Abstract
Val34Leu polymorphism of the A subunit of coagulation factor XIII (FXIII-A) is located in the activation peptide (AP) just 3 amino acids away from the thrombin cleavage site. This mutation has been associated with a protective effect against occlusive arterial diseases and venous thrombosis; however, its biochemical consequences have not been explored. In the current study it was demonstrated that the intracellular stability and the plasma concentration of FXIII of different Val34Leu genotypes are identical, which suggests that there is no difference in the rate of synthesis and externalization of wild-type and mutant FXIII-A. In contrast, the release of AP by thrombin from the Leu34 allele proceeded significantly faster than from its wild-type Val34 counterpart. By molecular modeling larger interaction energy was calculated between the Leu34 variant and the respective domains of thrombin than between the Val34 variant and thrombin. In agreement with these findings, the activation of mutant plasma FXIII by thrombin was faster and required less thrombin than that of the wild-type variant. Full thrombin activation of purified plasma FXIII of different genotypes, however, resulted in identical specific transglutaminase activities. Similarly, the mean specific FXIII activity in the plasma was the same in the groups with wild-type, heterozygous, and homozygous variants. Faster activation of the Leu34 allele hardly could be associated with its presumed protective effect against venous thrombosis. No such protective effect was observed in a large group of patients with familial thrombophilia.
Introduction
Blood coagulation factor XIII (FXIII) is a pro-transglutaminase of tetrameric structure (A2B2) consisting of 2 potentially active A (FXIII-A) and 2 inhibitory/protective B (FXIII-B) subunits. It becomes transformed into an active transglutaminase (FXIIIa) during the terminal phase of coagulation cascade by the proteolytic action of thrombin and Ca++. Thrombin removes an activation peptide (AP) of 37 amino acid residues from the N-terminal end of FXIII-A, then, in the presence of Ca++, FXIII-B dissociates and FXIII-A assumes an enzymatically active configuration. By catalyzing an acyl transfer reaction, transglutaminases, including FXIIIa, cross-link peptide-bound glutamine and lysine residues through isopeptide bonds. The main physiological task of FXIII is to form fibrin γ-chain dimers and high Mr α-chain polymers and to cross-link α2-plasmin inhibitor to fibrin. By these mechanisms it strengthens the mechanical stability of fibrin clot and protects newly formed fibrin from the prompt elimination by the powerful fibrinolytic mechanism (see Greenberg et al1 and Muszbek et al2).
FXIII is essential for maintaining hemostasis, and severe deficiency of FXIII-A results in severe bleeding diathesis.3,4 In more than 30 unrelated patients the genetic defect(s) responsible for severe FXIII deficiency have been identified.5,6 In addition to pathogenic mutations, a number of polymorphisms have been identified in the amino acid sequence of FXIII-A.7-10 The biochemical effect of these polymorphisms has not been explored. One of them, a Val to Leu change, occurs at position 34 in the AP, just 3 amino acid residues away from the thrombin activation site (Arg37-Gly38).9 A protective effect against myocardial infarction11-13 and thrombotic cerebral artery occlusion14 has been attributed to the Leu34 allele. Protection against venous thrombosis by the Leu34 allele has also been reported.15 16 The biochemical consequences of FXIII-A Val34Leu polymorphism are unknown, and no scientifically sound hypothesis for the biochemical mechanism of such protective effect has been offered. In theory, a polymorphism may influence the plasma level of FXIII, the process of activation, and the specific transglutaminase activity of FXIIIa. In the current study it was shown that neither the plasma level nor the specific activity of fully activated FXIII was influenced by the presence of Leu34 allele. In contrast, thrombin-induced release of AP from the mutant plasma protein was significantly accelerated, and, consequently, the activation of Leu34 plasma FXIII occurred at a considerably higher rate. A possible mechanism for the accelerated cleavage of Leu34 FXIII-A by thrombin is offered by molecular modeling. In our study, no protective effect of the Leu34 allele against venous thrombosis was observed in the patients with familial thrombophilia.
Patients, materials, and methods
Materials
Chemicals, unless specified otherwise, were purchased from Sigma (St Louis, MO). Plasma FXIII was purified from the plasma of healthy volunteers of wild-type, heterozygous, and homozygous FXIII-A Val34Leu genotype, as described by Lorand et al.17 Taq DNA polymerase was the product of Amersham Pharmacia Biotech (Little Chalfont, UK). Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). The dodecapeptide substrate used in the FXIII assay was synthesized in our laboratory according to the Merrifield solid-phase method.
Selection of patients with thrombophilia and controls
The patient group consisted of consecutively enrolled, unrelated persons who had their first thrombotic episode before the age of 45 years. Further inclusion criteria were positive family history of venous thrombosis or recurrent thrombosis or thrombosis at an unusual location (eg, mesenteric, portal, cerebral). Patients with acquired underlying disease of thrombophilic character (malignancy, myeloproliferative disease, anti-phospholipid antibody syndrome, and so on) were excluded. Diagnoses of deep vein thrombosis were confirmed by color Doppler ultrasonography or venography. At the time of diagnosis, 26.8% of the patients had an acquired risk factor (use of oral contraceptives, postpartum, surgery, immobilization) that could serve as a trigger for venous thrombosis. The mean age (± SD) at the first thrombotic event was 27.7 ± 8.1 years. Recurrent thrombosis developed in 11% of the patients, and 9.7% of the patients had thrombosis at unusual locations. The control group consisted of apparently healthy volunteers without any personal or family history of venous thromboembolism, arterial disease (myocardial infarction, stroke, angina or peripheral vascular disease), or malignancy. Controls and patients were from the same geographic region, were of the same ethnic background, and did not differ significantly according to age and gender. In the control and the patient groups, respectively, 24.6% and 22.2% of the women used oral contraceptives. Patient and control data are summarized in Table 1. Approval was obtained from the Ethics Committee of the Medical and Health Science Center, University of Debrecen (Debrecen, Hungary).
DNA amplification and genotyping for FXIII Val34Leu polymorphism
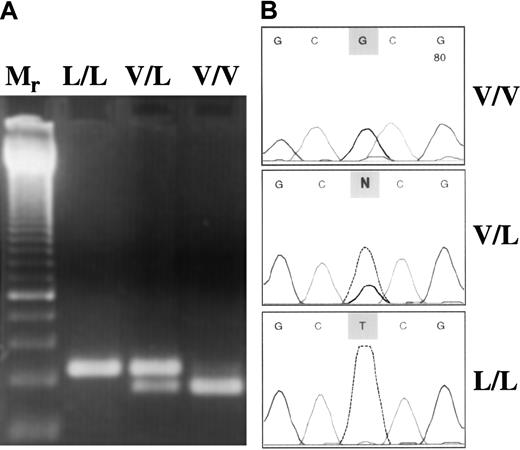
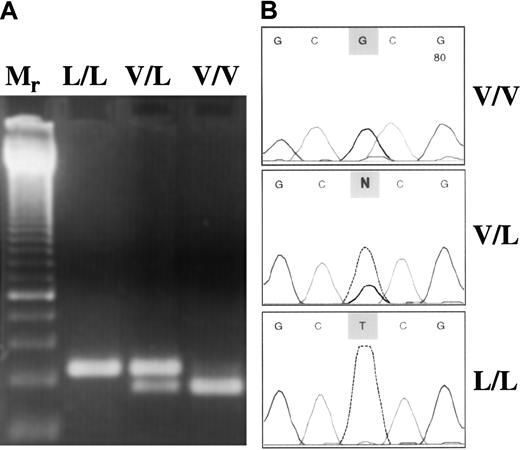
Anticoagulated whole blood was the source of genomic DNA. DNA was isolated from the buffy coat by QiaAmp Blood Kit (Qiagen, Hilden, Germany). FXIII Val34Leu polymorphism was analyzed with a newly developed method based on the principle of amplification-created restriction site. Oligonucleotide sequences used in polymerase chain reaction (PCR) amplification were 5′-ACTTCCAGGACCGCCTTTGGAGGC-3′ (forward) and 5′-GTTGACGCCCCGGGGCACCG-3′ (reverse). The underlined G represents a mismatch at the 3′ end of the reverse primer (the original nucleotide is A) to introduce a CfoI restriction site in the normal sequence (GCGC), which is lost in the presence of Leu allele (GCTC). The PCR reaction mix (50 μL) consisted of 0.1 μg genomic DNA, 5 pmol forward and reverse primer, and 1.0 U Taq DNA polymerase in 10 mmol/L Tris-HCl, 50 mmol/L KCl, pH 9.0 buffer containing 1.5 mmol/L MgCl2 and 200 μmol/L of each dNTP. The following amplification protocol was used: initial denaturation for 5 minutes at 95°C, 40 cycles of denaturation (1 minute at 95°C), annealing (1 minute at 55°C), extension (1 minute at 72°C), and a final extension for 10 minutes at 72°C. The PCR product was 114 bp long. After digestion with CfoI the digested fragment was 94 bp long in the wild genotype samples. In Leu/Leu homozygotes, the PCR products remained undigested, whereas in heterozygotes both the intact PCR product and the 94-bp fragment were visible in the 3% agarose gel stained with ethidium bromide (Figure1A). The PCR-based method described above was evaluated on randomly selected samples by fluorescent DNA sequencing using a BigDye Terminator Ready Reaction Kit on an ABI 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems, Foster City, CA) (Figure 1B). Results obtained by restriction digestion and sequencing were in 100% concordance. Two genetic alterations associated with familial thrombophilia, Factor V Leiden and prothrombin 20210A mutations, were analyzed by standard molecular genetic methods.18 19
Molecular genetic analysis of FXIII Val34Leu polymorphism.
(A) Restriction digestion pattern of PCR products. Mr, 50-bp ladder with a double-intensity band of 250 bp. Because the homozygous mutant (L/L) genotype has no restriction site forCfoI, only the nondigested 114-bp PCR product could be seen. In the V/L heterozygous sample, both the intact product (114 bp) and the 94-bp fragment were present. In the wild-type (V/V) sample, the 94-bp single band demonstrates complete digestion. (B) Evaluation of the PCR–restriction digestion method based on amplification created a restriction site by fluorescent DNA sequencing. Representative electropherograms corresponding to the respective genotype are shown.
Molecular genetic analysis of FXIII Val34Leu polymorphism.
(A) Restriction digestion pattern of PCR products. Mr, 50-bp ladder with a double-intensity band of 250 bp. Because the homozygous mutant (L/L) genotype has no restriction site forCfoI, only the nondigested 114-bp PCR product could be seen. In the V/L heterozygous sample, both the intact product (114 bp) and the 94-bp fragment were present. In the wild-type (V/V) sample, the 94-bp single band demonstrates complete digestion. (B) Evaluation of the PCR–restriction digestion method based on amplification created a restriction site by fluorescent DNA sequencing. Representative electropherograms corresponding to the respective genotype are shown.
Expression construct for a recombinant FXIII-A Val34Leu
The FXIII-A cDNA clone was a gift from Dr Dominic Chung, University of Washington (Seattle, WA). The coding region of the cDNA was subcloned in SVpoly expression vector by usingHindIII/XbaI restriction enzymes. The Val34Leu mutation was created into the cDNA by the Chameleon mutagenesis kit (Stratagene, La Jolla, CA). The following mutation-specific oligonucleotides were used: GAG CTT CAG GGC CTG GTG CCC CGG. Mutant clones were selected by the solid-phase mini-sequencing method.12,20 21
Expression of FXIII-A and pulse-chase analysis in COS cells
COS-1 cells were seeded at 70% confluence. They were then transfected with 2 mg/mL wild-type FXIII-A cDNA or FXIII-A Leu34 variant cDNA, with the DEAE–dextran–chloroquine method or Ca3(PO4)2 precipitation and cultured for 72 hours in Dulbecco modified Eagle medium containing penicillin–streptomycin and 10% bovine serum albumin. The intracellular stability of the FXIII-A Val34Leu variant was studied by pulse-chase analysis. FXIII-A Met242Thr was used as a positive control to demonstrate a FXIII-A mutation resulting in an unstable protein.22 After 68 hours, the cells were starved in a methionine-free medium for 15 minutes and then exposed to a pulse with 200 μCi 35S-methionine for 1 hour. Thereafter, the cells were monitored at variable chase periods (4, 8, and 24 hours). Mutant and wild-type FXIII-A were immunoprecipitated and run on 10% SDS-PAGE. Radiolabeled polypeptides were visualized by autoradiography, and FXIII-A was measured by scanning densitometry.
Determination of FXIII mass concentration and activity
The concentration of the tetrameric complex (A2B2) FXIII in the plasma or in FXIII preparations was measured by a newly designed, highly specific 1-step sandwich enzyme-linked immunosorbent assay (ELISA).23 A UV photometric assay was used for the determination of transglutaminase activity of plasma and purified FXIII.24 25 In the assay, FXIII was activated by thrombin and Ca++. A synthetic dodecapeptide corresponding to the N-terminal part of the α2-plasmin inhibitor and the glycine ethylester was used as transglutaminase substrates. Ammonia released by FXIIIa was continuously monitored by the NADPH-dependent glutamate dehydrogenase reaction. A Gly-Pro-Arg-Pro tetrapeptide was used to prevent fibrin polymerization during the measurement. Absorbance was measured at 340 nm after a lag period of 5 minutes. The change in absorbance per minute was calculated, and the results were expressed as U/L. In the assay for the determination of FXIII in the plasma, FXIII was fully activated by 20 U/mL thrombin during the lag phase of the reaction. For the determination of the activation of purified plasma FXIII at different thrombin concentrations, FXIII was first activated by thrombin for 5 minutes at 37°C, then the effect of thrombin was terminated by hirudin and the transglutaminase activity of formed FXIIIa was measured as described above. The assays were carried out in a Cobas Mira Plus (Roche, Basel, Switzerland) automatic analyzer.
Quantitative determination of FXIII-AP by high-performance liquid chromatography
The rate of the release of AP was determined on purified plasma FXIII of all 3 genotypes. Purified FXIII (720 μg/mL) was activated by 0.5 U/mL thrombin in 50 mmol/L Tris-HCl, 1.5 mmol/L CaCl2, pH 7.4, at 37°C. After 0, 2.5, 5, 10, and 20 minutes of incubation, the samples were boiled for 4 minutes. Heat-precipitated proteins were removed by centrifugation, and the supernatant was filtered through a disposable centrifuge filter device of 0.2-μm pore size (Whatman, Maidstone, UK). FXIII-AP in the filtrate was determined by a gradient HPLC method26 on a Shimadzu (Kyoto, Japan) device using a 5-μm Jupiter C-18 (Phenomenex, Torrance, CA), 300 Å reverse-phase column (250 × 4.6 mm). For the gradient a stock solution of 0.05 mol/L ammonium acetate was brought to pH 6.0 with diluted phosphoric acid. This stock solution was diluted 1:1 with water for solvent A and 1:1 with acetonitrile for solvent B. Elution gradient was 30% to 60% solvent B over 27 minutes. Elution profile was monitored at 210 nm on an SPD-M10A diode array detector (Shimadzu). The amount of AP released at full proteolytic activation of FXIII by 40 U/mL thrombin (for 30 minutes at 37°C) was taken as 1 mol AP/1 mol FXIII-A molecule, and the results were calculated accordingly. Full release of AP from FXIII-A in this condition was also verified by immunoblotting for FXIII-A.
Molecular modeling of FXIII–thrombin interaction
Molecular modeling studies were carried out to understand the reason for the different rates of activation of wild-type and mutant (Val34Leu) FXIII as thrombin substrates. In the calculations, the thrombin molecule and the oligopeptide containing 11 amino acids (from Gln32 to Gln42) around the thrombin cleavage site were involved. The starting 3-dimensional structure for thrombin27 was obtained from the protein data bank. Among the 3D structures of FXIII-A reported in the literature,28-30 the x-ray structure published by Weiss et al30 was the most complete around the Arg37-Gly38 peptide bond, and this structure was used in our study. Because no structural study has been published for the FXIII-A–thrombin complex, the optimal orientation for such a complex had to be determined. This was achieved by docking the wild-type substrate oligopeptide to the substrate binding valley of thrombin and performing molecular mechanics minimization (to 0.05 kJ/A per mol gradient-norm) followed by short dynamics simulations (100 ps with 1-fs time steps using SHAKE constraints on X-H bonds only). A few selected structures from the molecular dynamics trajectories were re-minimized, and the lowest energy conformers were accepted for each orientation. For modeling the thrombin–mutant oligopeptide interaction, the Val34Leu mutation was made on the model that represented the lowest energy conformer. Several side-chain conformations were generated, and the enzyme-substrate complexes were re-minimized. The geometry that corresponded to the lowest energy was accepted as the most likely structure of enzyme–(mutant)substrate complex. All molecular mechanics and dynamics calculations were carried out using the AMBER force field31 within the Macromodel (Schrödinger, Portland, OR) package.32 The VMD (University of Illinois, Urbana, IL)33 and Spock (Jon A. Christopher and Texas A&M University, Austin, TX)34packages were used for molecular graphics. The final rendering was made using the Raster3D (University of Washington, Seattle, WA) package.35
Statistical methods
Allele frequencies and odds ratios (OR) were calculated, and the data were presented with their 95% confidence intervals (CI). Power curves were constructed, and calculations were determined according to Campbell et al.36 To compare the distribution of FXIII plasma concentrations and specific activities with the expected normal distribution, the Kolmogorov-Smirnov and the Shapiro-Wilk tests were used (STATISTICA software; StatSoft, Tulsa, OK). FXIII concentrations and specific activities measured in the 3 groups of different genotype were compared using the Student ttest.
Results
Intracellular stability of FXIII-A Val34Leu variant
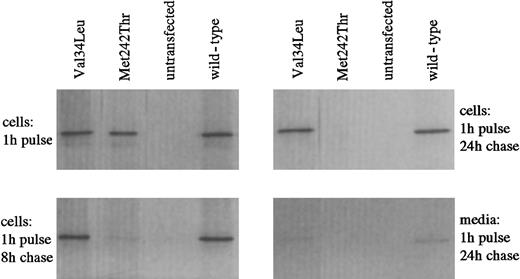
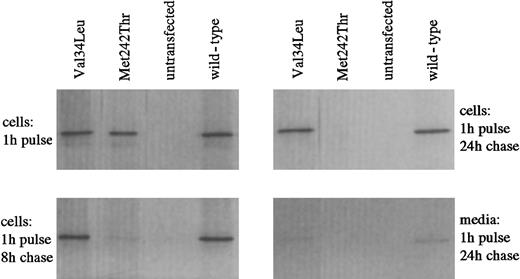
To address the effect of the Val34→Leu amino acid change on the protein, the intracellular stability of recombinant Leu34 FXIII-A expressed in COS-cells was studied by pulse-chase analysis. The intracellular level of FXIII-A Leu34 variant remained similar to that of wild-type FXIII-A over a chase period of 24 hours (Figure 2). In contrast, the unstable Met242Thr mutant, which results in FXIII deficiency, was totally degraded within 24 hours. After 24 hours, some leakage of both wild-type and Val34Leu FXIII proteins into the medium was detected. The leakage occurred in parallel with the release of lactate dehydrogenase, suggesting that FXIII wild-type and Leu34 variants were partially released from COS cells during cell death, as shown earlier.22
Investigation of the intracellular stability of wild-type and Val34Leu mutant recombinant FXIII-A by pulse chase analysis.
Metabolic labeling of COS cells transfected with wild-type or mutant FXIII-A was performed by incubating the cells with35S-methionine. The presence of labeled FXIII antigen was analyzed after variable chase periods (shown are the 8-hour and 24-hour periods) by immunoprecipitating the cell lysates with FXIII A-subunit antibody and subjecting the precipitated proteins to 10% SDS-PAGE. Radiolabeled FXIII-A was visualized by autoradiography. Met242Thr represents a mutation associated with instability of the protein and FXIII deficiency.
Investigation of the intracellular stability of wild-type and Val34Leu mutant recombinant FXIII-A by pulse chase analysis.
Metabolic labeling of COS cells transfected with wild-type or mutant FXIII-A was performed by incubating the cells with35S-methionine. The presence of labeled FXIII antigen was analyzed after variable chase periods (shown are the 8-hour and 24-hour periods) by immunoprecipitating the cell lysates with FXIII A-subunit antibody and subjecting the precipitated proteins to 10% SDS-PAGE. Radiolabeled FXIII-A was visualized by autoradiography. Met242Thr represents a mutation associated with instability of the protein and FXIII deficiency.
FXIII concentrations in the plasma of persons with different FXIII Val34Leu genotype
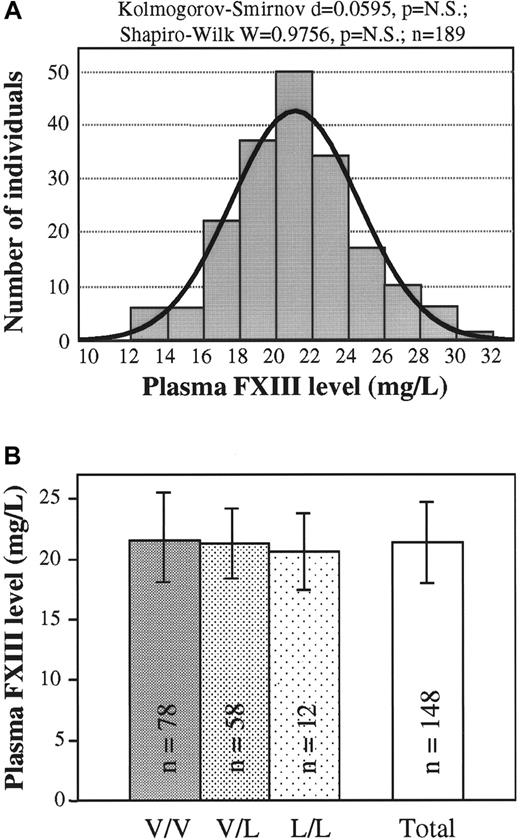
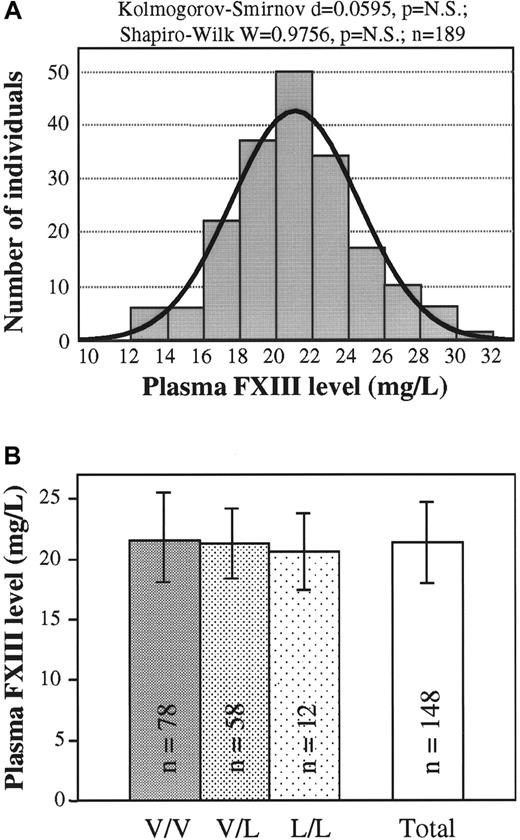
The use of the newly developed 1-step ELISA made the exact measurement of mass concentration of plasma FXIII possible. The normal frequency distribution of plasma FXIII concentration was proven by 2 different statistical methods (Figure3A). This finding suggested that the investigated controls represented a continuous single population not partitioned according to FXIII genotypes. In agreement with this finding, it was shown that there were no significant differences in the plasma FXIII concentrations of wild-type, heterozygous, and homozygous Val34Leu groups (Figure 3B).
Plasma FXIII mass concentration in healthy populations with different FXIII Val34Leu genotypes.
(A) Frequency distribution of plasma FXIII concentration (n = 189). Normal distribution was verified by 2 different statistical tests. The ideal normal distribution is indicated. (B) Means and standard deviations of FXIII concentrations in groups of healthy controls with different FXIII Val34Leu genotypes and in the total population. There were no statistically significant differences among the groups with different genotypes or between each of these groups and the total population.
Plasma FXIII mass concentration in healthy populations with different FXIII Val34Leu genotypes.
(A) Frequency distribution of plasma FXIII concentration (n = 189). Normal distribution was verified by 2 different statistical tests. The ideal normal distribution is indicated. (B) Means and standard deviations of FXIII concentrations in groups of healthy controls with different FXIII Val34Leu genotypes and in the total population. There were no statistically significant differences among the groups with different genotypes or between each of these groups and the total population.
Specific activity of purified and plasma FXIII of different genotype
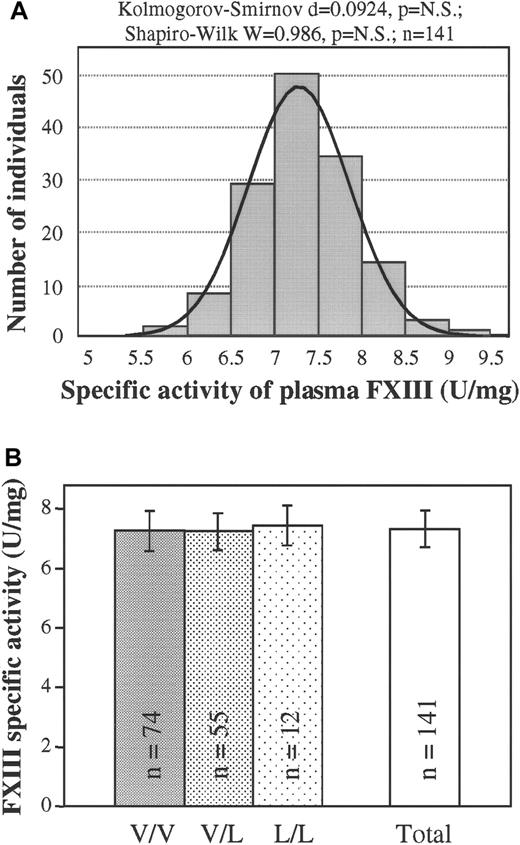
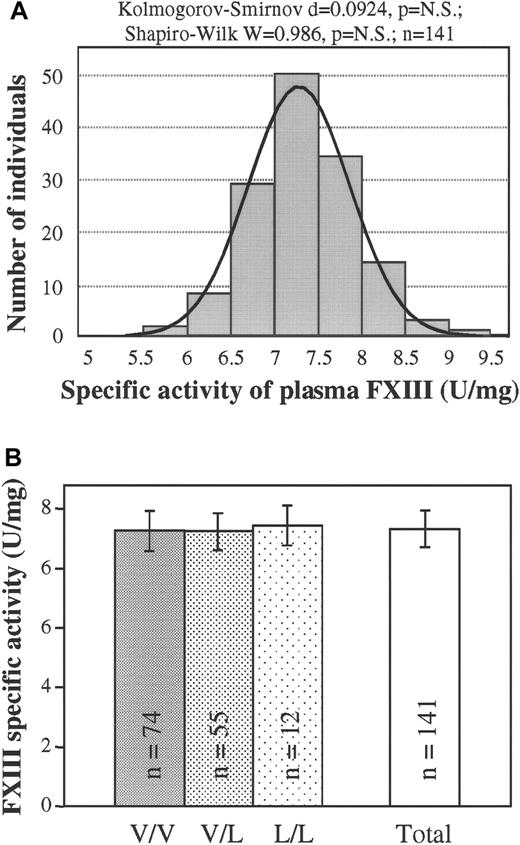
As was the frequency distribution of plasma FXIII concentration, the frequency distribution of the specific transglutaminase activity of FXIII in the plasma was demonstrated to be gaussian (Figure4A). Again, no statistically significant difference could be detected among the specific activities of groups of patients with different FXIII Val34Leu genotypes (Figure 4B). To support the above conclusion, specific activities were also determined on highly purified FXIII prepared from the plasma of patients with wild-type, heterozygous, or homozygous Val34Leu mutations. Nearly identical specific activities (5.70 ± 0.20 U/mg, 5.80 ± 0.16 U/mg, and 5.59 ± 0.09 U/mg, respectively; n = 3) were measured for the 3 genotypes.
Specific transglutaminase activity of plasma FXIII in healthy populations with different FXIII Val34Leu genotypes.
(A) Frequency distribution of the specific activity of plasma FXIII (n = 141). Normal distribution was verified by 2 different statistical tests. The ideal normal distribution is indicated. (B) Means and standard deviations of FXIII specific activity in groups of healthy controls with different FXIII Val34Leu genotypes and in the total population. There were no statistically significant differences among the groups with different genotypes or between each of these groups and the total population.
Specific transglutaminase activity of plasma FXIII in healthy populations with different FXIII Val34Leu genotypes.
(A) Frequency distribution of the specific activity of plasma FXIII (n = 141). Normal distribution was verified by 2 different statistical tests. The ideal normal distribution is indicated. (B) Means and standard deviations of FXIII specific activity in groups of healthy controls with different FXIII Val34Leu genotypes and in the total population. There were no statistically significant differences among the groups with different genotypes or between each of these groups and the total population.
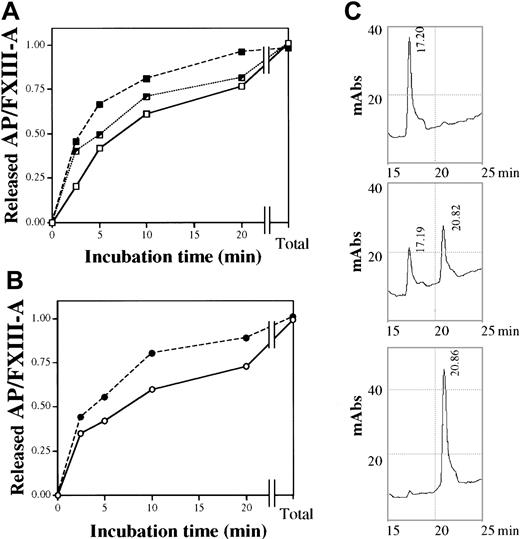
Cleavage of activation peptide from purified plasma FXIII of different Val34Leu genotype
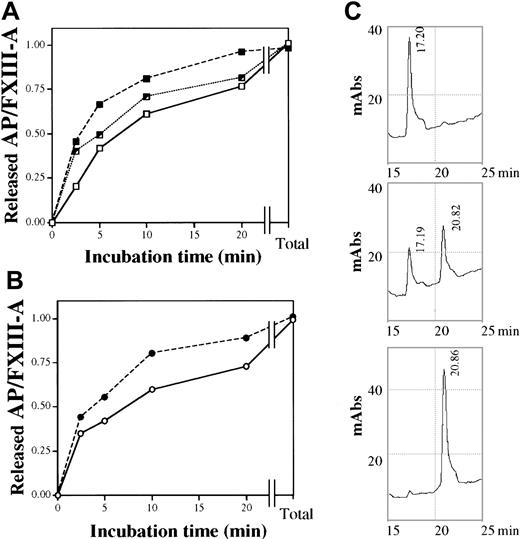
The time course of the thrombin-induced release of AP from FXIII was measured by HPLC. Wild-type and mutant AP were eluted at different retention times (Figure 5C), and AP released from heterozygous FXIII emerged as 2 distinct peaks. At high thrombin concentration (40 U/mL) and long incubation time (30 minutes), when the FXIII of all 3 genotypes was fully activated, the amounts of released AP were identical. At a much lower thrombin concentration (0.5 U/mL), it was clearly demonstrated that the rate of AP release from the homozygous Leu/Leu protein was significantly faster than from wild-type Val/Val FXIII (Figure 5A). At the initial stage of thrombin activation (2.5 minutes), the amount of AP cleaved from the mutant protein was double the amount cleaved from wild-type FXIII. The rate of release of AP from FXIII prepared from a heterozygous control was intermediate. Distinct retention times of wild-type and mutant AP made it possible to monitor the release of the 2 types of AP from heterozygous (Val/Leu) FXIII (Figure 5B). As expected, the rate of AP release from the FXIII-A subunit containing Leu34 was higher than from the subunit containing Val34.
Time course of the thrombin-induced release of AP from purified plasma FXIII of different Val34Leu genotype.
(A) The release of AP by 0.5 U/mL thrombin from wild-type (Val/Val, ■——■), heterozygous (Val/Leu, ┘ · · · · · · · ┘), and homozygous (Leu/Leu, ▪- - -▪) plasma FXIII. Total values represent the amounts released by 40 U/mL thrombin in 30 minutes at 37°C. (B) The release of 34Val (○——○) and 34Leu (●- - -●) AP from purified FXIII of a heterozygous control. (C) Elution profile of AP released from wild-type, heterozygous, and homozygous (upper, middle, and lower chromatogram, respectively) plasma FXIII by 0.5 U/mL thrombin in 20 minutes at 37°C. Retention times of wild-type and mutant AP are indicated. The elution profile was monitored at 210 nm.
Time course of the thrombin-induced release of AP from purified plasma FXIII of different Val34Leu genotype.
(A) The release of AP by 0.5 U/mL thrombin from wild-type (Val/Val, ■——■), heterozygous (Val/Leu, ┘ · · · · · · · ┘), and homozygous (Leu/Leu, ▪- - -▪) plasma FXIII. Total values represent the amounts released by 40 U/mL thrombin in 30 minutes at 37°C. (B) The release of 34Val (○——○) and 34Leu (●- - -●) AP from purified FXIII of a heterozygous control. (C) Elution profile of AP released from wild-type, heterozygous, and homozygous (upper, middle, and lower chromatogram, respectively) plasma FXIII by 0.5 U/mL thrombin in 20 minutes at 37°C. Retention times of wild-type and mutant AP are indicated. The elution profile was monitored at 210 nm.
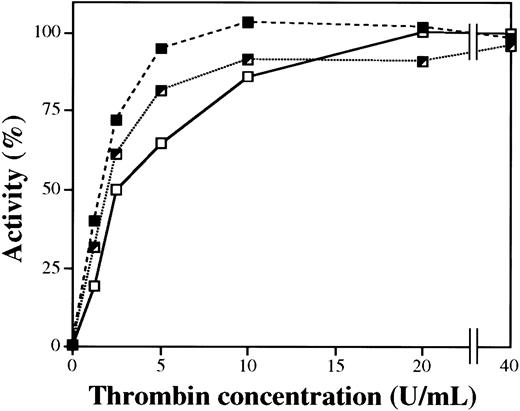
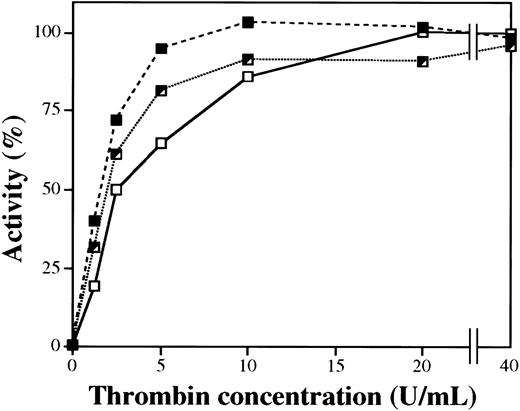
Activation of purified plasma FXIII of different Val34Leu genotypes by thrombin
Figure 6 demonstrates the dependence of FXIII activation on thrombin concentration. At saturation thrombin concentrations, the activities of FXIII of different genotypes were identical. However, at lower thrombin concentrations with 5 minute-activation times, the activity of homozygous Leu/Leu FXIII was significantly higher than the activity of wild-type Val/Val FXIII. Up to 10 U/mL thrombin, the activity of heterozygous Val/Leu FXIII was between the homozygous and wild-type variants. The time kinetics of the transformation of FXIII of different genotypes into an active transglutaminase during thrombin activation was also investigated. In complete accordance with the higher rate of release of Leu34 FXIII-AP by thrombin, the appearance of transglutaminase activity during thrombin-induced activation of the Leu34 variant also occurred at a higher rate (not demonstrated).
Activity of purified plasma FXIII of different Val34Leu genotype as the function of thrombin concentration during the activation period.
Purified plasma FXIII was activated by thrombin for 5 minutes at 37°C. Thrombin was blocked by hirudin, and FXIII activity was measured by the UV spectrophotometric method. Experiments were carried out with wild-type (Val/Val, ■——■), heterozygous (Val/Leu, ┘ · · · · · · · ┘), and homozygous (Leu/Leu, ▪- - -▪) plasma FXIII.
Activity of purified plasma FXIII of different Val34Leu genotype as the function of thrombin concentration during the activation period.
Purified plasma FXIII was activated by thrombin for 5 minutes at 37°C. Thrombin was blocked by hirudin, and FXIII activity was measured by the UV spectrophotometric method. Experiments were carried out with wild-type (Val/Val, ■——■), heterozygous (Val/Leu, ┘ · · · · · · · ┘), and homozygous (Leu/Leu, ▪- - -▪) plasma FXIII.
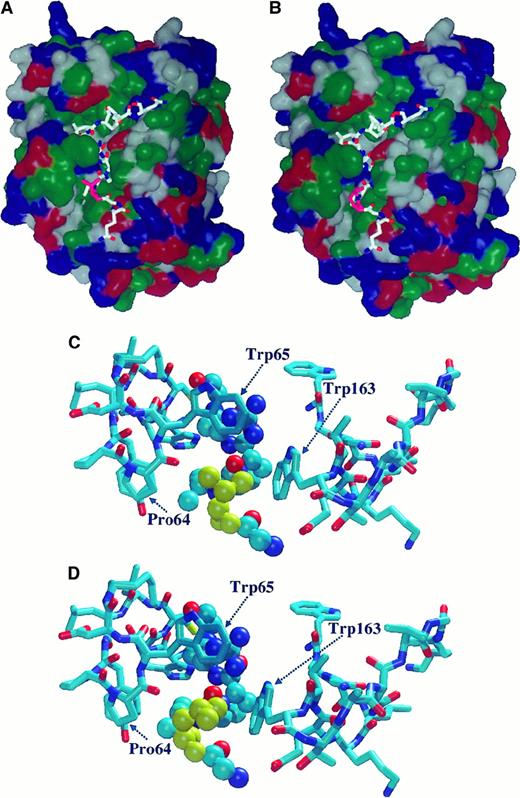
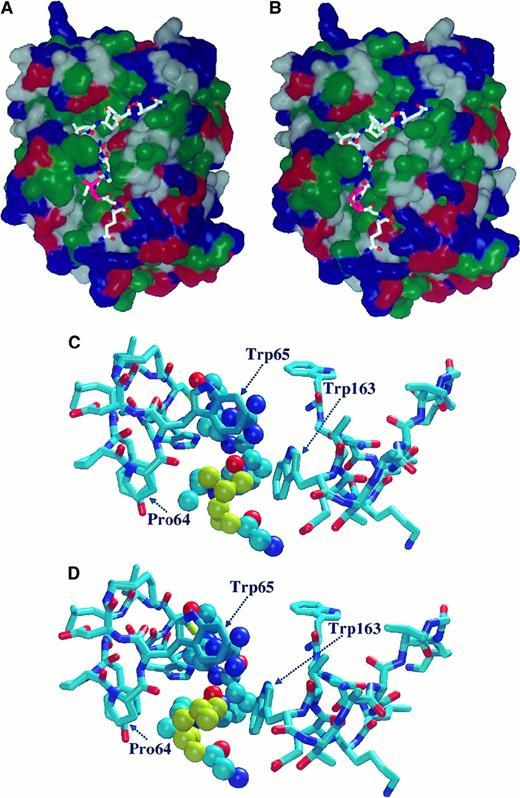
Molecular modeling of the FXIII–thrombin interaction
Using molecular mechanical minimization and dynamic simulations, one particular orientation was found to be far more stable than all the others. The most stable conformer at this orientation was found at least 30 kcal/mol more stable than any conformers from other orientations. The relatively large energy differences strongly suggested that a good approximation of the real enzyme-substrate orientation and geometry were found. In this orientation, the substrate peptide chain lay in the substrate binding valley in such a way that the hydrophobic side chains of Val34 and Leu34 were in a hydrophobic environment (Figure 7 A,B). The Val34/Leu34 side-chains had hydrophobic interactions with the Trp65, Pro64, and Trp163 side-chains. (Macromodel residue numbers correspond to Trp413, Pro412, and Trp511 in the prothrombin sequence.) The geometry for such interaction, however, was found to be more favorable (by 2 kcal/mol) for the Leu34 variant than for the wild-type oligopeptide. The side-chain of Leu34 fit better to the cavity formed by these amino acids (Figure 7C-D), which means that in the Leu34 mutant, the interaction energy was larger than with the wild-type oligopeptide. Because of the exponential relationship between the activation energy and the reaction rate, this relatively small energy difference seemed to be sufficient to accelerate the reaction by shifting the E + S ⇄ ES equilibrium in the direction of the enzyme-substrate (ES) complex.
Molecular modeling of the complex formed between thrombin and a model oligopeptide corresponding to the Gln32-Gln42 sequence of FXIII-A.
The oligopeptide contains the thrombin cleavage site (Arg37-Gly38) and the Val34Leu mutation site. (A, B) Orientation of model oligopeptides representing the wild-type Val34 (A) and the mutant Leu34 (B) FXIII-A on the surface of thrombin. Hydrophobic domains are shown in green. Blue and red represent basic and acidic regions, respectively. Val34 and Leu34 are shown in pink. (C, D) Optimized geometry of complex formed by thrombin and Val34 (C) or Leu34 (D) model oligopeptides. Val34 (C) and Leu34 (D) residues are shown in yellow. Thrombin residue numbers Pro64, Trp65, and Trp163 were obtained from the macromodel data file and correspond to Pro412, Trp413, and Trp511, respectively, in the prothrombin sequence. For the oligopeptide the Corey-Pauling-Kultun rendering was applied. For the demonstration of thrombin residues the liquorice rendering was used.
Molecular modeling of the complex formed between thrombin and a model oligopeptide corresponding to the Gln32-Gln42 sequence of FXIII-A.
The oligopeptide contains the thrombin cleavage site (Arg37-Gly38) and the Val34Leu mutation site. (A, B) Orientation of model oligopeptides representing the wild-type Val34 (A) and the mutant Leu34 (B) FXIII-A on the surface of thrombin. Hydrophobic domains are shown in green. Blue and red represent basic and acidic regions, respectively. Val34 and Leu34 are shown in pink. (C, D) Optimized geometry of complex formed by thrombin and Val34 (C) or Leu34 (D) model oligopeptides. Val34 (C) and Leu34 (D) residues are shown in yellow. Thrombin residue numbers Pro64, Trp65, and Trp163 were obtained from the macromodel data file and correspond to Pro412, Trp413, and Trp511, respectively, in the prothrombin sequence. For the oligopeptide the Corey-Pauling-Kultun rendering was applied. For the demonstration of thrombin residues the liquorice rendering was used.
Genetic studies in patients with familial thrombophilia and healthy controls
In a normal Hungarian population (n = 399; 205 men, 194 women; mean age, 28.7 years; SD, 10.1 years), a frequency of 25.4% was found for the Leu allele. Twenty-one (5.3%) had homozygous, 161 (40.3%) had heterozygous, and 217 (54.4%) had wild-type variants. These values are comparable to the data reported in the literature.9,37As shown in Table 1, 10.4% of the controls and 24.2% of the patients with familial thrombophilia carried the FV Leiden mutation in the heterozygous form. The high percentage of FV Leiden in the control group represents the high frequency of FV Leiden allele in the Hungarian population.38 Odds ratios associated with the FV Leiden mutation and the prothrombin 20210A allele were significant and fit well with the data available in the literature. Among these patients, 4.9% had protein S or protein C deficiency. The distribution of inherited risk factors verified that the selection of patients and controls was based on correct criteria. The analysis for FXIII Val34Leu polymorphism revealed practically identical allele frequencies and almost the same distribution of the 3 genotypes in the control and patient groups. Odds ratios calculated for the combined homozygous and heterozygous groups versus the wild-type group and for the homozygous group versus the wild-type group were 1.09 (95% CI, 0.78-1.52) and 1.04 (95% CI, 0.53-2.06), respectively—that is, no protective effect of the mutation could be observed.
The relatively high number of apparently healthy controls with the FV Leiden mutation allowed us to investigate the possible effect of FXIII Val34Leu polymorphism in this group. In the group of patients with FV Leiden and familial thrombophilia (n = 66; mean age, 33.4 years; SD, 10.2), the Leu allele occurred with a frequency of 28.79%, whereas a frequency of 26.67% was observed among controls with the FV Leiden mutation (n = 30; mean age, 31.4 years; SD, 13.3). The frequencies of the Leu34 allele in controls with and without (25.78%; n = 258) the FV Leiden mutation were practically identical. Finally, patients with thrombophilia and healthy controls were divided into 2 age groups. The frequency of the Leu allele among patients between 30 and 45 years (mean age, 41.0 years; SD, 6.94; n = 148) and age-matched controls (mean age, 41.0 years; SD, 7.89; n = 110) did not differ significantly (27.6% and 24.3%, respectively). Similarly, among patients younger than 30 years (mean age, 22.8 years; SD, 6.2, n = 125) and respective controls (mean age, 22.4 years; SD, 2.92; n = 178), no difference was found in Leu34 allele frequencies (26.0% and 27.8%, respectively).
Discussion
A new ELISA method designed for the determination of plasma FXIII made it possible to measure the level of tetrameric complex plasma FXIII and to express the results in mass concentration.23In previous studies concerning the influence of genetic polymorphism on FXIII level, FXIII-A and FXIII-B subunits were determined separately, and the results were expressed as a percentage of the antigen concentration measured in “normal” pooled plasma.39,40Because FXIII-A and FXIII-B antigen levels may change independently of each other and of the concentration of tetrameric plasma FXIII, only the determination of the tetrameric complex reflects the true plasma level of FXIII. Mass concentrations are not influenced by the composition or the FXIII content of plasma pools, and they make the calculation of true specific activities possible. Val34Leu polymorphism failed to influence the mass concentration of plasma FXIII, and the frequency distribution of FXIII level was normal. In a previous study,23 we demonstrated that there were no gender differences in the plasma FXIII level. The identical intracellular stability of Val34 and Leu34 recombinant FXIII-A variants is in good agreement with the finding that plasma FXIII concentration is independent of Val34Leu genotype.
Considering the closeness of the site of Val34Leu polymorphism to the thrombin activation site, one could suspect that the rate of proteolytic cleavage of Arg37Gly38 peptide bond, ie, the rate of the release of AP, is influenced by the Val-to-Leu substitution. Indeed, it was clearly demonstrated by direct HPLC measurement of AP that cleavage at the activation site by thrombin is accelerated by the presence of a Leu residue at position 34. It was interesting that even in the purified heterozygous FXIII preparation, the release of AP from the mutant A subunit was faster than from wild-type FXIII-A. The Val/Leu34 residue is positioned in a loop not clearly defined in the available crystal structures.28 41 Therefore, we attempted to approach the problem by molecular modeling. The larger interaction energy of the thrombin–Leu34 variant complex, as suggested by molecular modeling, provides at least a partial explanation for the accelerated reaction rate.
In accordance with the differences in the proteolytic release of AP, the formation of FXIIIa—that is, the increase of transglutaminase activity during FXIII activation—proceeded faster with the Leu34 variant than with wild-type FXIII. Consequently, at relatively lower thrombin concentrations, when FXIII is only partially activated, higher transglutaminase activities were measured with the homozygous Leu34 mutant than with wild-type FXIII. Activities measured with the heterozygous plasma factor were intermediate. Similar results have been obtained with platelet FXIII.42 Although the rate of activation of the Leu34 FXIII variant is higher than in wild-type FXIII, the specific transglutaminase activity of fully activated FXIII is independent of the Val34Leu genotype. The differences reported earlier37,39 40 must have been observed during incomplete thrombin activation.
The finding that the Leu34 variant becomes activated earlier might have some important biologic implications. Earlier activation of FXIII may result in faster fibrin cross-linking. Indeed, in separate experiments,42 we have shown with wild-type and mutant cellular FXIII that the rate of fibrin γ-chain dimerization and α-chain polymerization proceeds faster with the Leu34 variant than with its wild-type counterpart. It remains to be seen to what extent such an accelerated cross-linking reaction could influence the features of newly formed thrombi.
The frequency distribution of specific FXIII activity was perfectly normal. Similar results have been published recently.39 However, our results demonstrate a much narrower distribution range (x ± SD = 7.3 ± 0.59 U/mg) than reported. The difference might result from the expression of FXIII activity in relative percentage and from the use of FXIII-A antigen concentrations instead of complex plasma FXIII concentrations for calculation in the above report and, perhaps, from other methodological differences.
To date, there have been 2 studies analyzing the possible association of FXIII Val34Leu polymorphism with venous thrombosis. Catto et al15 demonstrated that the Leu34 allele represents a significant protection against venous thrombosis. Franco et al16 reported that the protective effect was restricted to homozygotes. On the basis of the current results, no scientifically sound hypothesis could be offered for explaining the association of a protective effect against venous thrombosis and the accelerated rate of FXIII activation. This fact and the partial discrepancy of the above results tempted us to investigate the distribution of Val34Leu polymorphism in a group of patients with familial thrombophilia. Although the frequency of FV Leiden mutation and the prothrombin 20210A allele demonstrated the correct selection of patient group, no difference in the frequency of the Leu34 allele or in the Val34Leu genotype distribution could be observed. When the combined group of homozygote and heterozygote mutation carriers were compared to those with the wild-type variant, our study of 273 patients (45.1% of the controls exposed to the mutation; type 1 error, 0.05) had 80% and 90% power to detect ORs of 1.62 and 1.74, respectively. In an earlier study involving 221 patients with venous thromboembolism, an OR of 0.56, the reciprocal of which is 1.79, was associated with Val/Leu plus Leu/Leu genotypes.15 It is clear that our study had more than adequate power to detect an effect of the same magnitude. If the Leu/Leu genotype was taken as exposure and compared to the Val/Val genotype, 80% and 90% power was attained when aiming to detect ORs of 2.33 and 2.59, respectively. Franco et al16 demonstrated an OR of 0.16 (reciprocal, 6.25). Such an effect would have been detected in our study with near certainty. No protective effect of the Leu34 allele against venous thrombosis could be detected in patients with thrombophilia who were younger than 30 or between 30 and 45 years, and no interactive effect with the FV Leiden mutation was found. In conclusion, the Val34Leu mutation represented a neutral polymorphism in our patient group.
Supported by the Hungarian National Research Fund (OTKA 030406), the Hungarian Ministry of Health (ETT 457/2000), and the Hungarian Academy of Sciences.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
László Muszbek, Department of Clinical Biochemistry and Molecular Pathology, University of Debrecen, Medical and Health Science Center, PO Box 40, Debrecen, H-4012, Hungary; e-mail: muszbek@jaguar.dote.hu.