Abstract
Erythroid protein 4.1 (4.1R) is an 80-kd cytoskeletal protein that stabilizes the membrane-skeletal network structure underlying the lipid bilayer. Using the carboxyl terminal domain (22/24 kd) of 4.1R as bait in a yeast 2-hybrid screen, we isolated cDNA clones encoding a polypeptide of eIF3-p44, which represents a subunit of a eukaryotic translation initiation factor 3 (eIF3) complex. The eIF3 complex consists of at least 10 subunits that play an essential role in the pathway of protein translation initiation. Northern blot analysis revealed that eIF3-p44 (approximately 1.35 kb) is constitutively expressed in many tissues. The essential sequence for this interaction was mapped to the carboxyl-terminus of 4.1R (residues 525-622) and a region (residues 54-321) of eIF3-p44. The direct association between 4.1R and eIF3-p44 was further confirmed by in vitro binding assays and coimmunoprecipitation studies. To characterize the functions of eIF3-p44, we depleted eIF3-p44 from rabbit reticulocyte lysates either by anti-eIF3-p44 antibody or by GST/4.1R-80 fusion protein. Our results show that the eIF3-p44 depleted cell-free translation system was unable to synthesize proteins efficiently. The direct association between 4.1R and elF3-p44 suggests that 4.1R may act as an anchor protein that links the cytoskeleton network to the translation apparatus.
Erythroid protein 4.1 (4.1R) was originally identified as an 80-kd cytoskeletal protein (4.1R-80) in red blood cells. It is believed to maintain red cell morphology and mechanical strength by linking membrane integral proteins such as glycophorin C and band 3 to the spectrin–actin-based cytoskeletal network. Deficiency of 4.1R in red blood cells leads to assembly of an unstable cytoskeleton structure that manifests as hereditary elliptocytosis, a disease characterized by the loss of normal discoid morphology and the presence of oval or elliptical red cells with unstable membranes.1 Furthermore, erythrocytes from 4.1R-null mice exhibit erythroid membrane skeleton abnormalities, which further suggests that the loss of 4.1R compromises membrane skeleton assembly in erythroid cells.2
The protein 4.1 family encompasses a group of structure-related proteins. The prototypical erythroid membrane skeletal protein 4.1 (4.1R) is encoded by a complexly spliced gene located on human chromosome 1p33-p34.2.3,4 Heterogeneity of 4.1R isoforms in size and subcellular localization has been noted. These isoforms are generated by complex alternative splicing of 4.1R pre-mRNA and by posttranslational modification of 4.1R proteins.5-7Western blot analysis revealed 4.1R isoforms ranging in size from 30 to 175 kd among various mammalian nonerythroid cells.8 The erythroid 4.1R is located mainly beneath the peripheral membrane of mature red cells. However, the immunoreactive epitopes of 4.1R in nonerythroid cells have been identified in the cytoplasm, stress fibers,9 nucleus,10,11centrosomes,12 and cell–cell contact regions.13
In addition to the prototypical erythroid 4.1R, 3 new 4.1-like genes have been identified that reveal high sequence homology with the 30-, 10-, and 22/24-kd domains of 4.1R. These include 2 homologues that are highly expressed in the brain and neurons (4.1B, 4.1N) and another that is generally expressed throughout the body (4.1G).14,15These 4.1-like genes appear to map to distinct chromosomes in humans and mice.16
Limited chymotryptic digestion of erythroid 4.1R-80 (80 kd) generates 4 structural domains (30, 16, 10, and 22/24 kd). The 30-kd domain mediates the attachment of 4.1R to the plasma membrane by binding to the cytoplasmic domains of the transmembrane proteins, glycophorin C,17-19 and band 3.20,21 It also interacts with p55,18,19 calmodulin,22 and pICln, a protein involved in cellular volume regulation.23 The 16-kd domain has phosphorylation sites for protein kinase C and protein kinase A (PKA),24,25 whereas the 10-kd domain contains an exon 16-encoded peptide that is important for interaction with spectrin and actin complexes.26-28
The physiological function of the 22/24-kd domain is less well characterized. Recent reports showed that the carboxyl terminal domain (22/24 kd) of the 135-kd 4.1R (4.1R-135) isoform interacts with the nuclear mitotic apparatus protein (NuMA), suggesting that some 4.1R isoforms may play roles in organizing the nuclear architecture and the mitotic spindle.29 To further elucidate the possible functions of the 22/24-kd domain in nonerythroid cells, we searched for proteins that bind to this particular domain. We performed a yeast 2-hybrid screen using the 22/24-kd domain as bait. Several positive clones including NuMA, eIF3-p44, Sec 14-like protein, and 26S proteasome subunit p55 were isolated. One of these clones, encoding eIF3-p44, a subunit of eukaryotic translation initiation factor 3 (eIF3),30,31 was further analyzed. eIF3 is a large translation initiation complex that contains at least 10 subunits. It plays a central role in the binding of the initiator methionyl–tRNA and mRNA to the 40S ribosomal subunit to form the 40S initiation complex.32 In this study, we show that 4.1R associates with eIF3-p44 both in vitro and in vivo, which suggests possible interactions between the cytoskeleton network and the translation apparatus.
Materials and methods
Yeast 2-hybrid screen
Yeast 2-hybrid screening was conducted using the yeast strain Y190, which contains the HIS3 and lacZ reporter genes required for GAL4-dependent transcriptional activation. The carboxyl 22/24-kd domain (residues 462-641) of 4.1R (4.1R-22) was subcloned in frame to the GAL4 DNA binding domain (GAL4-BD) in the pAS2-1 vector (Clontech, Palo Alto, CA). This construct was used as bait to screen human lymphocyte and testis cDNA libraries (Clontech) fused to a GAL4 activation domain (GAL4-AD) in the pACT2 vector. The 2 plasmids were then cotransformed into Y190 yeast. The transformants were selected on SD minimal medium containing 25 mmol/L 3-amino-1,2,4-triazole, but without the addition of leucine (−Leu), tryptophan (−Trp), and histidine (−His), as described by the manufacturer (Clontech). Positive colonies (ie, Leu+, Trp+, His+) were further tested for β-galactosidase activity using colony-lift and liquid assays, as described by the manufacturer (Clontech).
To identify the minimum regions of 4.1R and eIF3-p44 that bind to each other, constructs containing various portions of 4.1R were subcloned into the pAS2-1 vector (Figure 1A), and different eIF3-p44 constructs (Figure 1B) were inserted into the pACT2 vector. Yeast cells (Y187) were simultaneously transformed with the 2 constructs and assayed for β-galactosidase activity.
Interaction between regions of 4.1R and eIF3-p44 in the yeast 2-hybrid system.
The eIF3-p44 clone (p44-1) was first isolated by a yeast 2-hybrid screen from a human lymphocyte cDNA library using the 22/24-kd domain of 4.1R (4.1R-22) as bait. (A) Schematic diagram of the various portions of 4.1R that interact with eIF3-p44 in a yeast 2-hybrid screen. The constructs containing various portions of 4.1R fused to the DNA binding domain of Gal4 (Gal4-BD) were cotransformed with an eIF3-p44/pACT2 clone (p44-1) that expressed eIF3-p44 (residues 54-321) fused to the activation domain of Gal4 (Gal4-AD). (B) Schematic diagram of various portions of eIF3-p44 that interact with 4.1R. A series of eIF3-p44 deletion mutants, fused to Gal-AD in the pACT2 vector, were cotransformed with 4.1R-22/pAS2-1 in Y187 cells. +, expression of the lacZ reporter gene using the colony-lift assay; −, nonexpression of the reporter gene. Right-hand column represents results of the liquid assay for β-galactosidase activity.
Interaction between regions of 4.1R and eIF3-p44 in the yeast 2-hybrid system.
The eIF3-p44 clone (p44-1) was first isolated by a yeast 2-hybrid screen from a human lymphocyte cDNA library using the 22/24-kd domain of 4.1R (4.1R-22) as bait. (A) Schematic diagram of the various portions of 4.1R that interact with eIF3-p44 in a yeast 2-hybrid screen. The constructs containing various portions of 4.1R fused to the DNA binding domain of Gal4 (Gal4-BD) were cotransformed with an eIF3-p44/pACT2 clone (p44-1) that expressed eIF3-p44 (residues 54-321) fused to the activation domain of Gal4 (Gal4-AD). (B) Schematic diagram of various portions of eIF3-p44 that interact with 4.1R. A series of eIF3-p44 deletion mutants, fused to Gal-AD in the pACT2 vector, were cotransformed with 4.1R-22/pAS2-1 in Y187 cells. +, expression of the lacZ reporter gene using the colony-lift assay; −, nonexpression of the reporter gene. Right-hand column represents results of the liquid assay for β-galactosidase activity.
Northern blot analysis and cDNA library screening
A blot filter containing 2 μg poly (A)+ RNA derived from multiple human tissues was purchased from Clontech. The blot was hybridized to a cDNA probe derived from the 0.9-kb fragment ofeIF3-p44 corresponding to amino acid residues 54-321 (eIF3-p44-1) under previously described conditions.5 The same blot was stripped and reprobed with human β-actin cDNA to quantify RNA loading.
To obtain the full-length eIF3-p44 cDNA, the same probe was used to screen a human testis cDNA library (Clontech). The conditions for screening and isolation were previously described.5
Plasmids and antibodies
eIF3-p44-1 (residues 54-321), originally identified from a yeast 2-hybrid screen (Figure 1B), and eIF3-p44 (residues 1-321), a full-length eIF3-p44 cDNA isolated from a cDNA library, were used as templates to generate GST/eIF3-p44-1 and GST/eIF3-p44 constructs, respectively. The cDNA fragments spanning the coding regions of eIF3-p44-1 and eIF3-p44 were polymerase chain reaction-amplified and fused in frame to GST in the pGEX-2T expression vector (Pharmacia, Uppsala, Sweden). GST/eIF3-p44-1 and GST/eIF3-p44 fusion proteins were generated and purified on glutathione–agarose beads (Sigma, St. Louis, MO) as previously described.23 GST/eIF3-p44-1 fusion protein was used to produce polyclonal antibodies, and GST/eIF3-p44 fusion protein was used for the in vitro binding assay. Polyclonal antibodies against eIF3-p44 were raised in rabbits and in mice, as previously described.33 Briefly, eIF3-p44-1 fusion protein was mixed with complete Freund's adjuvant (Sigma) and injected subcutaneously into New Zealand White rabbits or ICR mice. After 4 weeks, the animals were boosted with GST/eIF3-p44-1 antigen mixed with incomplete Freund's adjuvant (Sigma). Two weeks later, sera were collected and precipitated with ammonium sulfate. The precipitated IgG fractions were dialyzed against 0.1 mol/L phosphate-buffered saline (PBS), pH 8.0.
The cDNA fragments encoding the N-terminal headpiece domain (residues 55-198) of the 135-kd 4.1R and the C-terminal 22/24-kd domain (residues 462-641) of the 80-kd 4.1R isoform were individually subcloned into the pGEX-2T vector. The recombinant proteins were expressed in E. coli and purified on glutathione-agarose beads as previously described.23 The immunization and generation of hybridomas (anti-N-4.1R antibody) against the N-terminal headpiece of the 135-kd 4.1R isoform were performed as previously described.33Polyclonal anti-C-4.1R antibodies against the C-terminal 22/24-kd domain were raised in rabbits and mice as described above. Anti-FLAG monoclonal antibody (mAb) was purchased from Sigma, and anti-β-actin mAb was purchased from Boehringer (Indianapolis, IN).
In vitro binding assay
The cDNA encoding an 80-kd 4.1R isoform was constructed in the pSG5 vector (Stratagene, La Jolla, CA). Synthetic sense-capped mRNA was generated from the T7 promoter within the vector. Sense mRNA was translated in a coupled in vitro transcription/translation system (TNT rabbit reticulocyte lysate; Promega, Madison, WI) in the presence of35S-methionine to radiolabel newly synthesized proteins.
Equal amounts of the labeled 80-kd 4.1R protein were incubated with affinity-purified GST or GST/eIF3-p44 fusion proteins previously immobilized on glutathione–agarose beads as described.23After incubation, the immobilized complexes were washed 3 times with bead binding buffer (50 mmol/L potassium phosphate, pH 7.5, 150 mmol/L KCl, 1 mmol/L MgCl2, 10% glycerol, 1% Triton X-100, 1 mmol/L phenylmethylsulfonyl fluoride). Bound protein complexes were analyzed on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography.23
Cell culture, transfection, and Western blot analysis
Molt-4 cells (a human lymphoid cell line) were maintained in RPMI-1640 medium, as previously described.34 SiHa cells (a human cervical carcinoma cell line) were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. The cDNA encoding human eIF3-p44-1 (residues 54-321) was inserted in frame into a cytomegalovirus promoter-driven FLAG epitope-tagged expression vector (Kodak, New Haven, CT), designated FLAG-eIF3-p44-1. SiHa cells (1 × 106) were transiently transfected with or without 10 μg FLAG-eIF3-p44-1 cDNA, as previously described.23
Twenty-four hours after transfection, cells were washed in PBS and lysed in 1 mL ice-cold EBC buffer (50 mmol/L Tris-HCl, pH 8.0, 120 mmol/L NaCl, 0.5% NP-40, 1 μg aprotinin, 1 μg leupeptin, and 2 mmol/L phenylmethylsulfonyl fluoride) for 30 minutes on ice, and the lysate was cleared by centrifugation. Solubilized proteins (10 μg) were separated on a 10% SDS-PAGE, blotted onto a polyvinylidene difluoride membrane, and probed with anti-eIF3-p44 rabbit polyclonal antibody, anti-FLAG mAb, or anti-β-actin mAb. Secondary antibodies were goat antirabbit or goat antimouse IgG conjugated with horseradish peroxidase (Promega). Immunoreactive polypeptides were then detected by the Western Exposure Chemiluminescent Detection System (Pierce, Rockford, IL) as previously described.23
Coimmunoprecipitation assay
Coimmunoprecipitation of 4.1R and eIF3-p44 was performed using Molt-4 cell extracts. Cells were lysed in EBC buffer, and the lysate was centrifuged at 10 000g for 10 minutes at 4°C. The supernatant (approximately 5 mg) was precleared with protein A–Sepharose beads (Sigma), immunoprecipitated with preimmune serum (approximately 40 μg) or anti-eIF3-p44 antibody (approximately 40 μg) overnight at 4°C, and incubated with protein A–Sepharose beads for another 2 hours. Immunoprecipitates were collected by centrifugation at 5000g for 5 minutes at 4°C and washed 3 times with EBC buffer. Samples were resuspended in 20 μL SDS sample buffer (62.5 mmol/L Tris-HCl, pH 6.8, 2% SDS, 5% β-mercaptoethanol, and 10 μg/mL bromophenol blue) and were boiled at 95°C for 5 minutes. The samples were then centrifuged at 10,000g for 10 minutes at 4°C, and the supernatants were separated on 7.5% SDS-PAGE. After transfer to a polyvinylidene difluoride membrane, the immunoreactive proteins were detected by anti-N-4.1R or anti-C-4.1R antibodies, as described above. For reverse immunoprecipitation, the cell lysates were immunoprecipitated with anti-C-4.1R antibodies and analyzed by immunoblotting with anti-eIF3-p44 antibodies.
In vitro depletion assay
To deplete eIF3-p44, rabbit reticulocyte lysates (12.5 μL) were preincubated for 2 hours at 4°C with preimmune serum (2 μg), anti-eIF3-p44 antibody (2 μg) previously coupled to protein A–Sepharose beads, immobilized GST (10 μg), or GST/4.1R-80 fusion proteins (10 μg) previously conjugated to glutathione–agarose beads. Samples were centrifuged at 5000g for 5 minutes at 4°C to remove the bound complexes, and the supernatant was then assayed for in vitro translation activity using theluciferase gene as the template. The35S-methionine–labeled luciferase proteins were analyzed on 10% SDS-PAGE and visualized by autoradiography. To test translation efficiency in the presence of GST/4.1R-80 fusion protein, the indicated amounts (0.25 to approximately 2 μg) of soluble GST/4.1R-80 fusion protein or GST protein (2 μg) were mixed with rabbit reticulocyte lysate for 2 hours at 4°C. Samples were examined for in vitro translation activity as described above.
Results
4.1R and eIF3-p44 interact in a yeast 2-hybrid assay
We previously showed that pICln (a protein involved in cellular volume regulation) binds to the 30-kd domain of 4.1R in a yeast 2-hybrid system.23 Using a similar approach, we set out to identify proteins that interact with the 22/24-kd domain of 4.1R (4.1R-22). The cDNA encoding the 22/24-kd domain, fused in frame to the Gal4 DNA-binding domain (Gal4-BD), was used as bait to screen human lymphocyte and testis cDNA libraries fused to the Gal4 activation domain (Gal4-AD). We screened approximately 3 × 106transformants, and 63 positive clones were obtained.
A search of the GenBank database revealed that at least 4 known genes (NuMA, eIF3-p44, Sec 14–like protein, and 26S proteasome subunit p55) were found from the screen. Among these known proteins, we further examined NuMA and eIF3-p44. NuMA is a nuclear mitotic apparatus protein that had previously been isolated in a yeast 2-hybrid assay using the full-length 135-kd 4.1R as bait.29 This clone was also independently isolated 17 times in our screen. The other known gene that interacts with the 22/24-kd domain of 4.1R (residues 462-641; Figure 1A) is eIF3-p44. The interaction of 4.1R and eIF3-p44 in the yeast 2-hybrid assay appears to be specific: eIF3-p44-1/pACT2 did not bind to the empty vector pAS2-1 containing only unfused Gal4-BD (Figure 1A). The 4.1R-22/pAS2-1 also did not interact with the unfused Gal4-AD vector, pACT2 (Figure 1B). It has been suggested that mRNA and the translation apparatus are associated with the cytoskeleton.35 36 Our finding raises the interesting possibility that 4.1R may act as a linker between the translation apparatus and the cytoskeleton.
Cloning and Northern blot analysis of eIF3-p44
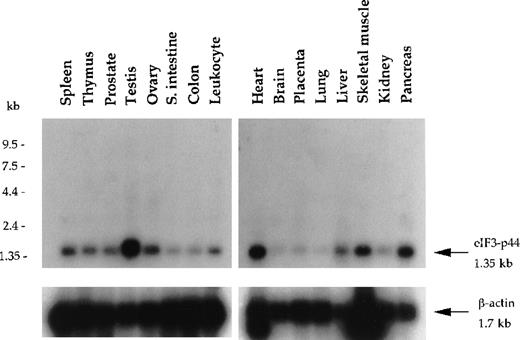
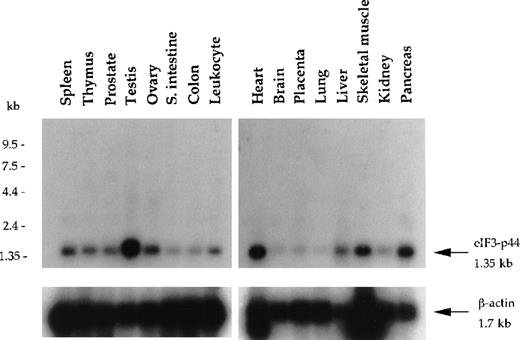
Using yeast 2-hybrid screening, we isolated a cDNA clone (p44-1; Figure 1B) encoding a eukaryotic translation initiation factor 3 subunit (eIF3-p44).30 31 Clone p44-1 represents part of the coding sequence and the 3′-untranslated region (UTR) of eIF3-p44, including the stop codon TAA followed by the polyadenylation signal AATAAA. Northern blot analysis revealed that the eIF3-p44transcript (1.35 kb) was detectable in all tissues examined and was predominantly expressed in testis, heart, pancreas, and skeletal muscle (Figure 2). To obtain the cDNA that covers the entire coding region of eIF3-p44, we screened a human testis cDNA library using eIF3-p44-1 cDNA as a probe. Several full-length clones were isolated, and their sequences were confirmed on both strands.
Northern blot analysis of eIF3-p44 in various human tissues.
Blot filters containing 2 μg poly (A)+RNA were hybridized with a 32P-labeled cDNA fragment (0.9 kb) encoding amino acid residues 54-321 of human eIF3-p44. The same blot was stripped and reprobed with β-actin to quantify RNA loading. eIF3-p44 mRNA is abundantly expressed in human tissues including testis, skeletal muscle, pancreas, and heart.
Northern blot analysis of eIF3-p44 in various human tissues.
Blot filters containing 2 μg poly (A)+RNA were hybridized with a 32P-labeled cDNA fragment (0.9 kb) encoding amino acid residues 54-321 of human eIF3-p44. The same blot was stripped and reprobed with β-actin to quantify RNA loading. eIF3-p44 mRNA is abundantly expressed in human tissues including testis, skeletal muscle, pancreas, and heart.
Interaction between 4.1R and eIF3-p44 occurs through the 22/24-kd domain of 4.1R and amino acids 54-321 of eIF3-p44
To determine the region of 4.1R that interacts with eIF3-p44, constructs containing different regions of 4.1R in the pAS2-1 vector and eIF3-p44-1 in the pACT2 vector were cotransformed into the yeast Y187 strain and plated on SD medium lacking tryptophan and leucine. After incubation for 2 days, the colonies were subjected to a liquid assay for β-galactosidase activity. Figure 1, panel A shows that eIF3-p44-1 interacted with 4.1R-80 (80 kd), 4.1R-32 (10 kd + 22/24 kd), 4.1R-22 (22/24 kd), and 4.1R-2 (residues 525-622), but not with 4.1R-30 (30 kd), 4.1R-1 (residues 462-531), 4.1R-3 (residues 608-641), or the pAS2-1 vector alone. Similar results were obtained using the colony-lift assay (data not shown) to examine for positive interactions. Thus, the 98 amino acids of 4.1R derived from the 22/24-kd domain (4.1R-2) are required and sufficient for interaction with eIF3-p44.
To map the region of eIF3-p44 that binds to 4.1R, we expressed different domains of eIF3-p44/pACT2 clones and cotransformed them into Y187 with 4.1R-22/pAS2-1. As shown in Figure 1B, only p44 (encoding the entire coding region of eIF3-p44) and p44-1 (residues 54-321) showed substantial binding to 4.1R-22, whereas p44-2 (residues 54-185) revealed a weak interaction. In contrast, peptide segments of eIF3-p44 encompassing amino acids 185-321 (p44-3) or 231-321 (p44-4), which contain the RNA recognition motif, did not interact with 4.1R-22. Therefore, we believe that the entire structure of eIF3-p44, particularly the region spanning amino acids 54-321, are necessary and important for the maintenance of the correct conformation for eIF3-p44 binding to 4.1R. Taken together, these results indicate that the interacting regions of these 2 proteins are restricted to the amino acids residues 525-622 of 4.1R and 54-321 of eIF3-p44.
4.1R interacts with eIF3-p44 in vitro
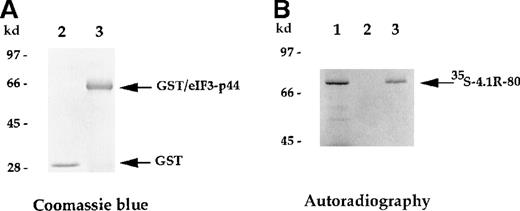
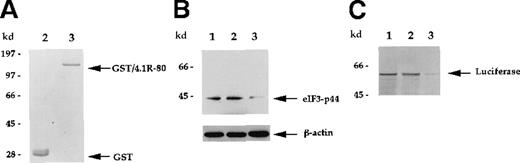
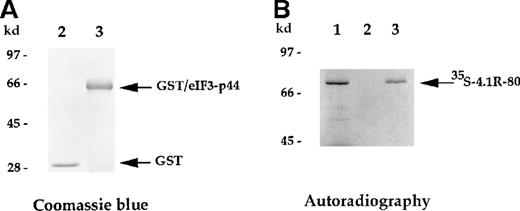
The direct interaction between 4.1R and eIF3-p44 was further analyzed using an in vitro binding assay. The full-lengtheIF3-p44 cDNA was constructed in frame into the pGEX2T vector. The GST/eIF3-p44 fusion protein was expressed and purified with glutathione–agarose beads (Figure 3A, lane 3). Immobilized GST/eIF3-p44 protein was incubated with35S-methione-labeled 4.1R (80-kd isoform), and retention on the beads was analyzed by SDS-PAGE. As shown in Figure 3B, GST/eIF3-p44 fusion protein interacted with the labeled 80-kd 4.1R (Figure 3B, lane 3). In contrast, the control GST fusion protein failed to bind to labeled 4.1R (Figure 3B, lane 2). Consistent with our results from the yeast 2-hybrid assays, these results further confirm that eIF3-p44 specifically interacts with 4.1R in vitro.
In vitro binding assay indicates that eIF3-p44 binds to 4.1R.
The eIF3-p44 cDNA was subcloned into the pGEX-2T vector and expressed as a GST/eIF3-p44 fusion protein in Escherichia coli. (A) Purified GST (lane 2) and GST/eIF3-p44 fusion protein (lane 3) were visualized by Coomassie blue staining. (B) Binding of GST/eIF3-p44 to 4.1R-80 (80-kd isoform) in vitro.35S-methionine–labeled 4.1R-80 (lane 1) was incubated with affinity-purified GST (lane 2) or GST/eIF3-p44 fusion protein (lane 3) previously coupled to glutathione–agarose beads. After incubation, the bound complexes were analyzed by SDS-PAGE and autoradiography. Radiolabeled 4.1R-80 bound to GST/eIF3-p44 fusion protein (lane 3) but not GST alone (lane 2).
In vitro binding assay indicates that eIF3-p44 binds to 4.1R.
The eIF3-p44 cDNA was subcloned into the pGEX-2T vector and expressed as a GST/eIF3-p44 fusion protein in Escherichia coli. (A) Purified GST (lane 2) and GST/eIF3-p44 fusion protein (lane 3) were visualized by Coomassie blue staining. (B) Binding of GST/eIF3-p44 to 4.1R-80 (80-kd isoform) in vitro.35S-methionine–labeled 4.1R-80 (lane 1) was incubated with affinity-purified GST (lane 2) or GST/eIF3-p44 fusion protein (lane 3) previously coupled to glutathione–agarose beads. After incubation, the bound complexes were analyzed by SDS-PAGE and autoradiography. Radiolabeled 4.1R-80 bound to GST/eIF3-p44 fusion protein (lane 3) but not GST alone (lane 2).
4.1R interacts with eIF3-p44 in vivo
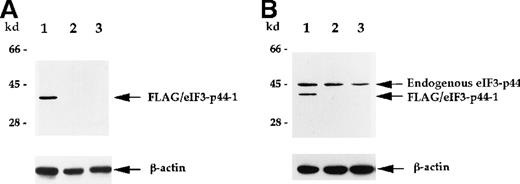
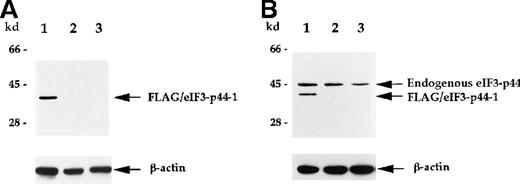
Polyclonal antibodies against eIF3-p44 were raised to further characterize the interaction between 4.1R and eIF3-p44. To test the specificity of this antibody, we transiently transfectedFLAG-eIF3-p44-1 into SiHa cells. The cell extract was prepared 24 hours after transfection and analyzed by anti-FLAG mAb or anti-eIF3-p44 polyclonal antibodies. Figure4 shows that FLAG-eIF3-p44-1 (residues 54-321) protein was recognized by both anti-FLAG (Figure 4A, lane 1) and anti-eIF3-p44 (Figure 4B, lane 1) antibodies, whereas, the endogenous eIF3-p44 in both transfected (Figure 4B, lane 1) and untransfected (Figure 4B, lane 2) SiHa and Molt-4 (Figure 4B, lane 3) cells was only detected by anti-eIF3-p44 antibody. Because the SiHa cells were transfected with an N-terminal–truncated clone, FLAG-eIF3-p44-1 (residues 54 to 321), a low molecular weight band was detected in transfected cells (Figure 4B, lane 1).
Characterization of anti-eIF3-p44 antibody.
The production of antibody against eIF3-p44 is described in “Materials and methods.” SiHa cells were transiently transfected with a FLAG-tagged eIF3-p44-1 plasmid. Cell lysates (10 μg) prepared from transfected cells (lane 1), untransfected cells (lane 2), and Molt-4 cells (lane 3) were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with anti-FLAG antibody (A) or anti-eIF3-p44 antibody (B). The same blot was reprobed with an antibody against β-actin as a control. Positions of the molecular weight markers are shown on the left.
Characterization of anti-eIF3-p44 antibody.
The production of antibody against eIF3-p44 is described in “Materials and methods.” SiHa cells were transiently transfected with a FLAG-tagged eIF3-p44-1 plasmid. Cell lysates (10 μg) prepared from transfected cells (lane 1), untransfected cells (lane 2), and Molt-4 cells (lane 3) were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with anti-FLAG antibody (A) or anti-eIF3-p44 antibody (B). The same blot was reprobed with an antibody against β-actin as a control. Positions of the molecular weight markers are shown on the left.
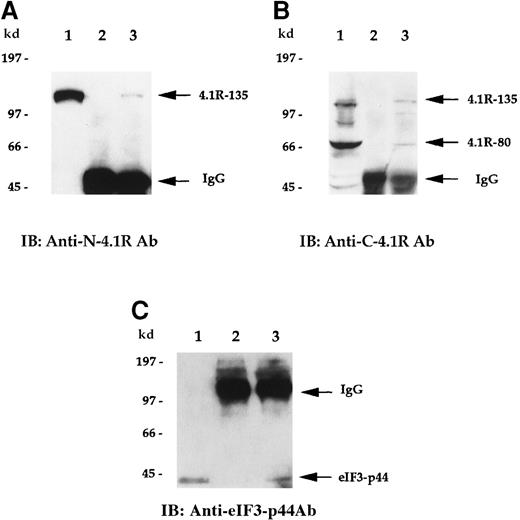
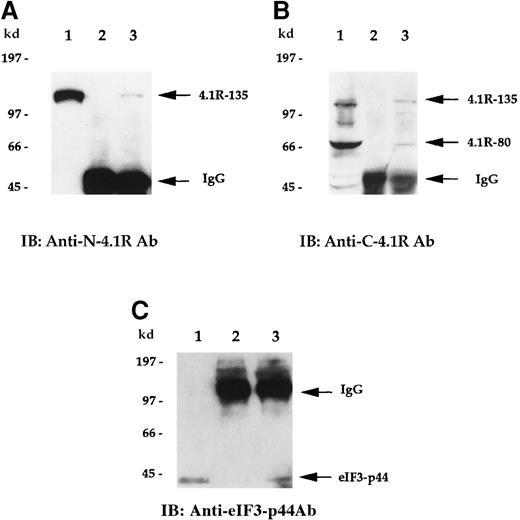
To examine whether endogenous 4.1R is associated with eIF3-p44 in vivo, we performed a co-immunoprecipitation assay. Cell lysates prepared from Molt-4 cells were first immunoprecipitated with either preimmune serum or anti-eIF3-p44 antibodies. Coprecipitated proteins were then detected by anti-N-4.1R (against the headpiece 209 amino acids of the 135-kd isoform) (Figure 5A) or anti-C-4.1R (against the C-terminal 22/24-kd domain) (Figure 5B) antibodies. As shown in Figure 5, panels A and B (lane 2), no endogenous 135-kd or 80-kd 4.1R was detected when preimmune serum was used for the immunoprecipitation experiment. In contrast, the 135-kd isoform of 4.1R was coprecipitated with anti-eIF3-p44 antibodies and was detected by anti-N-4.1R antibody (Figure 5A, lane 3). Similarly, both 135-kd and 80-kd 4.1R isoforms were coprecipitated with anti-eIF3-p44 and immunoreacted with anti-C-4.1R antibody (Figure 5B, lane 3). The in vivo association of 4.1R and eIF3-p44 was further confirmed by reverse immunoprecipitation (Figure 5C). Cell lysates were first immunoprecipitated with either preimmune serum (Figure 5C, lane 2) or anti-C-4.1R antibody (Figure 5C, lane 3), and this was followed by immunoblotting of co-precipitated proteins with anti-eIF3-p44 antibody. Because the heavy chains of the immunoglobulins comigrate with eIF3-p44 under reducing conditions on SDS-PAGE, immunoprecipitates were dissolved in sample buffer lacking β-mercaptoethanol to maintain the disulfide bonds between the light and heavy chains of the immunoglobulins. As shown in Figure 5, panel C, eIF3-p44 was pulled down by anti-C-4.1R antibody (lane 3) but not by pre-immune serum (lane 2). Taken together, these results indicate that eIF3-p44 interacts with 4.1R in vivo.
4.1R interacts with eIF3-p44 in vivo.
Cell lysates prepared from Molt-4 cells were immunoprecipitated with preimmune serum (lane 2) or anti-eIF3-p44 antibody (lane 3). Bound protein complexes were analyzed by immunoblotting with antibodies against the N-terminal portion of 135-kd 4.1R (A, anti-N-4.1R) or against the C-terminal 22/24-kd domain of 4.1R (B, anti-C-4.1R antibody). Lane 1 was loaded with Molt-4 cell lysates (10 μg) and immunoblotted with anti-N-4.1R (A), anti-C-4.1R (B), or anti-eIF3-p44 antibody (C). The 135-kd 4.1R isoform was specifically detected by anti-N-4.1R antibody (A, lane 3), whereas 2 alternative splicing isoforms of 4.1R (135-kd and 80-kd) were recognized by anti-C-4.1R antibody (B, lane 3). (C) Reverse immunoprecipitation. Cell lysates prepared from Molt-4 cells were immunoprecipitated with preimmune serum (lane 2) or anti-C-4.1R antibody (lane 3) and analyzed by immunoblotting with anti-eIF3-p44 antibody. Lane 1 represents a positive control in which Molt-4 cell extracts were immunoblotted with anti-eIF3-p44 antibody. Samples were dissolved in SDS sample buffer containing β-mercaptoethanol (A, B) or lacking β-mercaptoethanol (C, lanes 2 and 3).
4.1R interacts with eIF3-p44 in vivo.
Cell lysates prepared from Molt-4 cells were immunoprecipitated with preimmune serum (lane 2) or anti-eIF3-p44 antibody (lane 3). Bound protein complexes were analyzed by immunoblotting with antibodies against the N-terminal portion of 135-kd 4.1R (A, anti-N-4.1R) or against the C-terminal 22/24-kd domain of 4.1R (B, anti-C-4.1R antibody). Lane 1 was loaded with Molt-4 cell lysates (10 μg) and immunoblotted with anti-N-4.1R (A), anti-C-4.1R (B), or anti-eIF3-p44 antibody (C). The 135-kd 4.1R isoform was specifically detected by anti-N-4.1R antibody (A, lane 3), whereas 2 alternative splicing isoforms of 4.1R (135-kd and 80-kd) were recognized by anti-C-4.1R antibody (B, lane 3). (C) Reverse immunoprecipitation. Cell lysates prepared from Molt-4 cells were immunoprecipitated with preimmune serum (lane 2) or anti-C-4.1R antibody (lane 3) and analyzed by immunoblotting with anti-eIF3-p44 antibody. Lane 1 represents a positive control in which Molt-4 cell extracts were immunoblotted with anti-eIF3-p44 antibody. Samples were dissolved in SDS sample buffer containing β-mercaptoethanol (A, B) or lacking β-mercaptoethanol (C, lanes 2 and 3).
Cell-free translation system deficient in eIF3-p44 was unable to synthesize proteins efficiently
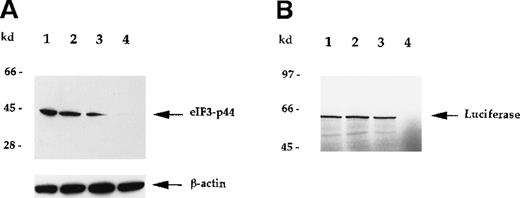
To verify that eIF3-p44 plays a role in protein biosynthesis,30 31 we attempted to deplete eIF3-p44 activity in reticulocyte lysates using anti-eIF3-p44 antibodies (Figure6) or GST/4.1R-80 fusion protein (Figure7). As seen in Figure 6, eIF3-p44 was completely removed from the reticulocyte lysates using immobilized antibodies against eIF3-p44 (Figure 6A, lane 4), and immunodepletion of eIF3-p44 from reticulocyte lysates resulted in the loss of template mRNA (luciferase) translation (Figure 6B, lane 4). The preimmune serum, though it partially depleted eIF3-p44 from the reticulocyte lysates (Figure 6A, lane 3), did not significantly inhibit mRNA translation (Figure 6B, lane 3).
Immunodepletion of eIF3-p44 by anti-eIF3-p44 antibody results in the loss of mRNA translation.
Rabbit reticulocyte lysates were incubated without (lane 1) or with protein A–Sepharose beads that had been preincubated with PBS buffer (lane 2), preimmune serum (lane 3), or anti-eIF3-p44 antibody (lane 4) as described in “Materials and methods.” (A) After centrifugation, the proteins in supernatants were separated by SDS-PAGE and immunoblotted with anti-eIF3-p44 or anti-β-actin antibody. β-Actin was used as an internal control. (B) Treated lysates were tested for in vitro translation activity using the luciferasegene as a template. 35S-methionine–labeled luciferase was analyzed by SDS-PAGE and autoradiography.
Immunodepletion of eIF3-p44 by anti-eIF3-p44 antibody results in the loss of mRNA translation.
Rabbit reticulocyte lysates were incubated without (lane 1) or with protein A–Sepharose beads that had been preincubated with PBS buffer (lane 2), preimmune serum (lane 3), or anti-eIF3-p44 antibody (lane 4) as described in “Materials and methods.” (A) After centrifugation, the proteins in supernatants were separated by SDS-PAGE and immunoblotted with anti-eIF3-p44 or anti-β-actin antibody. β-Actin was used as an internal control. (B) Treated lysates were tested for in vitro translation activity using the luciferasegene as a template. 35S-methionine–labeled luciferase was analyzed by SDS-PAGE and autoradiography.
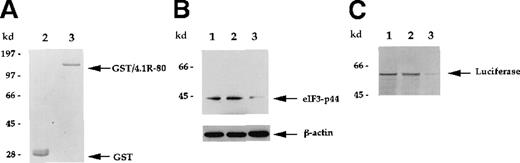
Depletion of eIF3-p44 using immobilized GST/4.1R-80 fusion protein.
(A) Purified, bacterially produced GST (lane 2) and GST/4.1R-80 fusion protein (lane 3) were visualized by Coomassie blue staining. (B) Rabbit reticulocyte lysates were incubated without GST (lane 1), with GST (lane 2), or with GST/4.1R-80 fusion protein (lane 3) coupled to glutathione–agarose beads as described in “Materials and methods.” After centrifugation, the supernatants were subjected to SDS-PAGE, and blots were probed with anti-eIF3-p44 or anti-β-actin antibodies. (C) Lysates without treatment (lane 1) or pretreated with affinity-purified GST (lane 2) or GST/4.1R-80 (lane 3) beads were tested for in vitro translation activity as described in Figure 6B.
Depletion of eIF3-p44 using immobilized GST/4.1R-80 fusion protein.
(A) Purified, bacterially produced GST (lane 2) and GST/4.1R-80 fusion protein (lane 3) were visualized by Coomassie blue staining. (B) Rabbit reticulocyte lysates were incubated without GST (lane 1), with GST (lane 2), or with GST/4.1R-80 fusion protein (lane 3) coupled to glutathione–agarose beads as described in “Materials and methods.” After centrifugation, the supernatants were subjected to SDS-PAGE, and blots were probed with anti-eIF3-p44 or anti-β-actin antibodies. (C) Lysates without treatment (lane 1) or pretreated with affinity-purified GST (lane 2) or GST/4.1R-80 (lane 3) beads were tested for in vitro translation activity as described in Figure 6B.
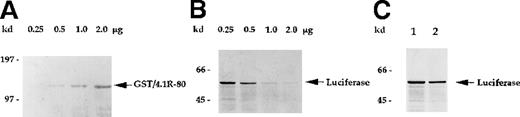
Similar effects were also observed when immobilized GST/4.1R-80 fusion protein was used to deplete eIF3-p44. As seen in Figure 7, the eIF3-p44 in reticulocyte lysate was significantly removed by the addition of 10 μg GST/4.1R-80 fusion protein (Figure 7B, lane 3) immobilized on glutathione–agarose beads, and the eIF3-p44-depleted lysate produced substantially decreased luciferase mRNA translation (Figure 7C, lane 3). In contrast, immobilized GST alone did not significantly remove eIF3-p44 (Figure 7B, lane 2), and the GST-treated lysate was not defective in mRNA translation (Figure 7C, lane 2). Furthermore, the addition of increasing amounts (0.25 to approximately 2 μg) of soluble GST/4.1R-80 fusion protein (Figure8A), but not GST protein (2 μg; Figure8C, lane 2), resulted in increasing loss of the protein translation efficiency of luciferase mRNA (Figure 8B). Taken together, these results suggest that eIF3-p44 can be depleted by either anti-eIF3-p44 antibodies or its interaction protein (4.1R) and that eIF3-p44-depleted or GST/4.1R-80 treated lysates do not efficiently synthesize proteins.
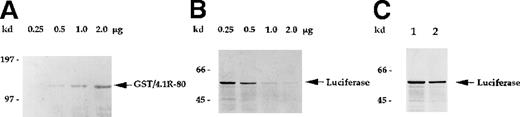
GST/4.1R-80–treated lysates exhibit reduced in vitro translation activity in a dose-dependent manner.
(A) Affinity-purified GST/4.1R-80 fusion protein (0.25 to approximately 2.0 μg) was visualized by Coomassie blue staining. (B) Rabbit reticulocyte lysates were incubated with the indicated amounts of soluble GST/4.1R-80 fusion protein for 2 hours at 4°C and tested for in vitro translation activity, as described in Figure6B. (C) Rabbit reticulocyte lysates were incubated with (lane 2) or without (lane 1) affinity-purified GST protein (2 μg) and tested for in vitro translation activity. In vitro translation activity was not inhibited by GST (C, lane 2) but was inhibited by the addition of GST/4.1R-80 fusion protein in a dose-dependent manner (B).
GST/4.1R-80–treated lysates exhibit reduced in vitro translation activity in a dose-dependent manner.
(A) Affinity-purified GST/4.1R-80 fusion protein (0.25 to approximately 2.0 μg) was visualized by Coomassie blue staining. (B) Rabbit reticulocyte lysates were incubated with the indicated amounts of soluble GST/4.1R-80 fusion protein for 2 hours at 4°C and tested for in vitro translation activity, as described in Figure6B. (C) Rabbit reticulocyte lysates were incubated with (lane 2) or without (lane 1) affinity-purified GST protein (2 μg) and tested for in vitro translation activity. In vitro translation activity was not inhibited by GST (C, lane 2) but was inhibited by the addition of GST/4.1R-80 fusion protein in a dose-dependent manner (B).
Discussion
We and others5,6,34 have shown that 4.1R is composed of multiple isoforms that are heterogeneous in size and subcellular localization.10-12 Many of these isoforms are present in nonerythroid cells3,5 or in erythroid cells at different developmental stages.6 Their biologic significance, however, has not yet been well characterized. In the current study we identified eIF3-p44, a subunit of the eukaryotic initiation factor 3 (eIF3) complex, as a binding target of 4.1R. The interaction between the carboxyl terminal domain (22/24 kd) of 4.1R and eIF3-p44 was first discovered using the yeast 2-hybrid system and then confirmed by in vitro binding, in vitro depletion, and coimmunoprecipitation studies.
The finding that eIF3-p44 directly interacts with a cytoskeletal protein (4.1R) was informative. The interaction between eukaryotic translation components during the early phase of protein synthesis has been well characterized.37 Eukaryotic translation initiation begins with the assembly of a preinitiation complex, including a small ribosome subunit (40S), eIF1A, eIF3, and eIF2-GTP-tRNA. Next this complex binds to mRNA in a reaction requiring adenosine triphosphate and the mRNA m7G-cap binding protein complex (eIF4A, eIF4B, eIF4E, and eIF4G) and then scans toward the 3′ end until it forms a stable complex at the first AUG initiation codon. eIF4G serves as a scaffold protein for the assembly of eIF4E and eIF4A and recruits Mnk1 to phosphorylate eIF4E.38
eIF3 is a large, multi-subunit complex (approximately 600 kd) that plays a central role in the initiation of translation. It was originally isolated from rabbit reticulocyte lysates, and it contains at least 10 different protein subunits ranging from 35 to 170 kd.32 Among these protein subunits, eIF3-p44 binds specifically to eIF3-p170 and contains a consensus RNA recognition motif near its carboxyl terminus.30 eIF3 binds to 40S ribosomal subunits, resulting in the dissociation of 80S ribosomes. eIF3 also stabilizes initiator methionyl-tRNA binding to 40S subunits and participates in mRNA binding through its interaction with eIF4G.32 Interestingly, 4.1R was reported to be an actin-associated protein that binds to the spectrin–actin complex through its 10-kd domain.26-28 Purified 4.1R also interacts with tubulin39 and myosin.40 In the current study, we show that eIF3-p44 binds to both 80-kd and 135-kd 4.1R isoforms (Figure 5) and that this interaction is through the carboxyl terminal 22/24-kd (residues 525-622) domain of 4.1R and amino acids residues 54-321 of eIF3-p44 (Figure 1). These findings suggest that 4.1R may serve as a bridging molecule that attaches the translational apparatus to the cytoskeletal scaffold.
Recent studies have shown that mRNA and polysomes are associated with the cytoskeleton and that this association may influence the transport, anchoring, and translation of mRNA.35,36 For example, the elongation factor 1 alpha (EF-1α) is abundantly expressed and constitutes 1% to 2% of the total proteins in normal growing cells. It catalyzes the GTP-dependent binding of aminoacyl-tRNA to ribosomes, and it regulates the faithfulness and rate of polypeptide elongation during translation.41 EF-1α is an actin-binding protein,42 and the involvement of EF-1α in translation is possibly regulated by its ability to dissociate from actin in response to local environmental changes in pH. The release of EF-1α from actin filaments would then facilitate polypeptide elongation by the F-actin–associated translational apparatus.43
Unlike EF-1α, whose involvement in translation is regulated in a pH-dependent manner, the association of 4.1R to the cytoskeleton and translational apparatus may be regulated by phosphorylation. It has been reported that 4.1R promotes a high-affinity association between spectrin and F-actin and that this association appears to be regulated by phosphorylation. 4.1R can be phosphorylated by various kinases, including protein kinase C and PKA.44 The phosphorylation of 4.1R by these kinases decreases the ability of 4.1R to bind to spectrin and inhibits the formation of the spectrin–actin–4.1R complex.45 We observed a significant inhibition of the protein translation activity when the reticulocyte lysates were incubated with 4.1R previously phosphorylated by PKA (unpublished data). This observation raises the possibility that the involvement of 4.1R in the cytoskeleton and translational apparatus may be regulated by phosphorylation. The PKA-phosphorylated amino acids of 4.1R have recently been identified as Ser-331 in the 16-kd domain and Ser-467 in the 10-kd spectrin–actin-binding domain.25 Awareness of such interaction may in turn lead to understanding the molecular mechanism of how phosphorylation regulates the 4.1R-linked cytoskeleton to the translational apparatus.
Depletion of eIF3-p44 from rabbit reticulocyte lysates using anti-eIF3-p44 antibodies (Figure 6) or immobilized GST/4.1R-80 fusion protein (Figure 7) resulted in a reduction of the lysates' ability to synthesize proteins efficiently. This is because eIF3-p44–depleted lysate may lose the entire eIF3 complex or the factors essential for protein translation. However, the preincubation of reticulocyte lysates with affinity-purified GST/4.1R-80 fusion protein without removing eIF3-p44 from the lysates caused a dose-dependent inhibition of mRNA translation (Figure 8). eIF3-p44 contains an RNA recognition motif near its C-terminus that can bind to both 18S rRNA and β-globin mRNA and that appears to be a nonspecific RNA binding protein.30 In the current study, we found that the 22/24-kd domain (residues 525-622) of 4.1R directly binds to eIF3-p44 (residues 54-321; Figure 1B). This finding suggests that the direct interaction of 4.1R with eIF3-p44 may sterically hinder the binding of eIF3-p44 to RNA. Therefore, the addition of increasing amounts of soluble GST/4.1R-80 fusion protein to lysates may result in a reduction of mRNA translation activity (Figure8).
Although the functional interactions of the 30-kd and 10-kd domains of 4.1R have been well documented, the roles of the 22/24-kd domain and the headpiece (209 amino acids) of the 135-kd 4.1R isoform remain unclear. Recently, several 4.1R- or 4.1G-associated proteins, including pICln,23 NuMA,29 P4.1-CAP (CPAP),46and FKBP13,15 have been isolated using yeast 2-hybrid screens. The different subcellular locations and uncommon features of these 4.1R-associated proteins implies the functional diversity and complexity of 4.1R. For example, the 30-kd domain of 4.1R mediates its binding to pICln, a protein involved in cellular volume regulation,23 whereas the 22/24-kd domain and the headpiece (209 amino acids) of the 135-kd isoform interact with a nuclear mitotic apparatus protein (NuMA)29 and a centrosome protein (CPAP),46 respectively. In the current study, we also show that the 22/24-kd domain of 4.1R interacts with eIF3-p44, a subunit of mammalian translation initiation factor 3. Further study of the physiologic implications of the interaction between 4.1R and associated proteins will eventually provide a more complete understanding of the various functional roles of 4.1R in nonerythroid cells.
Supported by grant NSC88-2314-B001-013 from the National Science Council, Republic of China, and an institutional grant from Academia Sinica, Taiwan, Republic of China.
Reprints:Tang K. Tang, Institute of Biomedical Sciences, Academia Sinica, 128 Yen-Chiu-Yuan Road, Sec. 2, Taipei 115, Taiwan; e-mail: tktang@ibms.sinica.edu.tw.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.