained by purifying B-lymphoid progenitors from normal human bone marrow, we selected an array system that uses radionuclide labeling of RNA and has a high degree of sensitivity.29,52 Thus, hybridization signals were clearly detectable in samples of 0.5 to 1 μg of total RNA. This system was previously shown to provide quantitation of RNA transcript levels similar to those of quantitative PCR.29 One potential limitation of all array-based assays is that transcript expression does not always predict levels of protein expression because of posttranscriptional regulation. This is well illustrated by our finding of abundant RNA transcripts of caldesmon and myeloid nuclear differentiation antigen in leukemic cells by array analysis, which was contrasted by a very weak expression of the corresponding proteins.
In summary, our results indicate that large-scale gene profiling can be applied to identify new markers for monitoring residual cancer cells. In acute leukemia, this strategy should ultimately lead to the establishment of simple antibody panels for universal monitoring of MRD.
We thank Dr B. Wieringa (University of Nijmegen, The Netherlands) for kindly providing anti–creatine kinase B monoclonal antibody, M. Hancock for statistical assistance, and S. Naron for editorial suggestions.
Supported by grants CA60419, CA52259, CA21765, and CA20180 from the National Cancer Institute; by the Rizzo Memorial Grant from the Leukemia Research Foundation; and by the American Lebanese Syrian Associated Charities (ALSAC).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dario Campana, Dept of Hematology-Oncology, St Jude Children's Research Hospital, 332 North Lauderdale, Memphis, TN 38105; e-mail: dario.campana@stjude.org.