Abstract
The ability to modify responses to type I interferons (IFNs) could alter processes such as hematopoiesis and immunity, which involve endogenous IFNs and responses to exogenous IFNs. The data presented here support a significant role for a recently identified soluble isoform of the murine type I IFN receptor, muIfnar-2a, as an efficient regulator of IFN responses. The messenger RNA (mRNA) transcript encoding muIfnar-2a is generally more abundant than that encoding the transmembrane isoform, muIfnar-2c. Furthermore, the ratio ofmuIfnar-2a:2c transcripts varied from more than 10:1 in the liver and other organs to less than 1:1 in bone-marrow macrophages, indicating independent regulation of the 2 transcripts encoding receptor isoforms and suggesting that the soluble muIfnar-2a levels are biologically relevant in some organs. Western blot analysis showed that soluble muIfnar-2 was present at high levels in murine serum and other biologic fluids and bound type I IFN. Recombinant muIfnar-2a competitively inhibited the activity of both IFNα and β in reporter assays using the L929 cell line and in antiproliferative and antiviral assays using primary cells. Surprisingly, using primary thymocytes fromIfnar-2−/− mice, recombinant muIfnar-2a formed a complex with IFN α or β and muIfnar-1 at the cell surface and transmitted an antiproliferative signal. These data indicate potential dual actions of soluble muIfnar-2 and imply that a signal can be transduced through the Ifnar-1 chain of the receptor complex in the absence of the cytoplasmic domain of Ifnar-2. Therefore, our results suggest that soluble Ifnar-2 is an important regulator of endogenous and systemically administered type I IFN.
Introduction
The type I interferons (IFNs) are a family of cytokines produced in physiologic and pathologic conditions. Their biologic activities include inhibition of macrophage and lymphocyte proliferation, differentiation of macrophages, activation of natural killer and dendritic cells, and increased survival of memory T cells in addition to their well-characterized antiviral and antitumor activities.1-6 These activities make type I IFNs important mediators of physiologic processes and disease pathogenesis and useful in the treatment of viral infections, some cancers (particularly those of hematopoietic origin), and other diseases, including multiple sclerosis.7
In both human and mouse, the type I IFNs include multiple IFNα subtypes with about 75% to 99% amino acid identity and a single IFNβ with about 30% amino acid identity to IFNα.8 All type I IFNs have similar biologic activities and compete for binding to a common cell surface receptor.9 The type I IFN receptor is composed of 2 known subunits: IFNAR-110 and IFNAR-2.11 In humans and mice, there is a single form of IFNAR-1.12 In contrast, multiple isoforms of human IFNAR-2 have been shown to result from alternative splicing of the same gene producing huIFNAR-2a (encoding a putative soluble isoform), huIFNAR-2b (encoding a putative transmembrane receptor with a truncated cytoplasmic domain), and huIFNAR-2c (encoding a putative full-length transmembrane receptor).13 HuIFNAR-2c reconstituted IFN signaling in the huIFNAR-2–deficient cell line (U5A), which was otherwise unresponsive to IFN, whereas huIFNAR-2b did not, indicating that the latter was a nonfunctional isoform.13 We recently cloned the murine orthologue of IFNAR-2 (muIfnar-2) and identified complementary DNAs (cDNAs) encoding full-length muIfnar-2c and soluble muIfnar-2a isoforms.14 Northern blots showed 2 transcripts of 1.5 and 4.0 kilobases (kb) encoding the soluble (2a) and transmembrane (2c) isoforms, respectively. Interestingly, no muIfnar-2b has been identified in the mouse to date.
The role of IFNAR-1 and IFNAR-2 in binding type I IFNs has not been fully defined. However, studies of cloned huIFNAR-1 and of mice with a null mutation in the Ifnar-1 gene15,16indicated that this component was essential for mediating antiviral responses to type I IFNs. In addition, studies using bone marrow–derived macrophages from muIfnar-1−/−mice showed that there was still considerable binding of IFN, presumably to muIfnar-2, but no detectable signaling.14This result was consistent with the studies of huIFNAR-2 expressed in cultured mammalian cells, which indicated the probable intrinsic binding activity of IFNAR-2.17 18 However, no direct studies have been reported on the ability of IFNAR-2a to bind IFN or modify biologic responses.
To obtain data on the expression of muIfnar-2a and its biologic activity, we have used the mouse model because the murine type I IFN system is similar to the human system, and the mouse can be genetically manipulated and is more amenable to study. We present the first direct evidence for an important function for the receptor isoform muIfnar-2a. The messenger RNA (mRNA) expression of muIfnar-2a varied in different tissues and was generally more abundant. Soluble muIfnar-2 was detected at high levels in murine serum and other biologic fluids and was able to bind type I IFN. We demonstrate, for the first time, that recombinant muIfnar-2a can inhibit IFNα and IFNβ signaling in the L929 cell line and in primary thymocytes and murine embryo fibroblasts. We also demonstrate that in the absence of muIfnar-2c, muIfnar-2a can form a complex with IFNα or IFNβ and muIfnar-1 and transduce an antiproliferative response in primary thymocytes. Thus, muIfnar-2a could be an important, but so far overlooked, in vivo regulator of type I IFN responses.
Materials and methods
Cell culture, interferons, and mice
COS-7 cells and murine L929 cells were grown in DMEM and RPMI 1640, respectively, and supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin (Life Technologies, Ontario, Canada). Thymocytes were cultured in RPMI supplemented with 5% FCS, 1% penicillin/streptomycin and 2.5 × 10−5 M β-mercaptoethanol. Recombinant murine IFNβ was obtained from Toray Industries, Tokyo, Japan; recombinant murine IFNα1 was produced inPichia pastoris and purified by reverse phase high-performance liquid chromatography (HPLC) (S.T., unpublished data, May 2000); recombinant murine IFNα4 produced in NS1 cells was a gift from D. Gewert. The antiviral activity of IFN preparations or murine embryo fibroblasts was determined by cytopathic effect reduction bioassay using L929 cells,16 Semliki Forest virus, and NIH international reference preparation (Ga02-901-511) as standard. The specific activity of muIFNα1 was 5.6 × 107 IU/mg.
Mice were maintained under standard conditions
Tissues for RNA extraction were from C57BL/6XCBA mice. Thymocytes were obtained from the muIfnar-2+/+ and −/− offspring of matings between mice that were heterozygous for a null mutation in the muIfnar-2 gene (P.J.H., unpublished data, May 2000).
Isolation and Northern analysis of poly(A)+messenger RNA
Poly(A)+ mRNA from normal mouse tissues, cell lines, and primary cells was prepared essentially as described.19Northern blots of poly(A)+ mRNA were prepared and hybridized as previously described14 with a32P-labeled muIfnar-2 cDNA14 and then washed using high-stringency conditions (0.2 X SSC, 0.1% SDS at 65°C). The blots were also probed with a 32P-labeled 1.2 kb PstI fragment of glyceraldehyde-3-phosphate (GAPDH) cDNA to normalize the different samples for RNA loading. To enable valid comparisons across separate filters, sets of RNA samples for quantification experiments were simultaneously electrophoresed, transferred to membranes, and probed with the same preparation of 32P-labeled probe. Northern blots were exposed to a phosphor screen, and a quantitative analysis was performed using a Fuji BAS 1000 PhosphorImage analyzer and MacBAS v2.5 software (Fuji, Melbourne, Australia).
Construction of plasmids
To generate recombinant muIfnar-2a protein using the bacterial expression vector pGEX-2T, a cDNA clone encoding muIfnar-2a14 was modified using Pfu polymerase (Stratagene, La Jolla, CA). A BamHI site was inserted at the 5′ end immediately after the predicted leader peptide and an EcoRI site after the predicted stop codon to enable cloning, resulting in the construct pGEXmuIfnar-2a.
To produce a recombinant extracellular domain (ECD) of muIfnar-2c, the 5′ end of a cDNA clone encoding muIfnar-2c14 was modified to encode a SnaBI site and an in-frame human c-myc 9E10 epitope (SMEQKLISEEDLN).20 The yeast α-factor signal peptide was substituted for the native muIfnar-2c signal peptide, and the N-terminus of the recombinant muIfnar-2c ECD was at amino acid position 28. The 3′ end was also modified by introducing 6 histidine residues for subsequent affinity purification and an in-frame stop codon immediately before the transmembrane domain after amino acid position 242.14 Introduction of anXbaI site at the 3′ end allowed cloning of the polymerase chain reaction (PCR)-modified fragment into the Pichia pastoris expression vector pPIC9, which was digested withSnaBI and AvrII. Construction of the modified muIfnar-2c (pmyc-Ifnar-2-His ECD) was carried out using a PCR-based technique21 and Pfu polymerase.
The nucleotide sequences of all PCR-generated constructs were confirmed by dideoxy sequencing using an ABI PRISM Ready Reaction sequencing kit (PE Applied Biosystems, Scoresby, Australia).
To generate plasmids encoding muIfnar-2 for expression in COS7 cells, a 3-kb EcoRV fragment containing the entire coding region ofmuIfnar-2c14 was ligated into the blunt-endedXbaI site of pEF-BOS.22 AHincII-SmaI fragment containing the entire coding region of muIfnar-2a14 was subcloned into theBstXI site of pEF-BOS after the addition of BstXI linkers.
To construct a type I IFN-responsive reporter plasmid, aXbaI-SpeI fragment containing the 2′-5′ oligoadenylate synthetase gene promoter was inserted into theNheI site of pGL3-Basic (Promega, Madison, WI). For stable cell line generation, a XhoI-SalI fragment containing the neomycin resistance gene from pMCINeo was inserted, resulting in the construct p2′5′OASLucNeo.
Expression and purification of recombinant muIfnar-2c extracellular domain in Pichia pastoris
To express recombinant muIfnar-2c ECD in P pastoris, the plasmid pmyc-Ifnar-2-His ECD was digested with eitherBglII or SalI to obtain recombinant clones with different methanol utilization phenotypes (His+Muts: methanol utilization slow, or His+Mut+: wild-type methanol utilization, respectively). Integration into host cells was accomplished by electroporation of his4 (GS115) P pastoris cells according to the manufacturer's recommendations (Invitrogen). Selection of yeast colonies expressing myc-muIfnar-2-His ECD (muIfnar-2 ECD, San Diego, CA) was carried out essentially as previously described23 using a 9E10 anti-mycantibody. The majority of the His+ transformants were 9E10 positive and were of the Mut+ phenotype. However, levels of protein expression appeared highest in the few 9E10+Muts positive clones, and hence, the highest-producing Muts clone was chosen for further experiments. This clone was cultured as previously described,23 except 1.5% methanol was added to induce the cells to express the heterologous protein. Expression of muIfnar-2 ECD was analyzed by SDS-PAGE of the culture supernatant, followed by Western blotting and detection with 9E10 antibody.
P pastoris expression supernatant containing recombinant muIfnar-2 ECD protein was dialyzed in 20 mM Tris and 100 mM NaCl (pH 8.0) for 24 hours at 4°C. Adsorption of muIfnar-2 ECD protein to TALON cobalt resin (Clontech, Palo Alto, CA) was performed under native conditions according to the manufacturer's recommendations. The purified protein was concentrated using Centricon-10 columns (Amicon, North Ryde, Australia) according to the manufacturer's recommendations, and the purity assessed by SDS-PAGE, followed by silver staining.24 The protein concentration in the preparation was determined against bovine serum albumin (BSA) standards using Bradford25 reagent (Biorad, Richmond, VA). Approximately 328 μg was purified from a 600-mL culture (0.55 mg/L).
Expression and purification of recombinant muIfnar-2a inEscherichia coli
Recombinant muIfnar-2a was expressed as a fusion protein with glutathione S-transferase (GST) using Escherichia coli DH10B cells transformed with the plasmid pGEXmuIfnar-2a essentially as described previously.26 Benzamidine sepharose 50% (wt/vol) (Pharmacia, Piscataway, NJ) was added to the thrombin (Sigma, St Louis, MO)–cleaved protein preparation and incubated for 2 hours at 4°C to remove the residual thrombin. The purity and concentration of the protein preparation was determined as previously described, with approximately 80 to 100 μg/mL of recombinant muIfnar-2a protein obtained per preparation. SDS-PAGE analysis demonstrated a band of approximately 29 kd (data not shown) that was consistent with the predicted size of 28.9 kd derived from the cDNA sequence.14
Antibody production
For the first immunization, an equal volume of Freund's complete adjuvant was added to approximately 150 μg of purified muIfnar-2 ECD produced in P pastoris, and the emulsified mixture injected subcutaneously into multiple sites on New Zealand White rabbits. For subsequent immunizations, incomplete Freund's adjuvant was added to the same amount of purified protein. After 3 immunizations, the rabbits were bled and their sera collected. An antibody titer of 10 000 was confirmed by enzyme-linked immunosorbent assay (ELISA) using recombinant muIfnar-2 ECD and muIfnar-2a proteins as antigens, the collected sera as the primary antibody, horseradish peroxidase–conjugated goat antirabbit Ig as the secondary antibody (Silenus, Melbourne, Australia), and ABTS (Sigma) as a colorimetric substrate.
Western blot analysis
Proteins separated by SDS-PAGE were transferred onto prewetted Hybond-C Extra membranes using a transfer buffer containing 25 mM Tris, 192 mM Glycine, 0.1% SDS, 20% (vol/vol) methanol in a Semiphor transfer system (Hoeffer, Piscataway, NJ) at 100 mA for 1 to 2 hours. After transfer, membranes were incubated in 1% BSA (Sigma)/0.1% Tween-20 (ICN, Costa Mesa, CA) in phosphate-buffered saline (PBS) containing no sodium azide, followed by incubation with anti–muIfnar-2 ECD antibody and then with horseradish peroxidase-conjugated goat antirabbit Ig antibody (Silenus). Recombinant proteins were visualized with enhanced chemiluminescence (ECL) reagents (Supersignal; Pierce, Birminghamshire, United Kingdom), followed by autoradiography. In competition experiments, the primary antibody and competitor protein were preincubated together for 1 hour at 37°C before being added to the membrane. To deglycosylate proteins with endoglycosidase F (400 mU/mL) or O-glycosidase (500 mU/mL; Boehringer Mannheim, Castle Hill, Australia), the protein sample (murine serum or muIfnar-2 ECD) was diluted in 0.1% SDS, heated at 95°C for 5 minutes, then incubated for 16 to 24 hours at 37°C in 0.2 M NaOAc, 0.2% Triton X-100 (ICN), and different concentrations of enzyme(s). Where endoglycosidase H (5 U/mL; Boehringer Mannheim) was used, the protein sample was treated as above and incubated in 0.2 M NaOAc. SDS-PAGE and Western blot analysis were then performed as described previously.
Binding assays
Recombinant muIfnar-2a.
Binding assays for recombinant muIfnar-2a were carried out as previously described.27,28 ConA Sepharose (Pharmacia) was diluted 1:4 in 100 mM NaOAc pH 6.0, 1 mM MgCl2, 1 mM MnCl2, 1 mM CaCl2, 0.02% Tween-20, and 0.02% NaN3. P pastoris–derived recombinant muIfnar-2 ECD at 5 ng/μL and 10 ng/μL was incubated at room temperature for 3 hours with 5 × 105 cpm of 32P-muIFNα1-P (S.T., C.M.O., and P.J.H., unpublished data, May 2000) and with or without a 100-fold excess of unlabeled muIFNα4 competitor. Control experiments showed that muIFNα1-P (containing a kinase site)29 (S.T., C.M.O., and P.J.H., unpublished data, May 2000) could be labeled with 32P without affecting antiviral specific activity (data not shown).32P-muIFNα1 bound to ConA Sepharose was separated from unbound 32P-muIFNα1 by centrifugation through a FCS gradient, and the radioactivity measured in a β-counter. Specific binding is defined as the difference between the amount of32P-muIFNα1 bound to the receptor and immobilized on ConA Sepharose in the presence and the absence of excess unlabeled muIFNα4 competitor.
Serum.
Murine serum was diluted to a final concentration of 1:100 in RPMI 1640 containing 10% FCS, 1% penicillin/streptomycin, 50 mM HEPES pH 7.4, 1 mM CaCl2, and incubated at room temperature for 1 hour with32P- muIFNα1, with or without a 100-fold excess of unlabeled muIFNα4 competitor. Bound and free32P-muIFNα1 was separated through a Sephadex G-50 column (Pharmacia), and total binding was measured in a β-counter.
Transient transfections
Transient expression of muIfnar-2a and muIfnar-2c into COS-7 cells was achieved by electroporation with plasmid DNA essentially as previously described.26 After 72 hours posttransfection, the culture medium was collected, cellular debris removed by centrifugation, and the supernatant concentrated 10-fold by using Centricon-10 columns before Western blot analysis.
Stable transfections and luciferase reporter assays
For the generation of stable cell lines, the murine fibroblast L929 cell line was transfected with 10 μg of aNotI-digested p2′5′OASLucNeo cDNA by electroporation. Cells were selected with 600 μg/mL Geneticin (Life Technologies) for 2 weeks, resulting in the isolation of 24 resistant clones. To analyze type I IFN responsiveness, 1 × 105 cells from each clone were incubated for 7 hours at 37°C (5% CO2) with 100 IU/mL recombinant muIFNα1, lysed directly in reporter lysis buffer (Promega), and luciferase light units analyzed on a Lumat LB9501 Luminometer (Berthold, Melbourne, Australia) by using a Promega Luciferase Assay kit. The clone (2′5′OASLuc L cells) showing the greatest IFN-induced luciferase activity was chosen for subsequent experiments.
Blocking of IFN-induced luciferase activity by recombinant soluble muIfnar-2
An aliquot of 1 × 105 2′5′OASLuc L cells was added to each well of a 24-well plate. After 24 hours, the media was removed and to each well was added in duplicate either 10, 1, or 0 IU/mL muIFNα1 or muIFNβ preincubated at 37°C for 1 hour with either 100, 10, 1, or 0 ng/mL recombinant muIfnar-2a/muIfnar-2 ECD, or muIfnar-2a/muIfnar-2 ECD, heat-inactivated at 95°C for 10 minutes. The plates were incubated at 37°C for 7 hours, then the cells in each well analyzed for luciferase activity as described previously.
Antiproliferative assays
Thymi were removed from C57BL/6XCBA or C57BL/6 age-matched mice, or mice with a null mutation of the muIfnar-2 gene (P.J.H., unpublished data, May 2000), their cells dispersed in PBS through a fine-mesh sieve and washed with PBS. Aliquots of 5 × 105 cells in medium were added to each well of a 96-well flat-bottomed plate, followed by the addition of 40 μg/mL phytohaemagglutinin (PHA; Sigma) and recombinant muIFN with or without purified recombinant muIfnar-2a in quadruplicate as above. After 30 hours incubation at 37°C, 0.0185 MBq (0.5 μCi)3H-thymidine (ICN) was added to each well. Preliminary experiments showed that 48 hours of PHA stimulation was the optimum time for maximum 3H-thymidine incorporation (a typical experiment gave 2200 cpm compared with a background of about 100 cpm). After a further 18 hours incubation, the thymocytes were harvested by using a Beckman 96-well Micromate cell harvester onto glass fiber disks (Packard, Sydney, Australia). These were placed into separate scintillation vials containing 1 mL scintillant (National Diagnostics, Somerville, NJ) and counted in a 1900TR Liquid Scintillation Analyzer (Packard). The antiproliferative activity of IFN was determined as the percentage reduction in 3H-thymidine incorporation relative to untreated PHA-stimulated thymocytes.
Statistical analyses
Differences between means were determined using a 2-tailed Student t test.
Results
Expression of muIfnar-2 messenger RNA isoforms in normal adult mouse tissues and during development
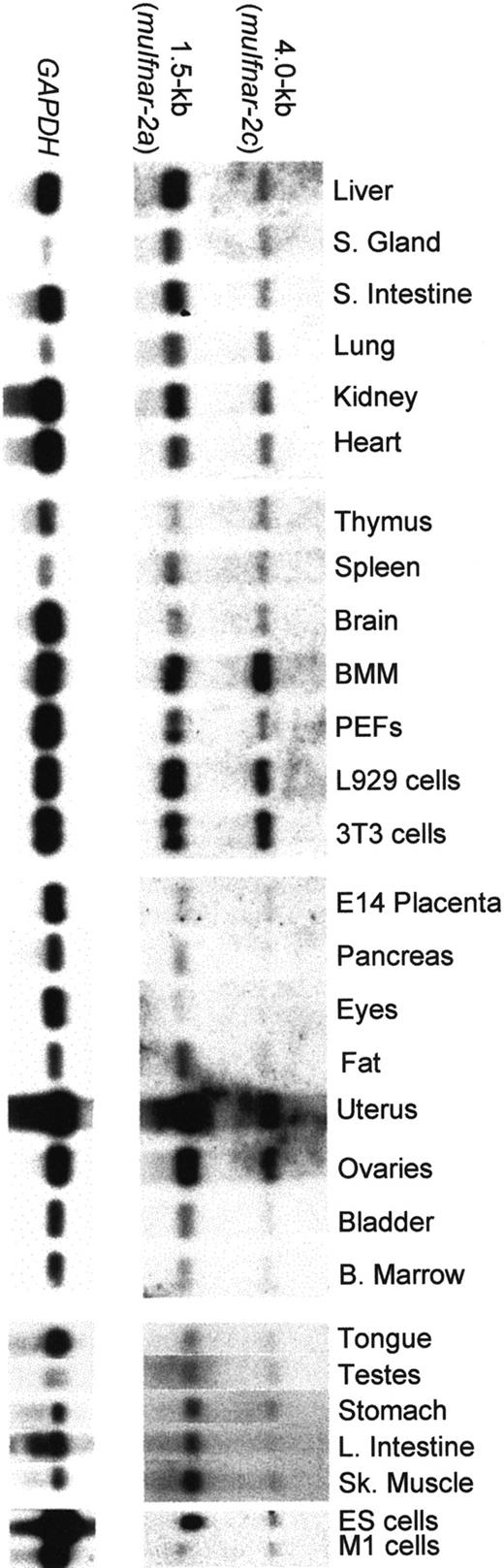
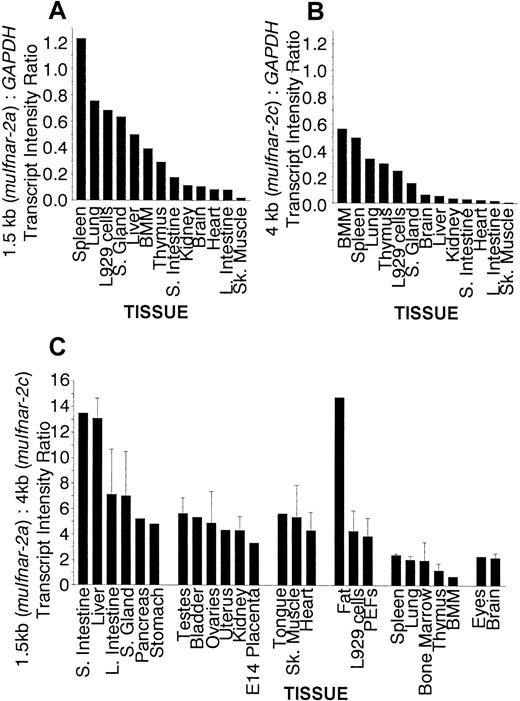
In order to investigate the in vivo function of muIfnar-2, an extensive panel of poly(A)+ mRNAs from normal adult mouse tissues, primary cells, and murine cell lines was tested for the expression of muIfnar-2 transcripts. Figure1 shows major transcripts of 1.5 and 4 kb in all the tissues, cells, and cell lines examined. In the majority of tissues tested, the 1.5-kb mRNA transcript encoding muIfnar-2a was more abundant than the 4-kb transcript encoding muIfnar-2c. The levels of each muIfnar-2 transcript relative to the levels of the housekeeping gene GAPDH were quantified using a PhosphorImager (Fuji). Figure 2A shows that there was a range of expression levels in the various tissues of the 1.5-kb transcript encoding muIfnar-2a relative to GAPDH. Similarly, the levels of the 4-kb transcript relative toGAPDH also showed a marked variation between tissues (Figure2B). The hierarchy of expression for both transcripts in the different mouse tissues was similar in some cases (Figure 2A-B). The most marked exception was the liver in which the ratio of the 1.5-kb transcript relative to GAPDH was high, whereas the relative levels of the 4-kb transcript were comparatively low. Figure 2C shows the ratio of the 1.5-kb transcript (muIfnar-2a) to that of the 4-kb transcript (muIfnar-2c) in a range of tissues. In tissues from the gastrointestinal and urogenital systems, muscle, and connective tissues, high-to-moderate ratios of the 1.5- to 4-kb transcripts were observed. Ratios of more than 10:1 were observed in the small intestine, liver, and fat. However, in the lung and in the tissues of hematopoietic origin, eg, thymus, bone marrow–derived macrophages (BMM), and bone marrow, the ratio of the 1.5- to 4-kb transcript was as low as 0.8:1. These data demonstrate that the expression of the mRNA transcript encoding the soluble receptor muIfnar-2a is greater than that encoding the transmembrane isoform muIfnar-2c in most tissues and that the expression of both isoforms varies markedly between tissues.
Northern blot analysis of poly(A)+ mRNA from normal adult mouse tissues, primary cells and cell lines showing the distribution of
muIfnar-2 mRNA isoforms. The blots were sequentially hybridized with a full-length muIfnar-2c cDNA probe and a GAPDH probe.
Northern blot analysis of poly(A)+ mRNA from normal adult mouse tissues, primary cells and cell lines showing the distribution of
muIfnar-2 mRNA isoforms. The blots were sequentially hybridized with a full-length muIfnar-2c cDNA probe and a GAPDH probe.
Quantitiation of
muIfnar-2 mRNA expression in normal adult mouse tissues, primary cells and cell lines. (A) Quantitative analysis of Northern blot expression of muIfnar-2a relative toGAPDH. Northern blots of poly(A)+ mRNA (3 μg) from a representative range of mouse tissues, primary cells, and cell lines were simultaneously electrophoresed then transferred to membranes. The blots were sequentially hybridized with a full-lengthmuIfnar-2c cDNA probe and a GAPDH probe and then exposed to a phosphor screen. The radioactivity in each band hybridizing to the muIfnar-2c and the GAPDHprobes were quantified using a Phosphorimage analyzer with MacBAS v2.5 software. (B) Quantitative analysis of Northern blot expression ofmuIfnar-2c relative to GAPDH. Northern blots of poly(A)+ mRNA (3 μg) from a representative range of mouse tissues, primary cells, and cell lines were simultaneously electrophoresed then transferred to membranes and analyzed as described previously. (C) Quantitative analysis of Northern blot expression ofmuIfnar-2a relative to muIfnar-2c. Northern blots of poly(A)+ mRNA (3 μg) from mouse tissues, primary cells, and cell lines were hybridized with full-lengthmuIfnar-2c cDNA probe and then exposed to a phosphor screen and quantitated as described previously. Where indicated by error bars, data are the mean ± SEM of at least 3 replicate experiments.
Quantitiation of
muIfnar-2 mRNA expression in normal adult mouse tissues, primary cells and cell lines. (A) Quantitative analysis of Northern blot expression of muIfnar-2a relative toGAPDH. Northern blots of poly(A)+ mRNA (3 μg) from a representative range of mouse tissues, primary cells, and cell lines were simultaneously electrophoresed then transferred to membranes. The blots were sequentially hybridized with a full-lengthmuIfnar-2c cDNA probe and a GAPDH probe and then exposed to a phosphor screen. The radioactivity in each band hybridizing to the muIfnar-2c and the GAPDHprobes were quantified using a Phosphorimage analyzer with MacBAS v2.5 software. (B) Quantitative analysis of Northern blot expression ofmuIfnar-2c relative to GAPDH. Northern blots of poly(A)+ mRNA (3 μg) from a representative range of mouse tissues, primary cells, and cell lines were simultaneously electrophoresed then transferred to membranes and analyzed as described previously. (C) Quantitative analysis of Northern blot expression ofmuIfnar-2a relative to muIfnar-2c. Northern blots of poly(A)+ mRNA (3 μg) from mouse tissues, primary cells, and cell lines were hybridized with full-lengthmuIfnar-2c cDNA probe and then exposed to a phosphor screen and quantitated as described previously. Where indicated by error bars, data are the mean ± SEM of at least 3 replicate experiments.
Because the expressions of type I IFNs are developmentally regulated,3 we examined the expression ofmuIfnar-2 at different stages of embryonic and neonatal development. Northern analysis of poly(A)+ mRNA from a range of tissues during development showed that transcripts encoding both muIfnar-2a and muIfnar-2c could be detected as early as embryonic day 10, in all tissues examined from embryonic day 14 (Figure3A), and in embryonic stem (ES) cells (Figure 1). Figure 3B shows that the ratio of the intensity of the 1.5-kb transcript (2a) to that of the 4-kb transcript (2c) was low and virtually unchanged in all the tissues examined during embryonic development up to the neonate. The 1.5-kb:4-kb transcript ratio was relatively constant between the neonate and the adult stages in the lung, spleen, and brain. In contrast, a dramatic increase in the 1.5-kb:4-kb ratio occurred between the neonatal and the adult stages in the liver and stomach, implying an increased requirement for muIfnar-2a in these tissues.
Quantitative analysis at different stages of development.
(A) Northern blot analysis of poly(A)+ mRNA from whole mouse embryos and embryonic organs at different stages of development and normal adult mouse tissues showing the distribution ofmuIfnar-2 mRNA isoforms. The blots were sequentially hybridized with a full-length muIfnar-2c cDNA probe and aGAPDH probe. (B) Quantitation of muIfnar-2 mRNA expression during murine development. Quantitative analysis of Northern blot expression of muIfnar-2a relative tomuIfnar-2c in whole mouse embryos and embryonic organs at different stages of development, and normal adult mouse tissues. Data are representative of replicate experiments.
Quantitative analysis at different stages of development.
(A) Northern blot analysis of poly(A)+ mRNA from whole mouse embryos and embryonic organs at different stages of development and normal adult mouse tissues showing the distribution ofmuIfnar-2 mRNA isoforms. The blots were sequentially hybridized with a full-length muIfnar-2c cDNA probe and aGAPDH probe. (B) Quantitation of muIfnar-2 mRNA expression during murine development. Quantitative analysis of Northern blot expression of muIfnar-2a relative tomuIfnar-2c in whole mouse embryos and embryonic organs at different stages of development, and normal adult mouse tissues. Data are representative of replicate experiments.
Characterization of muIfnar-2a protein
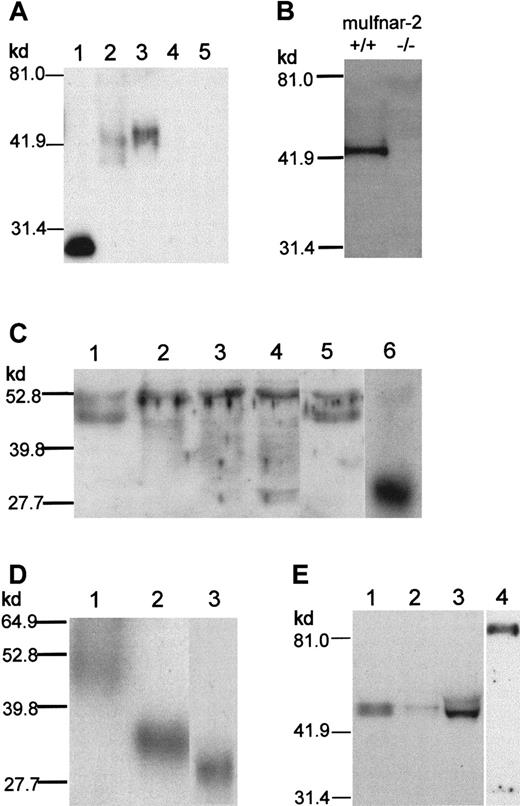
Because mRNA expression data implied that the regulation ofmuIfnar-2a was functionally important, it was necessary to characterize the muIfnar-2a protein. To address this question, we produced polyclonal antibodies to recombinant muIfnar-2 ECD produced inP pastoris. In Western blot analysis using the muIfnar-2 polyclonal antibodies, recombinant muIfnar-2a produced in E coli showed a band of approximately 29 kd (Figure4A, lane 1; 450 ng loaded), whereas purified muIfnar-2 ECD produced in P pastoris showed a band at approximately 42 kd (Figure 4A, lane 2; 30 ng loaded). Western blot analysis of the culture supernatant from COS-7 mammalian cells transfected with a muIfnar-2a expression construct revealed a band at approximately 45 kd (Figure 4A, lane 3). This protein was not present in the culture medium of COS-7 cells transiently transfected with either an expression construct encoding muIfnar-2c or pEF-BOS vector DNA alone (Figure 4A, lanes 4,5). However, a band of the expected size for the transmembrane form of muIfnar-2 was detected in the membrane fraction (data not shown). This latter result indicated that, in this case, the majority of soluble muIfnar-2 protein was not produced by proteolytic cleavage of muIfnar-2c protein at the cell surface, but translated from the muIfnar-2atranscript.
Characterization of anti–muIfnar-2 antibodies and Western blot analysis of muIfnar-2 proteins.
(A) Western blot analysis. Recombinant muIfnar-2 proteins produced inE coli (450 ng; lane 1, muIfnar-2a), P pastoris(30 ng; lane 2, muIfnar-2 ECD), COS-7 cell culture supernatant (1 μL of a 1:10 dilution; lanes 3,4,5 muIfnar-2a, muIfnar-2c, and pEF-BOS vector control, respectively) were separated by SDS-PAGE under reducing conditions, transferred to Hybond C-extra membranes and incubated with anti–muIFNAR-2 antibodies as described in “Materials and methods.” (B) Western blots of serum from mice wild-type (+/+) and homozygous (−/−) for the muIfnar-2 gene. (C) Western blots of 1:20 diluted serum that were untreated (lane 1), or treated with 0.4 mU (lane 2), 0.8 mU (lane 3), 1.6 mU (lane 4) endoglycosidase F or 2.0 mU O-glycosidase (lane 5), and 150 ng E coli-derived recombinant muIfnar-2a as a reference (lane 6). (D) Western blots of 150 ng muIfnar-2 ECD untreated (lane 1), treated with 7.5 μU endoglycosidase H (lane 2), and recombinant muIfnar-2a (lane 3). (E) Western blots of normal adult mouse serum (lane 1, 1:20 dilution), mouse peritoneal wash (lane 2, 1:2 dilution), mouse urine (lane 3), and mouse mouth wash (lane 4).
Characterization of anti–muIfnar-2 antibodies and Western blot analysis of muIfnar-2 proteins.
(A) Western blot analysis. Recombinant muIfnar-2 proteins produced inE coli (450 ng; lane 1, muIfnar-2a), P pastoris(30 ng; lane 2, muIfnar-2 ECD), COS-7 cell culture supernatant (1 μL of a 1:10 dilution; lanes 3,4,5 muIfnar-2a, muIfnar-2c, and pEF-BOS vector control, respectively) were separated by SDS-PAGE under reducing conditions, transferred to Hybond C-extra membranes and incubated with anti–muIFNAR-2 antibodies as described in “Materials and methods.” (B) Western blots of serum from mice wild-type (+/+) and homozygous (−/−) for the muIfnar-2 gene. (C) Western blots of 1:20 diluted serum that were untreated (lane 1), or treated with 0.4 mU (lane 2), 0.8 mU (lane 3), 1.6 mU (lane 4) endoglycosidase F or 2.0 mU O-glycosidase (lane 5), and 150 ng E coli-derived recombinant muIfnar-2a as a reference (lane 6). (D) Western blots of 150 ng muIfnar-2 ECD untreated (lane 1), treated with 7.5 μU endoglycosidase H (lane 2), and recombinant muIfnar-2a (lane 3). (E) Western blots of normal adult mouse serum (lane 1, 1:20 dilution), mouse peritoneal wash (lane 2, 1:2 dilution), mouse urine (lane 3), and mouse mouth wash (lane 4).
A major band of approximately 45 kd was readily detected in the serum of normal adult mice by Western blot (Figure 4B). This band was not detectable in the serum of mice with a null mutation of themuIfnar-2 gene (P.J.H., unpublished data, May 2000) (Figure 4B). Experiments using recombinant muIfnar-2a as a competitor showed that only the 45-kd band was specifically competed (data not shown). These results confirm the specificity of both the 45-kd band and the anti–muIfnar-2 polyclonal antibodies. In order to obtain an approximate estimate of soluble muIfnar-2 serum concentrations, a comparison of soluble muIfnar-2 protein levels in normal mouse serum to serial dilutions of recombinant muIfnar-2a protein by Western blot analysis indicated that it was present at approximately 20 to 50 μg/mL (data not shown).
In order to determine whether the molecular weight difference between E coli–derived recombinant muIfnar-2a and serum muIfnar-2 or muIfnar-2 ECD was due to glycosylation, serum and P pastoris–derived recombinant muIfnar-2 ECD were treated with various N- and O-glycosidases. Treatment of serum with endoglycosidase F resulted in the loss of the specific 45-kd band corresponding to soluble muIfnar-2 and the appearance of multiple bands, ranging in mobility from 44 to 30 kd, which is the observed mobility of E coli–derived muIfnar-2a (Figure 4C). These bands were not due to degradation, as they were not present in untreated serum and serum treated with O-glycosidase. The band at approximately 53 kd is nonspecific because it was unable to be specifically competed with recombinant muIfnar-2. The mobility of P pastoris–derived muIfnar-2 ECD could be reduced to within 3 kd of E coli–derived muIfnar-2a (Figure 4D); the additional 3 kd is due to the myc and His-epitopes present on muIfnar-2 ECD.
A range of adult mouse biologic fluids was examined for the presence of soluble muIfnar-2 by Western blot analysis with the polyclonal anti–muIfnar-2 ECD antibody. A 45-kd band corresponding to the muIfnar-2a protein was detected not only in serum, but also at a relatively lower abundance in the urine and a peritoneal wash of normal adult mice (Figure 4E, lanes 1,2,3, respectively). An immunoreactive protein, migrating at approximately 85 to 90 kd, was detected in murine mouth rinsings (Figure 4E, lane 4). The mobility of this protein is suggestive of a dimeric form of muIfnar-2a, although it was unable to be reduced to a lower Mr, even with 1 M DTT as the reducing agent (data not shown). The band corresponding to this protein was able to be competed with recombinant muIfnar-2a (data not shown). The detection of soluble muIfnar-2 in saliva is consistent with the highmuIfnar-2a:GAPDH ratio shown in Figure 2A. MuIfnar-2a protein could also be detected at very low levels in the culture media of the L929 and murine 3T3 cell lines (data not shown). Therefore, not only was the muIfnar-2a mRNA transcript expressed at high levels, but there were significant levels of muIfnar-2 protein circulating in vivo, thus providing an opportunity for it to systemically regulate the actions of type I IFNs.
Binding of IFN by soluble muIfnar-2
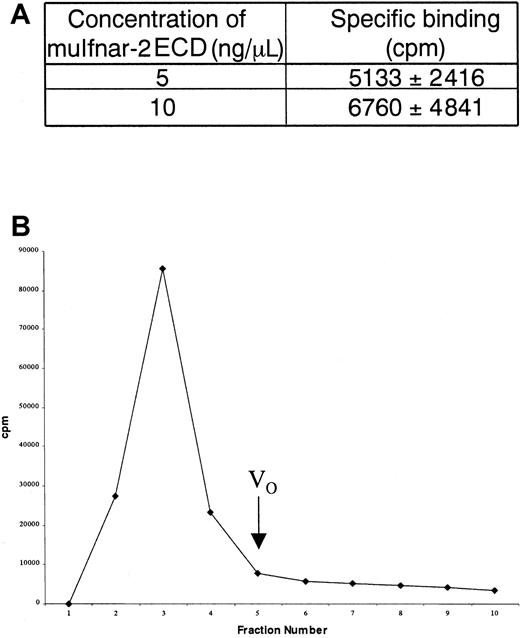
In order to determine whether the soluble receptor could bind IFN ligand, we used the ability of ConA Sepharose to precipitate glycosylated muIfnar-2 ECD bound to the unglycosylated muIFNα1. Significant amounts of 32P-muIFNα1 (5133 and 6760 cpm) were precipitated with soluble muIfnar-2 ECD (5 and 10 μg/mL, respectively). This could be competed with unlabeled muIFNα4, indicating specific binding of the ligand to receptor (Figure5A). In separate experiments, serum was mixed with 32P-muIFNα1, then applied to Sephadex G-50 gel filtration columns (Pharmacia) to determine the extent of high-molecular-weight complex formation. A significant proportion of32P-muIFNα1 was excluded from the column (Figure 5B), consistent with the presence of receptor-ligand complexes of Mr greater than or equal to 50 kd (where the Mrof IFNα1 and soluble muIfnar-2 ECD is 20 kd and 42 kd, respectively). The radioactivity in the high-molecular-weight complex could be competed with unlabeled muIFNα4, indicating that the binding was specific.
Binding of soluble muIfnar-2 to type I IFN.
(A) Table showing the specific binding of 32P-muIFNα1 to 5 ng/μL and 10 ng/μL recombinant muIfnar-2 ECD. Data are expressed as the mean ± SD cpm. (B) Histogram showing the elution profile of total 32P-muIFNα1 bound to high-molecular weight complexes in serum. The mean cpm of fractions 2 to 4 were used to determine total binding (mean = 80 403 cpm), specific binding (mean = 38 440 cpm) and nonspecific binding (mean = 41 963 cpm). VO indicates void volume.
Binding of soluble muIfnar-2 to type I IFN.
(A) Table showing the specific binding of 32P-muIFNα1 to 5 ng/μL and 10 ng/μL recombinant muIfnar-2 ECD. Data are expressed as the mean ± SD cpm. (B) Histogram showing the elution profile of total 32P-muIFNα1 bound to high-molecular weight complexes in serum. The mean cpm of fractions 2 to 4 were used to determine total binding (mean = 80 403 cpm), specific binding (mean = 38 440 cpm) and nonspecific binding (mean = 41 963 cpm). VO indicates void volume.
Inhibition of interferon-induced signaling by recombinant muIfnar-2a
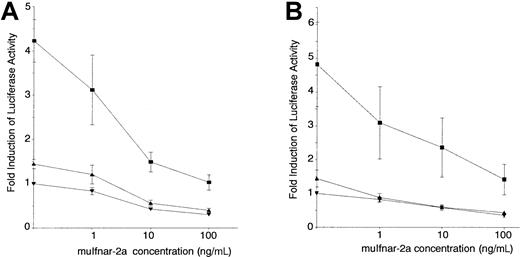
The ability of muIfnar-2a to perform a biologic function was tested in an in vitro assay system established to measure the ability of muIfnar-2a to act as a competitive inhibitor of type I IFN–induced signaling. Murine L929 cells were stably transfected with a reporter construct containing the promoter of the IFN-inducible gene 2′,5′-oligoadenylate synthetase (OAS) linked to the luciferase gene (2′5′OASLuc L cells). Recombinant muIFNα1 was able to induce 2′,5′-OAS promoter-driven luciferase activity in these cells in a dose-dependent manner (Figure6A). The addition of recombinant muIfnar-2a was able to significantly block the IFNα1-induced luciferase activity in a dose-dependent manner, with 70% inhibition of the activity of 10 IU/mL IFN occurring with as little as 10 ng/mL of muIfnar-2a (P < .005) (Figure 6A). These levels represent physiologic concentrations of IFN and approximately equimolar concentrations of muIfnar-2a protein and muIFNα1. A similar concentration of muIfnar-2a was able to inhibit muIFNβ-induced luciferase expression by 50% (P < .005) (Figure 6B). The amino acid sequence of IFNβ is the most divergent of the type I IFNs. A significant basal level of luciferase activity could be measured in the 2′5′OASLuc L cells when no exogenous IFNα1 was added, indicating some autocrine or paracrine type I IFN production and action in these cells. All concentrations of muIfnar-2a were also able to block this basal level of IFNα and β-induced luciferase activity (P < .025). Recombinant muIfnar-2 ECD had the same inhibitory effect on IFN-induced luciferase activity in the 2′5′OASLuc L cells as E coli–derived muIfnar-2a (data not shown), suggesting that this activity was not affected by the presence of the 11 muIfnar-2a-specific amino acids and that glycosylation was not necessary for this activity. Viable cell counts were not significantly altered by the addition of the muIfnar-2a protein preparation, indicating that the reduction in enzyme activity was not due to a cytotoxic effect (data not shown). Furthermore, the addition of a preparation of muIfnar-2a heat inactivated at 95°C for 5 minutes caused no significant reduction in the basal level of luciferase activity in the 2′5′OASLuc L cells, indicating that the inhibition was caused specifically by muIfnar-2a protein and not by another factor in the protein preparation. Therefore, muIfnar-2a can inhibit the function of type I IFNs by competing with muIfnar-2c for association in a receptor complex and subsequent signal transduction.
Inhibition of type I IFN-induced luciferase activity in 2′5′
OASLuc L cells by recombinant muIfnar-2a. Murine L929 cells were stably transfected with a cDNA encoding luciferase linked to the IFN responsive promoter 2′5′oligoadenylate synthetase (2′5′OASLuc L cells). Different concentrations of recombinant muIfnar-2a were preincubated with the indicated concentrations of type I IFN at 37°C for 1 hour: (A) 10 IU/mL (▪), 1 IU/mL (▴), or 0 (▾) recombinant muIFNα1 or (B) 10 IU/mL (▪), 1 IU/mL (▴), or 0 (▾) recombinant muIFNβ, and then added to 2′5′OASLuc L cells and incubated for a further 7 hours. The data are presented as the mean ± SEM (n = 8) fold induction of luciferase activity.
Inhibition of type I IFN-induced luciferase activity in 2′5′
OASLuc L cells by recombinant muIfnar-2a. Murine L929 cells were stably transfected with a cDNA encoding luciferase linked to the IFN responsive promoter 2′5′oligoadenylate synthetase (2′5′OASLuc L cells). Different concentrations of recombinant muIfnar-2a were preincubated with the indicated concentrations of type I IFN at 37°C for 1 hour: (A) 10 IU/mL (▪), 1 IU/mL (▴), or 0 (▾) recombinant muIFNα1 or (B) 10 IU/mL (▪), 1 IU/mL (▴), or 0 (▾) recombinant muIFNβ, and then added to 2′5′OASLuc L cells and incubated for a further 7 hours. The data are presented as the mean ± SEM (n = 8) fold induction of luciferase activity.
Inhibition of the antiproliferative effects of type I interferons on thymocytes by muIfnar-2a
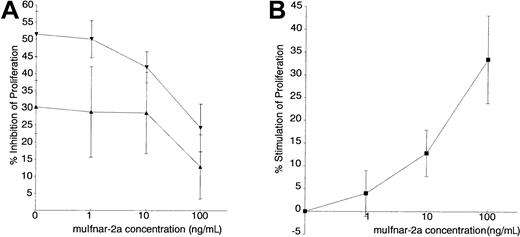
Primary murine thymocytes were chosen for these experiments because they are sensitive to the antiproliferative and immunoregulatory effects of type I IFNs. Addition of 10 and 100 IU/mL muIFNα4 inhibited the proliferation of PHA-stimulated thymocytes by 30% and 50%, respectively. The effect of recombinant muIfnar-2a on this biologic response was then examined. There was a dose-dependent inhibition of IFN activity by muIfnar-2a, with the addition of 100 ng/mL of recombinant muIfnar-2a inhibiting the IFNα4-induced antiproliferative action on these cells by about 50% (P < .01) (Figure 7A). Heat-inactivated muIfnar-2a had no significant effect on the PHA-stimulated thymocytes (data not shown), indicating that this effect was specific to active recombinant muIfnar-2a protein. Recombinant muIfnar-2a was also able to inhibit the antiviral activity of murine IFNα4 on Semliki Forest virus by 40% and 55% by using 300 and 500 ng/mL, respectively, in a cytopathic reduction effect bioassay (data not shown).
Inhibition of the antiproliferative effect of IFNα on primary thymocytes by recombinant muIfnar-2a.
DNA replication was measured by incorporation of3H-thymidine, and the data presented as the mean ± SEM, (n = 7) percentage inhibition of proliferation. (A) Thymocytes stimulated to proliferate with PHA (40 μg/mL) were incubated with 100 IU/mL (▾) and 10 IU/mL (▴) recombinant muIFNα4, and the indicated concentrations of muIfnar-2a. (B) Percentage stimulation of proliferation of PHA-stimulated thymocytes in the absence of exogenous IFN, by the indicated concentrations of recombinant muIfnar-2a.
Inhibition of the antiproliferative effect of IFNα on primary thymocytes by recombinant muIfnar-2a.
DNA replication was measured by incorporation of3H-thymidine, and the data presented as the mean ± SEM, (n = 7) percentage inhibition of proliferation. (A) Thymocytes stimulated to proliferate with PHA (40 μg/mL) were incubated with 100 IU/mL (▾) and 10 IU/mL (▴) recombinant muIFNα4, and the indicated concentrations of muIfnar-2a. (B) Percentage stimulation of proliferation of PHA-stimulated thymocytes in the absence of exogenous IFN, by the indicated concentrations of recombinant muIfnar-2a.
When recombinant muIfnar-2a was added to PHA-stimulated thymocytes in the absence of exogenous type I IFN, a dose-dependent stimulation of proliferation was observed (P < .005 at 10 and 100 ng/mL) (Figure 7B). This result suggested that autocrine or paracrine secretion and action of type I IFN by the primary thymocytes normally exert a basal level of growth inhibition that can be blocked by exogenous muIfnar-2a, resulting in increased proliferation. This is consistent with the reduction of basal reporter activity by recombinant muIfnar-2a (Figure 6A,B). Together, these results importantly demonstrate that muIfnar-2a has an inhibitory effect on the actions of type I IFNs on primary thymocytes.
Complementation of the type I interferon response inmuIfnar-2−/−thymocytes by muIfnar-2a
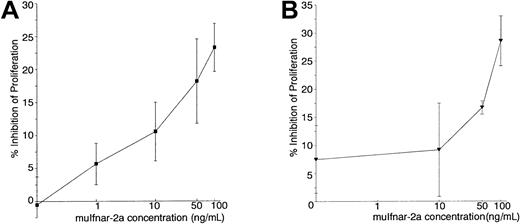
We next examined whether muIfnar-2a was able to interact with IFN and the other component of the type I IFN receptor, muIfnar-1, in the absence of muIfnar-2c to mediate a type I IFN signal. IFN had no antiproliferative effect on muIfnar-2−/−thymocytes, even up to 10 000 IU/mL (data not shown), but after the addition of 1 to 100 ng/mL recombinant muIfnar-2a plus 100 IU/mL muIFNα4 (Figure 8A) or muIFNβ (Figure8B), the antiproliferative response was restored in a dose-dependent manner. The inhibition by 100 IU/mL muIFNα4 or muIFNβ plus 100 ng/mL muIfnar-2a was up to 60% of the maximum inhibition of proliferation induced by 100 IU/mL muIFNα4 or muIFNβ on wild-type thymocytes (P < .005 and .025, respectively). Up to 100 ng/mL heat-inactivated muIfnar-2a did not significantly inhibit PHA-induced proliferation (data not shown), indicating that the effect was specific to active recombinant muIfnar-2a. These results suggest that muIfnar-2a can form a ligand-induced association with the muIfnar-1 chain of the type I IFN receptor to elicit a partial antiproliferative response without the requirement of the intracellular domain of the muIfnar-2c chain. These data also suggest that, in murine thymocytes, type I IFN–induced antiproliferative signals could be partially transmitted through the cytoplasmic domain of the muIfnar-1 chain of the type I IFN–receptor complex.
Complementation of type I IFN-induced antiproliferative activity in
muIfnar-2−/− thymocytes by recombinant muIfnar-2a. PHA-stimulated muIfnar-2−/−thymocytes were incubated with 100 IU/mL recombinant muIFNα4 (A) or 100 IU/mL recombinant muIFNβ (B), plus the indicated concentrations of muIfnar-2a. DNA replication was measured by incorporation of3H-thymidine, and the data are presented as the mean ± SEM (n = 4) percentage inhibition of proliferation.
Complementation of type I IFN-induced antiproliferative activity in
muIfnar-2−/− thymocytes by recombinant muIfnar-2a. PHA-stimulated muIfnar-2−/−thymocytes were incubated with 100 IU/mL recombinant muIFNα4 (A) or 100 IU/mL recombinant muIFNβ (B), plus the indicated concentrations of muIfnar-2a. DNA replication was measured by incorporation of3H-thymidine, and the data are presented as the mean ± SEM (n = 4) percentage inhibition of proliferation.
Discussion
The type I IFNs are important in vivo regulators of processes such as hematopoiesis, immune responses, growth, tumorigenesis, and responses to infection. In physiologic situations, IFN is not detectable in serum, but is produced locally and acts in an autocrine or paracrine manner, whereas in viral infections it can be found at high levels, up to 1000 IU/mL.30 In addition to the beneficial effects on the host, type I IFNs can also have adverse effects as seen after systemic administration of IFNα, including nausea, headaches, fever, leukopenia, and at times more severe sequelae such as autoimmune disease.7 Therefore, it is essential that the effects of IFN production and action be regulated so that the benefits to the host outweigh the adverse effects. One mechanism of regulating cytokine activity is by soluble receptors. However, the presence and action of a soluble receptor has not been considered in the type I IFN system until recently. A soluble IFNAR-2 was identified in human urine,11 but as yet no data are available on its tissue-specific expression and biologic activity.
The first step toward demonstrating the biologic significance of a soluble IFN receptor was to determine its in vivo expression relative to the transmembrane receptor. Northern blot analyses showed that there were 2 muIfnar-2 mRNA transcripts of 4.0 and 1.5 kb, the former encoding the transmembrane receptor (muIfnar-2c) and the latter encoding the generally more abundant, soluble isoform (muIfnar-2a). A subsequent study has described a 4.0-kb transcript encoding a second putative soluble receptor isoform (muIfnar-2a′).31However, this transcript is probably of low abundance, as it was only detectable after 60 cycles of PCR. The minor levels ofmuIfnar-2a′ mRNA in the 4.0-kb transcript would, if anything, underestimate the contribution of transcripts for soluble muIfnar-2. In the majority of mouse tissues and cell lines, the ratio of muIfnar-2a:muIfnar-2c (2a:2c) mRNA was about 5:1. However, high ratios (more than 10:1) in tissues such as the liver, small intestine, and fat imply an important function for muIfnar-2a. At the other extreme, there were tissues, particularly those involved in hematopoiesis in which the 2a:2c ratio was low (eg, less than 1:1 in BMM), implying that the production of a soluble receptor by these tissues would be a disadvantage or is unnecessary for the required effect of type I IFNs.
As previous studies have shown that the ability to produce and respond to type I IFNs is developmentally regulated,3 32 we examined the expression of muIfnar-2 transcripts during murine embryonic development. Both transcripts were present in all tissues examined throughout development, including ES cells, and the low 2a:2c ratio was maintained through to the neonate. Therefore, if regulated type I IFN responses are important during development, the regulation must occur at the level of IFN ligands or other response genes. Postnatally, some organs maintained the low 2a:2c ratios that existed throughout embryonic development, whereas in others, eg, the liver and small intestine, ratios were increased. The timing of the up-regulation of muIfnar-2a expression implies an environmental stimulus and that soluble muIfnar-2 is required to modulate IFN responses to these stimuli. The range of 2a:2c mRNA ratios in adult tissues and during development demonstrates that the expression levels of both isoforms are regulated, strongly implying a function for muIfnar-2a. Differential expression, splicing, or stability of mRNA transcripts may regulate the mRNA levels. This regulation might produce different stoichiometries of receptor components, resulting in specific responses to the type I IFNs in different organs or cell types.
Because the pattern of muIfnar-2a mRNA expression strongly indicated a function for this isoform, it was important to characterize the muIfnar-2a protein. Western blot analysis of mouse serum, using a well-characterized polyclonal antiserum, demonstrated that a 45-kd band representing glycosylated soluble muIfnar-2 was present. The level of soluble muIfnar-2 in the serum was estimated to be about 20 to 50 μg/mL. This level would be in considerable molar excess compared with the range of concentrations of serum type I IFN that is seen in an acute viral infection.30 Experiments demonstrated that the soluble muIfnar-2 in serum could bind 32P-labeled murine IFNα1. Soluble muIfnar-2 was also detected in peritoneal and mouth washes and in urine. The presence of soluble muIfnar-2 in murine body fluids could have a profound effect on modulating endogenous and exogenous IFNs.
Recombinant muIfnar-2a or muIfnar-2c ECD was expressed in E coli, P pastoris, or mammalian cells. The product(s) had a mobility of 29-kd nonglycosylated or 45-kd glycosylated, but our data indicated that glycosylation was not important for biologic activities as measured by in vitro assays. Soluble cytokine receptors can be generated by 2 mechanisms, which are not necessarily mutually exclusive: alternative splicing of the same gene as that encoding the transmembrane isoform, or proteolytic cleavage of the cell surface receptor.33 To address the mechanism of soluble receptor generation, a comparison of culture supernatants from cells transfected with muIfnar-2a or muIfnar-2c cDNAs demonstrated that soluble muIfnar-2 was found only in the former. This indicated, at least in this case, that soluble muIfnar-2 was the product of themuIfnar-2a transcript alternatively spliced from themuIfnar-2 gene, which also encodes muIfnar-2c (M.P.H., unpublished data, May 2000), rather than a proteolytically cleaved product of muIfnar-2c.
Because in other cytokine systems soluble receptors can act as agonists or antagonists of the activity of their cognate ligands,33we investigated whether muIfnar-2a could affect the biologic actions of type I IFNs. In murine L929 cells, muIfnar-2a inhibited the induction of an IFN-sensitive reporter in a dose-dependent manner. Interestingly, muIfnar-2a was an equally potent inhibitor of not only IFNα1 but also the most divergent of the type I IFNs, IFNβ; it is therefore likely to inhibit all type I IFNs. The inhibition by muIfnar-2a occurred with approximately equimolar concentrations of IFN and at lower than physiologic concentrations of soluble muIfnar-2a.
It was then necessary to determine whether muIfnar-2a could modulate a biologic effect of type I IFNs in primary cells. We demonstrated that muIfnar-2a was a potent, dose-dependent inhibitor of the antiproliferative effect of type I IFNs on primary thymocytes, and of the antiviral effect on primary murine embryo fibroblasts. Not only did the soluble muIfnar-2 inhibit exogenous IFN but also the autocrine or paracrine IFN that is produced by these cells and exerts some “basal” effects on cells such as suppression of proliferation. The partial block of this basal activity may have been due to several factors. First, not all of the basal activity may be the consequence of type I IFN activity, but may be due to the actions of other cytokines or endogenous levels of regulatory transcription factors. Secondly, it may be due to the low efficiency of blocking basal activity because of kinetics of interactions with cell-surface receptors; even high-affinity antibodies can be poor blockers of autocrine/paracrine ligand-receptor interactions.34 Therefore, soluble muIfnar-2 could be an important regulator of thymocyte development and responses in vivo.
Because some soluble cytokine receptors, such as the soluble IL-6 receptor, can bind the ligand and interact with the other chain of the receptor complex to transduce a signal,35 we determined whether this was possible for muIfnar-2a using thymocytes frommuIfnar-2−/− mice. Surprisingly, the data demonstrated that muIfnar-2a could bind type I IFN, form a complex, presumably with muIfnar-1 on the surface of themuIfnar-2−/− cells, and transmit an antiproliferative response. The intensity of that response using 100 ng/mL muIfnar-2a was over half that of a full response elicited by the same concentration of IFN on normal thymocytes. Thus, not only can soluble muIfnar-2 act as an inhibitor of IFN, but it could potentially also potentiate an IFN response.
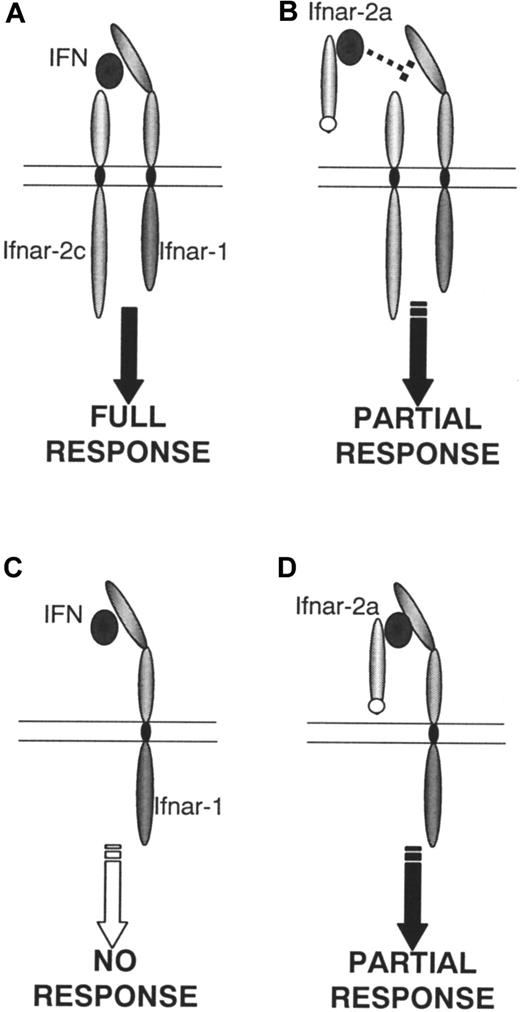
These results compel a re-evaluation of how the known components of the type I IFN receptor might interact with ligands and transduce their variety of signals. MuIfnar-2a can bind IFN and inhibit the interaction among ligand, cell surface muIfnar-1, and muIfnar-2c that is necessary for a full biologic response (Figure9A-B). Alternatively, the muIfnar-2a–ligand complex could also interact with cell surface muIfnar-1 to transduce a partial response (Figure 9D), whereas muIfnar-1 alone is unable to do so (Figure 9C). This result is consistent with the demonstration that human IFNAR-2b, a transmembrane isoform that lacks a complete cytoplasmic domain, can bind ligand and form a complex with huIFNAR-1, but is inconsistent with the lack of antiviral signaling or activation of the JAK/STAT pathway.17,36 One explanation may lie in the different biologic responses measured. Alternatively, it is possible that the muIfnar-2a:IFN:muIfnar-1 complex (Figure 9D) may activate a unique pathway responsible for antiproliferative responses in thymocytes. Interestingly, it was recently reported that type I IFN elicited an antiproliferative response in T cells that was independent of the JAK/STAT pathway and used the ZAP-70/lck/CD45 pathway.37The agonistic properties of muIfnar-2a have only been demonstrated inIfnar-2−/− cells, a situation that is unlikely to occur in vivo in wild-type animals. However, it is possible that some cells have an excess of surface Ifnar-1 overIfnar-2, in which case the agonist/signaling property of the soluble receptor could be operative. Therefore, the relative proportions of muIfnar-1, muIfnar-2c, and soluble muIfnar-2 in different organs or tissues may in part determine the extent of these 2 interactions (Figure 9A-D). This, in turn, could influence both the magnitude and nature of the biologic response to IFNs. Importantly, these data demonstrate that a signal can be transduced in the absence of the intracellular domain of Ifnar-2 (Figure 9D), possibly through Ifnar-1. The mechanism for this remains to be elucidated, but could involve ligand-mediated dimerization ofIfnar-1, or recruitment of another, unknown component of the receptor complex. In view of the potentially dual properties, agonistic or antagonistic, of an IFN:muIfnar-2a complex, it will be important to ascertain the impact of circulating muIfnar-2a on exogenously administered and endogenous IFN. Studies are currently in progress using genetic manipulation to determine the in vivo effects of endogenous muIfnar-2a.
Model of type I IFN receptor-ligand interactions in murine thymocytes.
When both receptor components are expressed (A) a full antiproliferative response occurs on stimulation with type I IFN. This response can be partially inhibited by muIfnar-2a (B). No response occurs when only muIfnar-1 (C) is present. However, when muIfnar-1, IFN and muIfnar-2a are present, a partial response is restored (D).
Model of type I IFN receptor-ligand interactions in murine thymocytes.
When both receptor components are expressed (A) a full antiproliferative response occurs on stimulation with type I IFN. This response can be partially inhibited by muIfnar-2a (B). No response occurs when only muIfnar-1 (C) is present. However, when muIfnar-1, IFN and muIfnar-2a are present, a partial response is restored (D).
Acknowledgments
The authors thank Dr S. Hwang for providing themuIfnar-2c cDNA cloned into pEF-BOS; L. Gullyan for skilled technical assistance.
Supported by a grant from the National Health and Medical Research Council of Australia. M.P.H. and S.T. are the recipients of Australian Postgraduate Awards.
M.P.H. and C.M.O. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Paul J. Hertzog, Center for Functional Genomics and Human Disease, Monash Institute of Reproduction and Development, 27-31 Wright St, Clayton, Victoria 3168, Australia; e-mail: Paul.Hertzog@med.monash.edu.au.