Abstract
To date, all of the chromosomal deletions that cause -thalassemia remove the structural genes and/or their regulatory element (HS –40). A unique deletion occurs in a single family that juxtaposes a region that normally lies approximately 18-kilobase downstream of the human cluster, next to a structurally normal -globin gene, and silences its expression. During development, the CpG island associated with the -globin promoter in the rearranged chromosome becomes densely methylated and insensitive to endonucleases, demonstrating that the normal chromatin structure around the -globin gene is perturbed by this mutation and that the gene is inactivated by a negative chromosomal position effect. These findings highlight the importance of the chromosomal environment in regulating globin gene expression.
The human α-globin gene cluster lies in the telomeric region of chromosome 16 (16p13.3). A 285-kb (kilobase) segment of DNA extending from the terminal (TTAGGG)n repeats of chromosome 16 has been fully sequenced and shown to be a GC- andAlu-rich region containing a high density of CpG islands and genes. The α cluster includes an embryonic gene (ζ) and 2 fetal/adult α genes (α2 and α1) arranged along the chromosome in the order in which they are expressed during development (Figure 1). Expression of these genes is critically dependent on a single erythroid-specific positive regulatory element (HS –40), which lies 60 kb from the α genes in the intron of an adjacent widely expressed gene.1
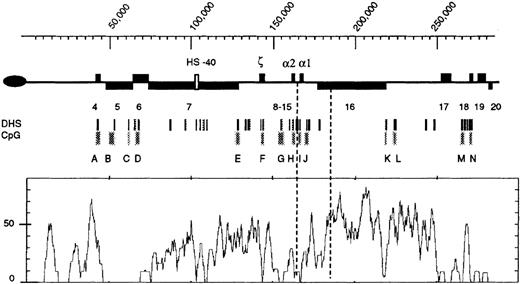
Structure of the terminal region (approximately 300 kb) of the human chromosome 16p.
The oval on the left represents the telomere. Previously described genes (4-203) are shown as black boxes above the line (transcribed toward the centromere) or below the line (transcribed toward the telomere). The α-globin regulatory element is shown as a white box (approximate coordinates 103 500-103 850). The embryonic (ζ) and fetal/adult (α) genes are indicated. Below the chromosome, the positions of previously characterized DNaseI hypersensitive sites (DHSs) and CpG islands (labeled A-N) are shown. Below this is a graph of the percent ofAlu sequences per 3 kb. Dashed vertical lines represent the 5′ and 3′ extents of the ZF deletion. The scale is in base pairs. Coordinate 1 is the first nucleotide in the chromosomal sequence described in Flint et al.3
Structure of the terminal region (approximately 300 kb) of the human chromosome 16p.
The oval on the left represents the telomere. Previously described genes (4-203) are shown as black boxes above the line (transcribed toward the centromere) or below the line (transcribed toward the telomere). The α-globin regulatory element is shown as a white box (approximate coordinates 103 500-103 850). The embryonic (ζ) and fetal/adult (α) genes are indicated. Below the chromosome, the positions of previously characterized DNaseI hypersensitive sites (DHSs) and CpG islands (labeled A-N) are shown. Below this is a graph of the percent ofAlu sequences per 3 kb. Dashed vertical lines represent the 5′ and 3′ extents of the ZF deletion. The scale is in base pairs. Coordinate 1 is the first nucleotide in the chromosomal sequence described in Flint et al.3
The relationship between chromosome structure and function has been well characterized in this region.2,3 It exists as a segment of “open,” transcriptionally active chromatin containing many DNase1 hypersensitive sites, punctuated by areas of apparently “closed” chromatin, including the most telomeric region and a long segment of nuclease-insensitive chromatin between coordinates approximately 180 000 to 218 000. The CpG islands (Figure1, A-N) in the 285-kb region are unmethylated in all cell types and tissues examined, but most of the interisland CpG sites appear to be densely methylated (D.R.H., unpublished data, 1999). Although the subtelomeric region replicates late in the cell cycle, the adjacent GC-rich region replicates early in all cell types examined.4
More than 50 natural deletions from this region have been identified, most of which were detected because they down-regulate the αgene expression and cause one of the well characterized hematologic phenotypes of α-thalassemia.5 All previously described mutations of this type either delete the α-globin genes or remove the remote regulatory element (HS –40), but the mutations cause no discernible changes in the patterns of methylation, DNase1 hypersensitivity, or replication. Here we describe an approximately 18-kb deletion that extends downstream of the α cluster. Although HS –40 and the α2 gene remain fully intact, α-globin expression from this chromosome is abolished. Changes in the pattern of methylation and DNase1 hypersensitivity within the α-globin cluster suggest that this deletion silences the α2 gene by a negative chromosomal position effect rather than by removing positive regulatory elements.
Materials and methods
Hematologic investigations
Full blood counts, examination of peripheral blood films, and identification of hemoglobin H (HbH) inclusions were performed as previously described in Weatherall et al.6
Cell culture
Epstein-Barr virus (EBV)–transformed cell lines were established for each family member. Interspecific hybrids containing the abnormal α−ZF chromosome were established by fusing EBV-transformed lymphocytes to mouse erythroleukemia cells as previously described in Higgs et al.1
Characterization of the chromosomal rearrangement
The α-globin genotypes were determined as previously described in Winichagoon et al.7Further mapping of the α−ZF breakpoint by Southern blot analysis was performed with a variety of restriction enzymes (Pst1, PvuII, StuI,HpaI, SacI, ScaI, BglII, andNcoI); a 0.5-kb HindIII)/PstI α-globin specific fragment representing the 3′ region of the α-globin genes (to determine the 5′ breakpoint); and HER50, a 1.1-kb HindIII/EcoRI fragment from approximate coordinate 193 000 (to determine the 3′ breakpoint), were used as probes. Known DNA sequences3 around the 5′ and 3′ breakpoints were used to design forward 280 (5′-GGTGCTGAACCATCCCCTGTC-3′) and reverse 279 (5′-CCCATTTCCTAAAAGTGTCCCTTC-3′) polymerase chain reaction (PCR) primers. These primers amplify a 928-bp breakpoint fragment that was sequenced directly using the same primers with incorporation of fluorescently labeled ddNTP (dideoxy nucleoside 5′-triphosphate) followed by analysis on an ABI 377 sequencer (Applied Biosystems, Foster City, CA). The α2-globin gene and the regulatory element (HS –40) from the abnormal α−ZF chromosome were amplified from DNA derived from the interspecific hybrid (ZF8), as described below, and directly sequenced. (The primers and DNA sequence are available on request.)
Analysis of the pattern of methylation
Patterns of methylation were analyzed in DNA from peripheral blood, EBV-transformed lymphocytes, and interspecific hybrids. DNA was also obtained from a sample of semen, supplementing the normal lysis buffer with 20 mmol/L dithiothreitol (DTT). DNA was initially cut with a restriction enzyme that is insensitive to methylation to generate specific fragments spanning the sequences of interest. These fragments were then digested with methylation-sensitive restriction enzymes (EagI, HpaII, or SstII and analyzed by Southern blot analysis. Many probes (Bg6.6, RA2.2, PL1, and IZHVR) have been previously described in Nicholls et al.8 In addition, we used several previously unreported probes including 510 × 511 (coordinates 66 093-66 785), 436 × 437 (coordinates 217 684-217 891), HR91 (approximate coordinate 232 000), HR23 (approximate coordinate 267 000), and HR22 (approximate coordinate 273 000). Methylation analysis using bisulphite-mediated genomic sequencing was adapted from Clark et al9 using DNA templates obtained from peripheral blood. After DNA modification, initial amplification of the α2-globin gene was achieved using the primers APS1 (5′-CAAAAAACAACACCATAATAAATTCTCTCT-3′) and APAS1 (5′-GGGGGTGTGGGTTGATTTTTTTTTT-3′). Second-round amplification and genomic sequencing were performed with the primers APS2 (5′-CCATAATAAATTCTCTCTAAATCTATAA-3′) and APAS2 (5′- GGGTTGATTTTTTTTTTTGTTAGGG-3′) using a Perkin Elmer Cycle sequencing kit (Perkin Elmer, Foster City, CA).
DNaseI and endonuclease sensitivity
DNaseI hypersensitive sites were analyzed as described in Higgs et al.1 In addition to previously described probes, the hypersensitive sites (HSs) at H and K were analyzed with the probes 382 × 383 (coordinates 158 487-159 287) and HR77 (approximate coordinate 218 000). Nuclei for assays of endonuclease sensitivity were prepared as described in Higgs et al1 and resuspended at approximately 107 nuclei per mL in the appropriate restriction enzyme digestion buffer. Increasing amounts (10-100 units) of enzyme were added to consecutive aliquots and incubated at 37°C for 60 minutes. DNA was subsequently extracted and analyzed by Southern blotting using the following probes: 0.5-kb HindIII/PstI, as described above; a probe corresponding to the α-globin regulatory element (approximate coordinate 103 000); and RA0.6 (approximate coordinate 67 000) at the 5′ end of the MPG gene.10
Studies of replication timing
Replication timing was analyzed throughout the entire terminal region of 16p13.3 using a modification of the fluoresence in situ hybridization (FISH)-based protocol described by Selig et al11 as set out in Smith and Higgs.4Briefly, interspecific somatic cell hybrid cell lines were grown to the mid log phase. To identify S-phase cells, 0.1 mmol/L BrdU was added to the culture 90 minutes prior to harvesting. The cells were collected by centrifugation, swollen in hypotonic solution, and fixed prior to making a batch of approximately 30 slides. To each slide 20-50 ng of nick-translated, Cot-1 suppressed cosmid probe was added and hybridized at 37°C overnight in 50% formamide, 2 × SSC, and 1% Tween 20. The following day, the slides were washed in 50% formamide and 2 × SSC, followed by 4 more changes of 2 × SSC. Stringent washing consisted of 3 changes of 0.1 × SSC at 60°C. S-phase cells were detected with anti-BrdU, and cosmid signals were detected with anti–DIG-FITC antibodies. A total of 200 nuclei were scored on each slide, and the proportion of duplicated signals was determined. For each batch of slides, the timing of replication was also determined for known early or late replicating control probes. The replication timing of 12 individual cosmids spanning the telomeric region of 16p3was analyzed for a normal homologue of chromosome 16p in an interspecific hybrid and the abnormal α−ZFchromosome.
Preparation and analysis of transgenic mice
The transgenic construct cDH2 (Figure2) was made by modification of Cos12, which contains the region from approximate coordinates 160 000-196 000 in the vector sCOS.12 The centromeric Not 1 site was inactivated by partially digesting with Not1, filling in with the Klenow enzyme, and by religating. The α-globin regulatory element was then inserted into the telomeric Not 1 site of this modified cosmid. A substantial part of the insert (approximate coordinates 160 000-185 660) linked to HS –40 was subsequently released withClaI. This fragment was prepared, and transgenic lines were produced as described in Higgs et al.1 The total RNA was isolated from embryonic blood and fetal liver (10.5-12.5 days post coitum) and from peripheral blood of adult transgenic mice. Human and mouse α-globin messenger RNA (mRNA) was detected using quantitative RNAse mapping as previously described.1
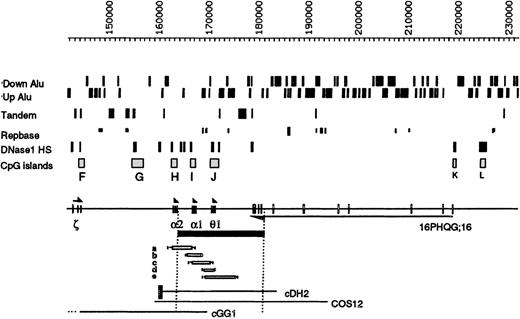
Details of the region around the −ZFbreakpoints.
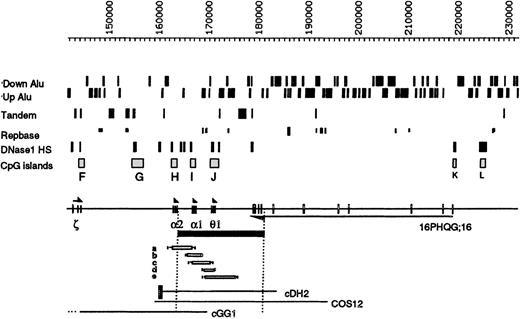
The positions of the Alu elements orientated toward (Up) or away from (Down) the telomere are shown. Below this, tandem repeats, other repeats, and DNaseI HS and CpG islands (labeled F-L) are shown. The ζ, α, and θ1genes are transcribed toward the centromere, and the gene16PHQG;16 is transcribed toward the telomere. The α−ZF deletion is shown as a black bar below the chromosome. Genomic mapping localized its 5′ breakpoint between an HpaI site (at coordinate 164 012) and a SacI site (at coordinate 164 356), beyond the polyA addition site of the α2-globin gene. Pulsed field gel electrophoresis of a Not1 α-specific fragment, approximately 380 kb, indicated that the deletion extends for approximately 18 kb (data not shown). The 3′ breakpoint was localized between a BglII site (coordinate 180 096) and a BamHI site (coordinate 182 417). Forward 280 and reverse 279 primers were designed from the breakpoint regions, and a 928-bp fragment spanning the breakpoint was amplified (data not shown) in the propositus (Z.F.) and his mother (H.F.). DNA sequence analysis demonstrated that the breakpoints lie between coordinates 164 044-45 and 182 395-96 and arose via an illegitimate recombination event (sequence available on request). Five previously described deletions that remove overlapping segments of this region are denoted a-e14; none of these silence α gene expression, although deletion a, −α3.7, only removes 1 α gene. Cosmids described in the text are shown at the bottom of the figure. The black box attached to cDH2 represents the α-globin regulatory element (HS –40).
Details of the region around the −ZFbreakpoints.
The positions of the Alu elements orientated toward (Up) or away from (Down) the telomere are shown. Below this, tandem repeats, other repeats, and DNaseI HS and CpG islands (labeled F-L) are shown. The ζ, α, and θ1genes are transcribed toward the centromere, and the gene16PHQG;16 is transcribed toward the telomere. The α−ZF deletion is shown as a black bar below the chromosome. Genomic mapping localized its 5′ breakpoint between an HpaI site (at coordinate 164 012) and a SacI site (at coordinate 164 356), beyond the polyA addition site of the α2-globin gene. Pulsed field gel electrophoresis of a Not1 α-specific fragment, approximately 380 kb, indicated that the deletion extends for approximately 18 kb (data not shown). The 3′ breakpoint was localized between a BglII site (coordinate 180 096) and a BamHI site (coordinate 182 417). Forward 280 and reverse 279 primers were designed from the breakpoint regions, and a 928-bp fragment spanning the breakpoint was amplified (data not shown) in the propositus (Z.F.) and his mother (H.F.). DNA sequence analysis demonstrated that the breakpoints lie between coordinates 164 044-45 and 182 395-96 and arose via an illegitimate recombination event (sequence available on request). Five previously described deletions that remove overlapping segments of this region are denoted a-e14; none of these silence α gene expression, although deletion a, −α3.7, only removes 1 α gene. Cosmids described in the text are shown at the bottom of the figure. The black box attached to cDH2 represents the α-globin regulatory element (HS –40).
Results
-Thalassemia resulting from a deletion extending downstream of the -globin cluster
Hematologically normal individuals have 4 α-globin genes (αα/αα), whereas most carriers of α-thalassemia have only 2 α-glob genes (αα/−−) or 3 α genes (αα/α−).5 α-Thalassemia trait was identified in 2 members (Z.F., the propositus, and H.F., the affected mother) of a Polish family (Table1). Provisional data demonstrated that a segment of more than 18-kb DNA extending downstream from between the α2 and α1 genes had been deleted from the affected chromosome (subsequently referred to as α−ZF), thereby completely removing the α1 gene.13 Further analysis (Figure 2 and legend) localized the breakpoints precisely between coordinates 164 044-45 and 182 395-96 and showed that the deletion results from an illegitimate recombination event that removes 18 352-bp DNA.
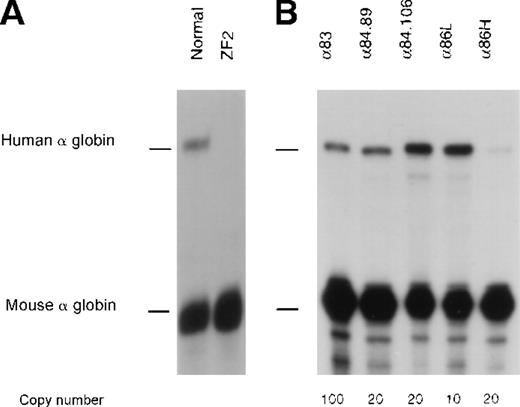
The hematologic phenotype in Z.F. (αα/α−ZF) is more severe than the phenotype normally seen in patients with 3 functional α genes (αα/α−), which suggests that the remainingα gene on the affected chromosome (α−ZF) might also be down-regulated by this mutation. The similar phenotype in H.F. (ααα/α−ZF) supports this hypothesis (Table 1, legend). To test this directly, the abnormal α−ZF chromosome 16 was isolated and analyzed in an interspecific hybrid after fusion of a lymphoblastoid cell line from Z.F. with a mouse erythroleukemia (MEL) cell line. Upon induction with HMBA and hemin, such hybrids terminally differentiate and normally express high levels of mouse and human globin mRNAs. The ratio of human/mouse α-globin mRNA in hybrids with a normal human chromosome 16 is 28.2% ± 15.1%.14 No human α-globin mRNA was expressed from the α−ZF chromosome in such hybrids, thereby confirming that the remaining α gene is completely inactive (Figure 3A).
Expression studies of cDH2.
Nuclease protection assays to analyze human -globin gene expression in (A) the abnormal interspecific hybrid (ZF2) and (B) adult peripheral blood from lines of transgenic mice carrying the cDH2 construct. Two samples from different individuals from line α84 are shown.
Expression studies of cDH2.
Nuclease protection assays to analyze human -globin gene expression in (A) the abnormal interspecific hybrid (ZF2) and (B) adult peripheral blood from lines of transgenic mice carrying the cDH2 construct. Two samples from different individuals from line α84 are shown.
The 5′ breakpoint of the α−ZF deletion lies 335- to 337-bp (coordinates 164 044-164 045) beyond the polyA addition site of the α2-globin gene, and all cis-acting sequences known to regulate expression of this gene remain intact on the α−ZF chromosome. To ensure that the α2 gene was down-regulated by the associated deletion and not as a result of any other mutation, the gene from the affected chromosome was amplified, and its sequence (coordinates 162 595-163 878) was shown to be normal. Similarly the sequence of the α-globin regulatory element (HS –40, coordinates 103 390-103 925) from this chromosome was entirely normal. Finally, when linked to HS –40, the α2 gene from the affected chromosome was expressed normally in stable transfectants of the MEL cells and transgenic mice (data not shown). Thus the 18 352-bp deletion not only removes the α1 gene but also inactivates the remaining α2-globin gene on the α−ZF chromosome.
The −ZF deletion does not remove a positive cis-acting element
These data suggest that the α−ZF deletion either removes a positive cis-acting element or juxtaposes negative sequences that down-regulate expression of the remaining α2-globin gene. To date, the only well characterized positive cis-acting sequences controlling α gene expression lie in the promoters and the remote regulatory element (HS –40). Removal of HS –40 by natural deletion5 or by specific “knock out”15reduces the α gene expression to less than 1% of normal, indicating that there are no other sequences in the 16p region which, on their own, are capable of up-regulating α gene expression.
To test this further, a construct (cDH2, Figure 2) containing HS –40 linked to a segment of DNA spanning approximate coordinates 159 000-185 660, including both α genes and the entire segment of DNA deleted in the α−ZF chromosome, was analyzed in 5 independent lines of transgenic mice (Table2 and Figure 3B). With this construct, the patterns and levels of human α-globin expression were not significantly different from those obtained with previously analyzed constructs completely lacking the down-stream region (Table 2, legend). As noted with other human α-globin transgenes, the level of human α-globin mRNA compared to endogenous mouse α-globin mRNA was lower in adult than embryonic erythroid cells.14 Therefore these findings confirm that the region within cDH2 does not contain any additional hitherto unidentified positive elements and show that the remaining α2 gene on the α−ZF chromosome is negatively regulated by the associated deletion.
Methylation of the -globin CpG island on the silenced −ZF chromosome
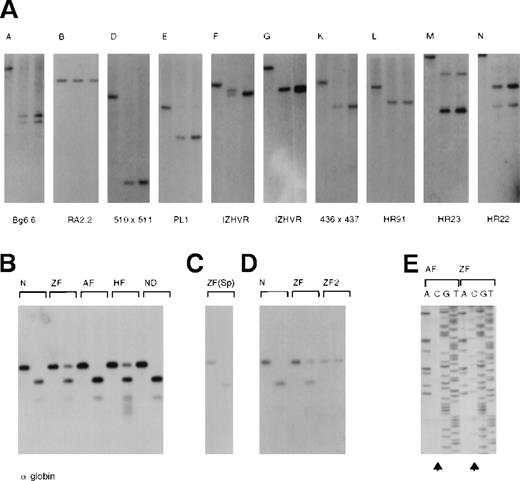
We have previously shown that all CpG islands in the 16p segment, with the exception of B (Figures 1 and4A), were unmethylated in all tissues and newly established cell lines that were analyzed. This applies to the islands at the α gene promoters (Figure 4B, H and I) even though theα genes are expressed in a tissue-specific manner.2 3 We examined the methylation state of islands on the α−ZF chromosome in spermatocytes (ZF), peripheral blood (ZF and HF), EBV-transformed lymphocytes (ZF and HF), and an interspecific hybrid carrying the α−ZFchromosome using the methylation-sensitive restriction enzymesEag1 (Figure 4B-D) and/or HpaII (data not shown).
Analysis of the pattern of methylation along the −ZF chromosome.
(A) CpG islands (A-N, see also Figure 1) were analyzed with a combination of enzymes and probes, which were previously described.2 53 In each panel, the left-hand lane is the chosen digest (eg, BglII) using DNA from the peripheral blood of an unaffected individual. The middle lane represents a double digest incorporating a methylation-sensitive enzyme (eg,BglII/SacII) using DNA from the peripheral blood of an unaffected individual. The third lane represents the same double digest (eg, BglII/SacII) using DNA from the peripheral blood of Z.F. The presence of 2 variably sized bands using IZHVR reflects the 2 different VNTR alleles detected with this probe. (B) Analysis of the CpG islands (H and I) associated with the α-globin genes using DNA from peripheral blood. In each case the left-hand lane is aPstI digest, and the right-hand lane is aPstI/EagI digest. (Only EagI cuts unmethylated sites.) N indicates an unaffected individual; ZF, the propositus; AF, the unaffected father; and HF, the affected mother. ND is a patient in whom an α gene was inactivated by a nondeletional form of α-thalassemia, demonstrating that such mutations do not alter the pattern of methylation. Note that both CpG islands (H and I) are examined in this assay. The different signal intensities of uncut DNA (methylated) to cut DNA (unmethylated) in Z.F. (αα/α−ZF) and H.F. (ααα/α−ZF) is explained by their different genotypes. Only the α−ZF chromosome is methylated at CpG island H. (C) Analysis of DNA from a sample of semen from Z.F. (D) Analysis of the CpG islands (H and I) in EBV-transformed lymphocyte lines from an unaffected individual (N), Z.F., and a MEL16 hybrid containing only the abnormal copy of chromosome 16 (ZF2). (E) Bisulphite modified sequence analysis of DNA from the peripheral blood of the propositus (ZF) and his unaffected father (AF). During bisulphite treatment of DNA, unmethylated cytosines were converted to uracil and subsequently PCR-amplified as thymidine. Methylated cytosines were resistant to this conversion and hence were amplified as cytosine. Arrows indicate that whereas all cytosines were converted to thymidines in A.F., many remain unconverted (methylated) in Z.F. Note that only the cytosines on one allele in Z.F. are methylated.
Analysis of the pattern of methylation along the −ZF chromosome.
(A) CpG islands (A-N, see also Figure 1) were analyzed with a combination of enzymes and probes, which were previously described.2 53 In each panel, the left-hand lane is the chosen digest (eg, BglII) using DNA from the peripheral blood of an unaffected individual. The middle lane represents a double digest incorporating a methylation-sensitive enzyme (eg,BglII/SacII) using DNA from the peripheral blood of an unaffected individual. The third lane represents the same double digest (eg, BglII/SacII) using DNA from the peripheral blood of Z.F. The presence of 2 variably sized bands using IZHVR reflects the 2 different VNTR alleles detected with this probe. (B) Analysis of the CpG islands (H and I) associated with the α-globin genes using DNA from peripheral blood. In each case the left-hand lane is aPstI digest, and the right-hand lane is aPstI/EagI digest. (Only EagI cuts unmethylated sites.) N indicates an unaffected individual; ZF, the propositus; AF, the unaffected father; and HF, the affected mother. ND is a patient in whom an α gene was inactivated by a nondeletional form of α-thalassemia, demonstrating that such mutations do not alter the pattern of methylation. Note that both CpG islands (H and I) are examined in this assay. The different signal intensities of uncut DNA (methylated) to cut DNA (unmethylated) in Z.F. (αα/α−ZF) and H.F. (ααα/α−ZF) is explained by their different genotypes. Only the α−ZF chromosome is methylated at CpG island H. (C) Analysis of DNA from a sample of semen from Z.F. (D) Analysis of the CpG islands (H and I) in EBV-transformed lymphocyte lines from an unaffected individual (N), Z.F., and a MEL16 hybrid containing only the abnormal copy of chromosome 16 (ZF2). (E) Bisulphite modified sequence analysis of DNA from the peripheral blood of the propositus (ZF) and his unaffected father (AF). During bisulphite treatment of DNA, unmethylated cytosines were converted to uracil and subsequently PCR-amplified as thymidine. Methylated cytosines were resistant to this conversion and hence were amplified as cytosine. Arrows indicate that whereas all cytosines were converted to thymidines in A.F., many remain unconverted (methylated) in Z.F. Note that only the cytosines on one allele in Z.F. are methylated.
Using Eag1, the CpG island (H) associated with the remainingα2 gene on the α−ZF chromosome was unmethylated in spermatocytes (Figure 4C), but it was methylated in peripheral blood, EBV lymphocytes, and the interspecific hybrid (Figure4B,D). Island H was analyzed further using HpaII. Although this island contains 16 sites (CCGG), none appeared to be cut in DNA from peripheral blood (data not shown). Finally, the CpG dinucleotides in the segment of H spanning the α2 promoter (coordinates 162 529-162 887) were analyzed using bisulphite-modified sequencing (Figure 4E and legend). In DNA from the peripheral blood of the unaffected father (AF), 1 of 47 CpGs were methylated, whereas in the propositus (ZF), 40 of 47 CpGs were methylated (Figure 4E).
Thus it seemed likely that at least part of the mechanism by which theα2 gene had been silenced involved methylation of the CpG dinucleotides. It was therefore of interest to know how far this effect propagated along the chromosome. All other CpG islands within the 16p segment were analyzed, as previously described, using methylation-sensitive restriction enzymes (Figure 4A), but none was inappropriately methylated. The closest CpG island (G) appeared unmethylated and the peripheral blood of Z.F. from all CpG dinucleotides analyzed in DNA from approximately 2 kb upstream of island H had the same pattern of methylation as a normal chromosome (data not shown).
The silenced -globin promoter shows reduced sensitivity to endonucleases
Although the CpG islands (H and I) associated with the αgenes are normally unmethylated in all tissues, the corresponding DNase1 HSs are expressed in a strictly erythroid-specific manner.16 Similarly, in vivo, these promoters are also sensitive to specific endonucleases (eg, HinfI), which cut within previously defined hypersensitive sites in erythroid cells (Figure 5 and D.R.H., unpublished data, 1999). DNase1 hypersensitivity and endonuclease sensitivity at the α promoters were analyzed in interspecific MEL hybrids containing either normal (αα, JY5-4) or abnormal (α−ZF, ZF2, and ZF8) copies of chromosome 16.
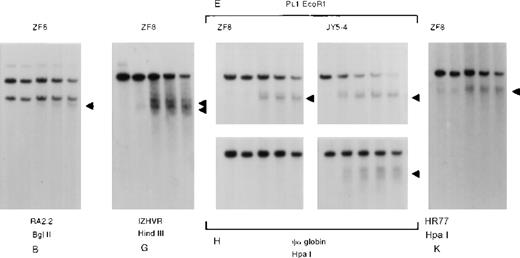
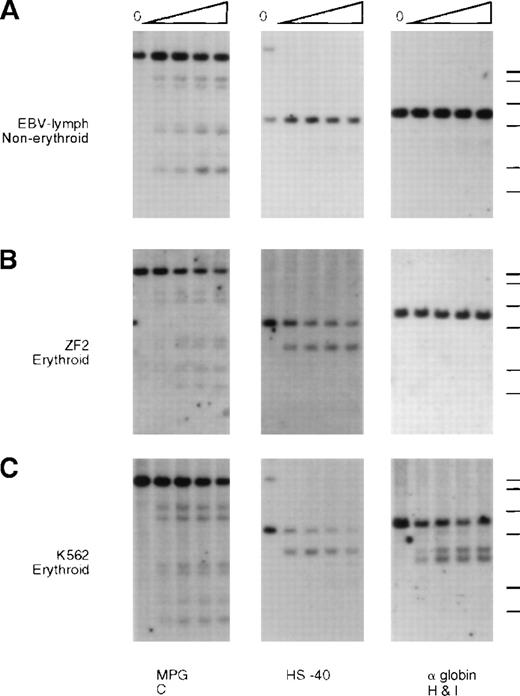
Analysis of endonuclease sensitivity in the −ZF chromosome.
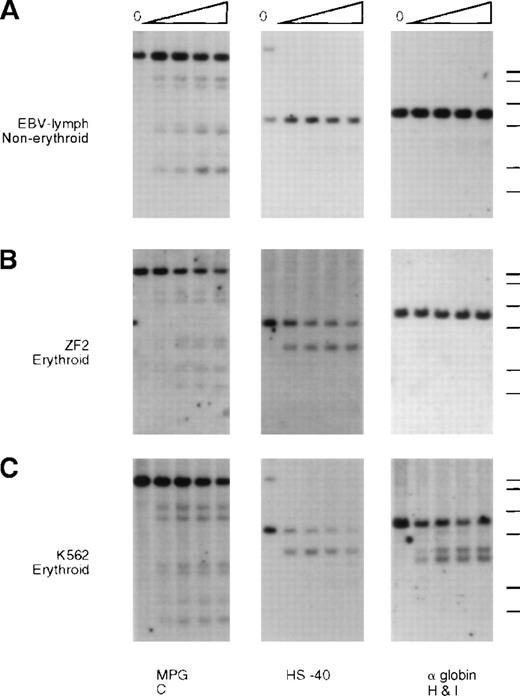
DNA from nuclei of (A) EBV lymphocytes, (B) an interspecific MEL hybrid containing the α−ZF chromosome (ZF2), and (C) the erythroid cell line K562. The DNA was incubated with increasing amounts of Hinf I (“Materials and methods”) and analyzed with probes around gene no. 6 (RA0.6, approximate coordinate 67 000), which encodes the ubiquitously expressed MPG gene, the α-globin regulatory element HS –40 (approximate coordinate 103 000), and the α-globin gene. Whereas the site associated with HS –40 is clearly present in ZF2, the sites are missing from the silenced α-globin promoter.
Analysis of endonuclease sensitivity in the −ZF chromosome.
DNA from nuclei of (A) EBV lymphocytes, (B) an interspecific MEL hybrid containing the α−ZF chromosome (ZF2), and (C) the erythroid cell line K562. The DNA was incubated with increasing amounts of Hinf I (“Materials and methods”) and analyzed with probes around gene no. 6 (RA0.6, approximate coordinate 67 000), which encodes the ubiquitously expressed MPG gene, the α-globin regulatory element HS –40 (approximate coordinate 103 000), and the α-globin gene. Whereas the site associated with HS –40 is clearly present in ZF2, the sites are missing from the silenced α-globin promoter.
Whereas the normal α promoters are usually sensitive to endonucleases in these erythroid cells (Figure 5, K562, and Figure6, JY5-4), the α2 promoter on the α−ZF chromosome was insensitive (Figure 5, ZF2, and Figure 6, ZF8). By contrast, the remote α-globin regulatory element (HS –40) was sensitive in both normal and abnormal chromosomes (Figure5, ZF2), adding further weight to previous studies15 16demonstrating that tissue-specific chromatin remodeling at HS –40 and the α promoters occurs independently (see “Discussion”).
Analysis of DNase1 hypersensitive sites along the −ZF chromosome.
DNA from the nuclei of an interspecific MEL hybrid containing the α−ZF chromosome (ZF8) or a normal chromosome (JY5-4) using probes and enzymes that detect the hypersensitive sites at the CpG islands associated with the α2-globin promoter (H) and the surrounding CpG islands B, G, E, and K (see Figure1). Analysis of HSs at E and H in normal hybrids (JY5-4) and abnormal hybrids (ZF8) shows that although the constitutive site at E is sensitive in both, the HS at the silenced α promoter is insensitive.
Analysis of DNase1 hypersensitive sites along the −ZF chromosome.
DNA from the nuclei of an interspecific MEL hybrid containing the α−ZF chromosome (ZF8) or a normal chromosome (JY5-4) using probes and enzymes that detect the hypersensitive sites at the CpG islands associated with the α2-globin promoter (H) and the surrounding CpG islands B, G, E, and K (see Figure1). Analysis of HSs at E and H in normal hybrids (JY5-4) and abnormal hybrids (ZF8) shows that although the constitutive site at E is sensitive in both, the HS at the silenced α promoter is insensitive.
As for the methylation pattern described previously, it was of interest to determine how far the abnormal pattern of reduced chromatin sensitivity extended from the α−ZFbreakpoints. Again it appeared that this effect was quite localized because the nearest known constitutive HSs lying upstream and downstream of the α2 promoter were sensitive as normal (Figure 6).
Silencing of the -globin gene is not associated with any change in the pattern of replication timing
Unlike the rest of the GC-rich chromosomal segment, the region downstream of the α−ZF deletion (coordinates 180 000-218 000) is devoid of both HSs and CpG islands. It therefore seemed possible that the α2 gene in the α−ZF chromosome was inactivated by juxtaposition next to this potentially repressive chromatin environment. There is evidence that such chromatin replicates late in the cell cycle17,18 and that multiprotein complexes at some origins of replication can silence gene expression.19 20 Therefore, it was of interest to study the effect of the α−ZFdeletion on the normal pattern of replication along this region of 16p.
Using a FISH-based assay to score duplicated loci in S-phase, we have recently shown that in MEL x chromosome 16 hybrids, the most telomeric region (approximately 20 kb) of 16p replicates late in the cell cycle, whereas the adjacent GC-rich region replicates early (Figure 7 and Smith and Higgs4). However, we noted that the Alu-dense region between approximate coordinates 180 000 and 218 000, where the 3′ breakpoint of the α−ZF chromosome lies, consistently replicates later than its flanking regions, albeit still relatively early in S-phase.4 Furthermore, in contrast to other segments of 16p, duplicated signals from this region, contained within Cos12 (Figure 2), often remain closely paired in many nuclei. This suggests that more time is taken for this region to separate during S-phase than for other regions along the duplicated chromatids.
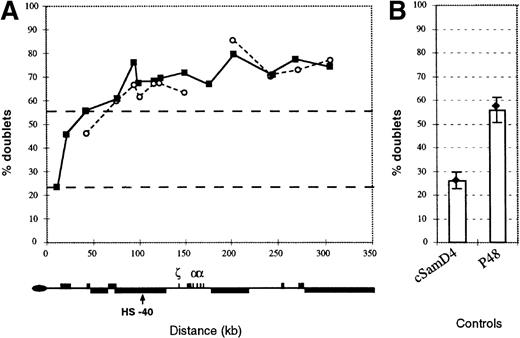
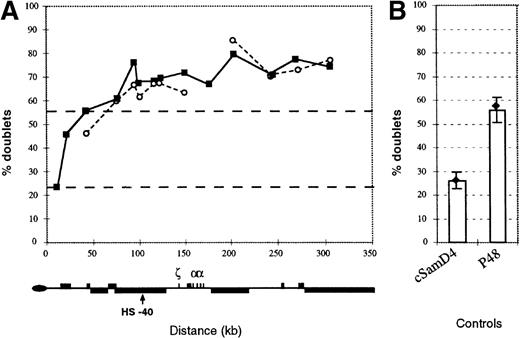
The pattern of replication along the −ZFchromosome.
(A) Replication of the human 16p region was assayed by FISH as set out in Smith and Higgs4 using a set of 12 cosmids spanning this region. The pattern in an interspecific hybrid containing a normal copy of chromosome 16 (closed boxes) is compared with a hybrid containing the α−ZF chromosome (open circles). In the α−ZF chromosome, there is no value for Cos 12 (midpoint at approximate coordinate 177 000) because this region is deleted. Black dashed lines indicate the mean percent doublet scores for the early replicating control cosmids (top line) and late replicating control cosmids (bottom line). Below, the key features of the 16p region are shown. (B) Replication timing of control cosmids. The mean value for all early (p48) and late (cSamD4) mouse controls4 are represented as columns. Values for controls measured in the hybrid containing the α−ZFchromosome are shown as a black diamond with lines representing plus or minus 1 SD.
The pattern of replication along the −ZFchromosome.
(A) Replication of the human 16p region was assayed by FISH as set out in Smith and Higgs4 using a set of 12 cosmids spanning this region. The pattern in an interspecific hybrid containing a normal copy of chromosome 16 (closed boxes) is compared with a hybrid containing the α−ZF chromosome (open circles). In the α−ZF chromosome, there is no value for Cos 12 (midpoint at approximate coordinate 177 000) because this region is deleted. Black dashed lines indicate the mean percent doublet scores for the early replicating control cosmids (top line) and late replicating control cosmids (bottom line). Below, the key features of the 16p region are shown. (B) Replication timing of control cosmids. The mean value for all early (p48) and late (cSamD4) mouse controls4 are represented as columns. Values for controls measured in the hybrid containing the α−ZFchromosome are shown as a black diamond with lines representing plus or minus 1 SD.
Using the FISH-based replication assay with 12 cosmids along the 300-kb 16p telomeric region, we found that this entire segment of the α−ZF chromosome, including the silenced αgene (corresponding to cGG1 in Figure 2), remains early replicating with a profile that is almost indistinguishable from a normal chromosome (Figure 7). This suggests that replication of the α−ZF chromosome initiates at the same origins at the same phase of the cell cycle as a normal chromosome and that silencing of gene expression in this case is not associated with any switch in the timing of replication.
The 3′ breakpoint lies in an Alu dense region and disrupts a widely expressed gene
The α−ZF deletion was further evaluated in the light of previously described structural and functional data relating to the region surrounding its breakpoints. We recently suggested that there may be some heterogeneity in chromatin structure across the 285-kb 16p segment.3 In particular, in the cell lines tested, we did not detect either the HSs or CpG islands in the most telomeric region (coordinates 1 to approximately 36 000) or in a region downstream of the α cluster (approximate coordinates 180 000-218 000). Both regions contain many repetitive DNA elements. The downstream region contains the highest density of Aluelements in the entire segment (Figure 1). Although neither of the α−ZF breakpoints lie within the Aluelements, the deletion juxtaposes the intact α2 gene next to an almost continuous block of Alu repeats (Figure 2).
There are several expressed sequence tags (ESTs) in the downstream region thought to represent transcripts originating from the CpG islands K and/or L (Figures 1 and 2). Reverse transcriptase–PCR (RT-PCR) and Northern blot analysis indicated that this region contains a gene (16pHQG;16) that is widely expressed in a variety of tissues (heart, brain, lung, liver, muscle, kidney, and pancreas) and cell lines (EBV-transformed B lymphocytes, K562, HEL, HepG2, and HT29). Its alternatively spliced RNA transcripts have been further characterized and shown to encode a putative RNA-binding protein expressed from at least 10 exons spanning coordinates 178 856-219 261 (C.T., unpublished data, 1999). The introns of this gene consist almost entirely of Alu elements. The gene is transcribed in the opposite orientation to the α-globin genes, with a polyA tract attached to a mRNA from the last exon (coordinates 178 856-179 220). Therefore, in addition to removing the α1 gene, the α−ZF deletion disrupts the 16pHQG;16 gene, removing its last 3 known exons (Figure 2).
Discussion
We have shown that an 18.3-kb deletion from the 3′ end of the human α-globin cluster stably silences α-globin gene expression from its otherwise natural chromosomal environment. Unlike all previously described deletions causing α-thalassaemia, this mutation does not remove any positive cis-acting elements. Therefore it appears to silence α gene expression by what is referred to as a negative chromosomal position effect, a term that covers a variety of mechanistically different phenomena in mammals,21,22Drosophila,22,23 plants,24 and yeast.25 Although such silencing mechanisms are thought to underlie several human genetic diseases and altered mouse phenotypes,21 26 there has been no detailed molecular characterization of negative position effects resulting from natural chromosomal rearrangements.
To date the best evidence that the mammalian “chromosomal environment” can influence gene expression comes from many observations demonstrating that genes on identical DNA segments may be expressed with variable patterns and at different levels when randomly integrated into the mouse genome. At the cellular level this often produces a variegated effect on gene expression. Some epigenetic characteristics (eg, methylation and DNaseI sensitivity), by which the chromosomal environment is currently defined, have been analyzed in such lines of transgenic mice with different conclusions.27-34 This suggests that there may be many ways in which chromosome position can affect gene expression.
The simplest explanation of our data would be that the α gene is silenced by its juxtaposition to the Alu-rich downstream region. It is known that stable, heritable gene silencing can occur when a euchromatic gene is juxtaposed to a region of heterochromatin. The region downstream of α-globin (approximate coordinates 180 000-218 000) may represent a relatively “heterochromatic” segment of DNA within the euchromatic 16p13.33 environment. The evidence for this is necessarily indirect. First, it contains a very high density of methylated repeat sequences (Alu and MER families), and it has been proposed that these features alone can be sufficient to create a hypoacetylated repressive chromatin environment.35,36 Second, in contrast to much of the surrounding chromosome, we did not detect DNaseI HSs or CpG islands in this region (Figure 1). Third, studies of replication timing using FISH analysis have shown that during the S-phase, the region within Cos12 (Figure 2, approximate coordinates 160 000-185 660) always replicates and separates later than its flanking regions.4
Although the silencing effect described here may be due to a single cis-element within the downstream region, preliminary sequence analysis did not identify significant homology to any previously described silencer elements. An alternative explanation is that multiple closely spaced Alu repeats act as a nucleation site for the formation of a stable repressive chromatin structure that extends, albeit a relatively short distance, to incorporate the juxtaposed αgene. The silencing phenomenon reported here may therefore be related to the previously described effects originating at the euchromatin/heterochromatin boundaries and polycomb response elements in Drosophila.
An unexpected finding was that a widely expressed gene encoding a putative RNA binding protein extends through this Alu dense region, transcribed in the opposite direction with respect to theα genes (C.T., unpublished data, 1999). It is possible that this gene represents an example of a human “heterochromatic gene” of the type described inDrosophila (eg, rolled and light), which are similarly embedded within regions containing high densities of repeats and transposons.37-39
Abnormal transcripts from this gene, extending across the breakpoint, may contribute to silencing and/or methylation of the juxtaposed α2-globin gene in the α−ZF chromosome, as recently proposed for regulation of some imprinted genes (eg,lgf2r, UBEA3, and LIT1)40-42 and the Xist gene, which is involved in X inactivation.43 Provisional experiments using RT-PCR suggest that such antisense transcripts are produced from the α−ZF chromosome (C.T., unpublished data, 1999) although the role of such transcripts in silencing is not yet clear. One hypothesis suggests that they directly interfere with transcription of the gene on the opposite DNA strand.44 Alternatively, antisense RNA might locally inactivate the chromosome by interacting with its chromatin in a similar way to the RNA product of Xist on the inactive X chromosome.45 46
Methylation is frequently associated with silencing of gene expression. For example, CpG islands normally become methylated during X-inactivation and genomic imprinting and may become abnormally methylated in tumorigenesis. Analysis of the CpG island associated with the α promoter on the α−ZF chromosome showed that although it is unmethylated in spermatocytes, it too becomes densely methylated during development.
Although methylation may play an important role in silencing it is not clear whether it is required to establish silencing or maintain silencing initiated by changes in chromatin structure. Recently, potentially important links between the processes of DNA methylation and histone deacetylation have been established. The protein MeCP2 specifically binds methylated DNA and has also been shown to interact with histone deacetylase and mammalian Sin3, providing a plausible connection between DNA methylation and the formation of a repressive chromatin environment.36,47 48Similarly, the protein MBD2, which also binds methylated DNA, can recruit the repressive NuRD complex, which contains the histone deacetylanes HDAC1 and HDAC2. At present, the order of events leading to hypoacetylation, methylation, and repression has not been established.
The α−ZF mutation adds to our understanding of the relationship between α gene regulation and its chromosomal environment. Despite their common ancestry, we have previously suggested that the α- and β-globin genes are regulated in a different manner, which may be related to their different chromosome environments.2,4,16 These genes may provide models for other genes located in contrasting regions of the genome.49The region of chromosome 11 containing the β-globin locus control region (β-LCR), which regulates expression of the entire β-cluster, influences long-range and local chromatin structure, the timing of replication, and the accumulated levels of the β-globin gene expression.50 51 When linked to β-LCR, the transgenes are protected from position effects.
By contrast the α-globin regulatory element, although absolutely required for α gene expression, has no discernible effect on chromatin structure, methylation, or replication timing.4,15 16 Transgenes linked to this element are always expressed but appear sensitive to their position of integration. Although HS –40 enhances the accumulated levels of α-globin mRNA expression, other cis-acting feature(s) must create the permissive chromosomal environment that allows this interaction to occur. All known positive cis-acting sequences are present on the α−ZF chromosome, and the regulatory element is active, as judged by the presence of the HS, yet the α gene is silenced. Clearly the HS –40/α interaction is insufficient to overcome the α−ZF position effect. Although we have not ruled out the presence of as yet unidentified specific elements, the normal permissive environment possibly results from an inherent, albeit unevenly distributed, feature of the 16p GC-rich chromosomal segment. The rearrangement described here may simply have changed a normally permissive microenvironment containing the α gene to a nonpermissive one.
In a normal chromosome the α genes are protected from the influence of this repressive environment that only lies approximately 12 kb away. Furthermore, some previously described deletions move theα genes to within approximately 6 kb of this region (Figure2) and yet have no effect on expression of the remaining αgene(s). One possibility is that the α genes are normally protected by a cis-acting element(s) which remains intact in these deletions but is removed in the α−ZF chromosome. The region of interest contains several Alu elements, a variable number tandem repeat (3′ hypervariable region) and a DNaseI hypersensitive site (approximate coordinates 178 500-179 000) at the end of the 16PHQG;16 gene. It has been suggested that boundary elements commonly flank chromosomal domains, protecting genes within them from position effects induced by surrounding heterochromatin or unrelated positive regulatory elements.52 However, the level and pattern expression of the cDH2 construct, which spans the entire region deleted in the α−ZF chromosome, does not provide evidence for such a boundary because the α genes in this construct were not consistently protected from position effects in transgenic mice.
In summary, juxtaposition of a sequence that normally lies 18 kb downstream of the α- globin complex next to the α2 gene silences its expression, thereby providing an entirely new mechanism that may cause α-thalassemia. Further work is required to determine the relative contributions of repressive chromatin environment, antisense transcripts, and the removal of a potential boundary element in causing this negative chromosomal position effect.
Acknowledgments
We are grateful to Professor D. J. Weatherall for his continued support and encouragement. We are also grateful to the late Professor T. H. J. Huisman, who originally brought our attention to the α−ZF deletion. We thank David Garrick, Jonathan Flint, and Veronica Buckle for their comments on the manuscript and Liz Rose and Milly Graver for preparation of the manuscript.
Reprints:D. R. Higgs, the MRC Molecular Haematology Unit, Institute of Molecular Medicine, John Radcliffe Hospital, Headington, Oxford, England; e-mail: drhiggs@molbiol.ox.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.