Cytogenetic abnormalities at chromosome 1q21 are among the most common lesions in diffuse large-cell lymphoma and have been associated with a poor prognosis. A novel cell line, SKI-DLCL-1, was established from ascitic fluid that carries a t(1;14)(q21;q32) chromosomal translocation. Using pulsed-field gel electrophoresis, the breakpoint on the IgH locus mapped to a gamma locus between C1 and C2. A cosmid library was prepared from SKI-DLCL-1, and Cγ-positive clones spanning the breakpoint were identified by screening with fluorescence in situ hybridization. The breakpoint occurs 860 bp downstream of the 3′ UTR of the MUC1 gene. The break appears to be a staggered double-strand break consistent with an error in immunoglobulin class switching. The MUC1 gene is highly transcribed and translated, and the protein is highly glycosylated. It is postulated that MUC1 expression is brought under the control of the 3′E enhancer. MUC1 lies in a region of chromosome 1 characterized by an unusually high density of genes, with 7 known genes in a region of approximately 85 kb. To determine whether there was a pleiotropic effect of the expression of genes in the region as a consequence of the translocation, the expression of 6 additional genes was assessed. None of the other genes in this region (CLK2, propin, COTE1, GBA, metaxin, and thrombospondin 3) are overexpressed in SKI-DLCL-1. Thus, the translocation t(1;14)(q21;q32) seen in both the primary tumor and the derived cell line results in the marked overexpression of MUC1 without affecting the expression of other genes in the region.
Nonrandom chromosomal translocations in malignancy have served as signposts identifying genes critical to the molecular pathogenesis of the disease. In the lymphomas, these nonrandom chromosomal translocations have frequently involved the immunoglobulin and T-cell receptor loci. It has been speculated that these nonrandom chromosomal translocations arise as errors in the process of immunoglobulin gene and T-cell receptor gene rearrangement or during immunoglobulin class switching. The genes identified at these translocation breakpoints have included those involved in critical cellular processes regulating cell growth, cell cycle, and apoptosis. Translocations involving the BCL1, BCL2, BCL6, and c-myc genes account for approximately 45% of cases of malignant lymphoma.1 2 However, there are a number of less common, nonrandom chromosomal translocations that have also been identified and have led to the identification of additional genes and that may well be critical to the pathogenesis of malignant lymphoma.
One of the most common such genetic abnormalities observed in malignant lymphoma occurs at chromosome 1q21.1,3 Abnormalities of this region have not been associated with a particular subtype of non-Hodgkin lymphoma, though in diffuse large-cell lymphoma (DLCL) they are associated with poor prognosis.1,4 Abnormalities include the recurrent translocations t(1;6)(q21;q23-q26), t(1;11)(q21;p15), and t(1;12)(q21;q24) together with the duplication dup(1)(q21;q32).3,4 Several genes have been identified at 1q21 and are thought to be involved in the pathogenesis of some cases of lymphoma, including BCL9 and genes homologous to Fcγ, MUM2, and MUM3.3 5
We derived a cell line from a patient with malignant ascites that had a complex karyotype including a translocation t(1;14)(q21;q32). We cloned the translocation breakpoint from this cell line and found it to lie 0.86 kb downstream of the MUC1 gene. The MUC1 gene translocated to der(14) chromosome is dramatically overexpressed in comparison to normal lymphoid tissue and other lymphoma cell lines. Interestingly, this dysregulated expression is confined to the MUC1 gene and does not affect the expression of 6 other genes that lie in a 75-kb region upstream of MUC1. The dysregulated expression of MUC1 may account for the unusual extranodal presentation of the index case.
Materials and methods
Cells and cell lines
After informed consent, abdominal ascites was obtained from a patient to establish the diagnosis of malignant lymphoma. The material was designated case 2385 by the cytogenetics laboratory. Excess material was frozen, and some was placed in culture in RPMI 1640 (Gibco-BRL, Bethesda, MD) supplemented with 20% fetal calf serum (FCS; HyClone, Logan, UT). After 24 days in culture, these cells appeared to be quiescent. However, in the fourth week, there was evidence of cellular proliferation. Subsequently, the cells proliferated at a brisk rate. Cytogenetic and flow cytometric analyses demonstrated that the cells were derived from the tumor clone in the malignant ascites.
Cell lines OCI-Ly8, JP-DLCL-1, FL318, and FL18 demonstrate cells derived from the transformation of low-grade lymphoma.6-8Cell lines Daudi and Raji represent Burkitt lymphomas.9MOLT-4 is derived from a T-cell lymphoma.9 They were routinely maintained in RPMI 1640 supplemented with 10% FCS, penicillin, streptomycin, and glutamine. Cells were grown at 37°C in humidified incubators supplemented with 5% CO2. MCF7 and HeLa were obtained from ATCC (American Type Culture Collection, Rockville, MD) and were maintained in Iscove's modified Dulbecco's medium supplemented with 10% FCS.
Cytogenetics and fluorescence in situ hybridization
Cytogenetics was performed by standard G-banding techniques as previously described.10 Fluorescence in situ hybridization (FISH) was performed with fluorescein isothiocyanate (FITC)-labeled probes and phycoerythrin-labeled, chromosome-specific centromeric probes. The conditions have been described.11
Pulsed-field gel electrophoresis and Southern blotting
Megabase DNA was embedded in InCert agarose (FMC, Rockland, ME) as previously described.12 DNA was resolved in a CHEF Mapper (Bio-Rad, Richmond, CA) using the auto-algorithm method or manually set pulse sequences as indicated in the figure legends. Gels were run in 0.5 × TBE buffer with a field strength of 6 V/cm at 14°C. DNA was transferred to nylon membranes (Hybond, Amersham, MA) as previously described with the following modification. Rather than performing vacuum transfer as previously described, DNA was transferred to immobilized membranes by traditional capillary action. DNA probes were prepared with random hexamer priming as recommended by the manufacturer (Stratagene, La Jolla, CA). Restriction endonuclease digestion was performed on agarose-embedded DNA as previously described.12 Blots were hybridized in conditions as previously described.12 Initial exposures of blots were performed on Fuji BAS PhosphorImager (Fuji Bio Imaging, Tokyo, Japan) to determine the appropriate length of exposure to film. Blots were subsequently exposed to film at −70°C using Lighting Plus intensifying screens (Dupont, Wilmington, DE).
Cosmid DNA cloning
DNA was extracted from the cell line SKI-DLCL-1 using standard techniques. Conditions were established for partial MboI digestion to result in approximately 40-kb DNA. This DNA was then ligated to the SuperCos cosmid vector (Stratagene) and packaged with GigaPak Gold (Stratagene). The library was plated at a density of approximately 50 000 clones per plate, replicated and screened as previously described.13 Cosmids were purified with multiple rounds of screening until they were homogenous.
Northern blot hybridization
Total RNA was extracted using TRIZOL (Gibco-BRL). Polyadenylated RNA was purified using the Fast Track RNA kit (Promega, Madison, WI). RNA was resolved in a 1% GTG agarose gel (FMC) containing 0.22 mol/L formaldehyde and 1 × MOPS. Gels were resolved at 9 V/cm and transferred to nylon membranes as described above for Southern blots. Blots were hybridized as for pulsed-field gel electrophoresis (PFGE).
Western blot hybridization
Cell lysates were prepared by adding lysis buffer (20 mmol/L Tris-HCl, pH 7.4, 20 mmol/L EGTA, pH 8.0, 80 mmol/L β-glycerophosphate, 1 mmol/L sodium vanadate, 50 mmol/L sodium fluoride, 15 mmol/L MgCl2, 1 mmol/L adenosine triphosphate, 25 μg/mL leupeptin, 25 μg/mL soybean trypsin inhibitor, and 1 mmol/L phenylmethylsulfonyl fluoride) to the cell pellets, followed by 8- to 10-second sonication (Sonic Disruptor; Tekmar, Cincinnati, OH). Cell debris was removed by centrifugation at 10 000 rpm for 3 minutes. All procedures were performed at 4°C or on ice. Protein concentration was determined spectrophotometrically using the Protein Assay Reagent (Bio-Rad). Protein extracts were denatured by boiling in sodium dodecyl sulphate (SDS) sample buffer (1.7% SDS, 5% 2-ME). Proteins were resolved in standard SDS–polyacrylamide gel electrophoresis.14
The protein was transferred by electroblotting to nitrocellulose membranes (Schleicher & Schuell, Keene, NH) as previously described.15 Nitrocellulose membranes were blocked with blocking buffer (5% low-fat powdered milk in 1 × PBS [140 mmol/L NaCl, 2.7 mmol/L KCl, 10 mmol/L Na2HPO4, 1.8 mmol/L KH2PO4, pH 7.3], 0.1% Tween-20) for 6 to 8 hours at 4°C and were washed with 1 × PBS with 0.1% Tween-20 (PBS-T). Protein was detected by overnight incubation with antibody in blocking buffer at 4°C. Blots were rinsed twice with PBS-T followed by washes of 1 × 15 minutes and 2 × 5 minutes in PBS-T. The second-step reagent, conjugated with horseradish peroxidase (HRP), was incubated at room temperature for 60 minutes. They were washed as above. Detection was accomplished with the ECL kit (Amersham) and exposure to film.
MUC1 was detected using the HMFG1 and HMFG2 monoclonal antibodies, which recognized the repeat region of MUC1, at a dilution of 1:600. The specificity of HMFG2 is for the repeat region of MUC1, and HMFG1 is dependent on proper glycosylation. The second step was a goat antimouse IgG conjugated with HRP (Promega) at a dilution of 1:5000.
DNA sequencing
DNA sequencing was performed on an ABI automated sequencer (ABI373; Applied Biosystems, Foster City, CA) and analyzed using Factura and Autoassembler software. BLAST searches were performed on the web-based application.16 Further DNA analysis was performed in MacVector (Oxford Molecular, Campbell, CA).
Results
Derivation of SKI-DLCL-1
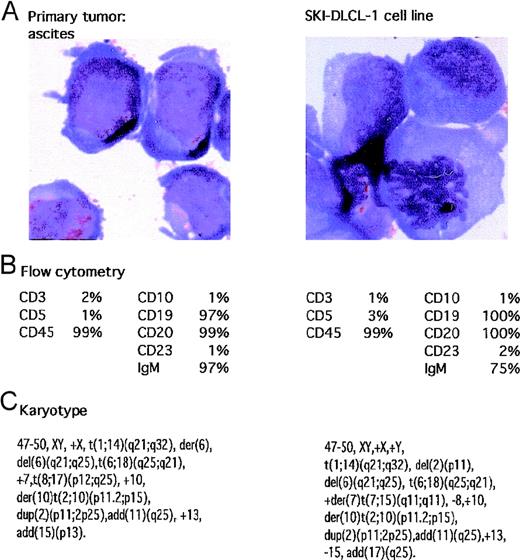
A patient had abdominal ascites. Cytologic analysis of the material suggested a large-cell lymphoma, which was confirmed by flow cytometry demonstrating a monoclonal IgMκ CD19+ and CD20+ population of cells. The cells were negative for CD5, CD10, and CD23. The fluid was placed in culture with RPMI 1640 supplemented with 20% FCS in a humidified incubator supplemented with 5% CO2. After a short period of quiescence, the cells demonstrated spontaneous growth, and the resultant cell line was named SKI-DLCL-1. The cells propagated in vitro demonstrated identical cytologic features to the original ascitic fluid. Of note, there were numerous cytoplasmic vacuoles (Figure 1). Cytogenetic analysis was performed on both the initial ascitic fluid and the cell line, which demonstrated identical complex cytogenetic abnormalities (Figure 1). It was also noteworthy that there was a reciprocal t(1;14)(q21;q32) chromosomal translocation. In addition, the cell line contained cytogenetically normal chromosomes 1 and 14.
Characteristics of malignant ascites and resultant SKI-DLCL-1 cell line.
(A) Cells were fixed and stained with May–Grünwald–Giemsa (magnification, ×100). SKI-DLCL-1 cells and cells from the original ascitic fluid demonstrate identical cytologic features consistent with a diagnosis of large-cell lymphoma. (B) Flow cytometric analysis of the ascites and the SKI-DLCL-1 with a panel of monoclonal antibodies. (C) Cytogenetic analysis of the diagnostic ascites and the resultant cell line demonstrates similar though nonidentical findings. This likely represents clonal selection by the process of establishing the cell line.
Characteristics of malignant ascites and resultant SKI-DLCL-1 cell line.
(A) Cells were fixed and stained with May–Grünwald–Giemsa (magnification, ×100). SKI-DLCL-1 cells and cells from the original ascitic fluid demonstrate identical cytologic features consistent with a diagnosis of large-cell lymphoma. (B) Flow cytometric analysis of the ascites and the SKI-DLCL-1 with a panel of monoclonal antibodies. (C) Cytogenetic analysis of the diagnostic ascites and the resultant cell line demonstrates similar though nonidentical findings. This likely represents clonal selection by the process of establishing the cell line.
Cloning of the t(1;14) chromosomal breakpoint
To identify the position of the chromosomal breakpoint, megabase DNA was digested with the restriction endonuclease NotI. This enzyme was selected because the entire immunoglobulin heavy chain locus (IGH) is subtended on a 750-kb NotI fragment. IGH gene rearrangements and chromosome breaks are identified as nongermlineNotI fragments.12 DNA from polyclonal peripheral blood lymphocytes (PBL) and the SKI-DLCL-1 cell line were sequentially hybridized with probes derived from the IGH locus. A probe from the joining region (JH) identified a germline 750-kb fragment in PBL, which was absent from this cell line. A single 600-kb band was detected with the JH probe (Figure2A). This was surprising because we expected a productively rearranged allele (the cell expresses a surface IgM) and a second allele involved in either a nonproductive rearrangement or in a germline configuration. This suggested that 1 IGH allele was at least partially deleted. The identical pattern was seen with a probe for the mu constant region (Cμ) (Figure 2B). Hybridization with a probe for the gamma constant region (Cγ) and alpha constant region (Cα) identified 3 fragments in the cell line that measured 140 kb, 600 kb, and 800 kb (Figures 2C, 2D). The 600-kb band comigrated with the band identified by the JH and Cμ probes. Therefore, this must have represented the productive allele from which the IgM transcript was expressed in this cell line. The identification of 2 additional bands with the Cα probe suggests that the deletion of the nonproductive allele does not extend through the entire IGH locus. Furthermore, the chromosome breakpoint likely occurred between the Cα1 and Cα2 genes (Figure 2D). The additional finding of an identical pattern with the Cγ probe suggested that the break would be upstream of Cγ4.
Characterization of the IGH rearrangements in SKI-DLCL-1 by PFGE.
High molecular weight DNA from peripheral blood lymphocytes (lane 1) and SKI-DLCL-1 cells (lane 2) was digested with NotI and resolved through PFGE with a window of resolution of 50 kb to 1500 kb (A-E) or 25 to 500 kb (F, G). The identical blot was sequentially hybridized with a series of probes, as indicated in A to E. A second blot was used (F, G) for hybridization by the indicated probes. The samples are as follows: lane 1, PBL; lane 2, ascitic fluid; lane 3, SKI-DLCL-1. Complete removal of the different probes after stripping was confirmed by exposure to film. ZNR, zone of no resolution.
Characterization of the IGH rearrangements in SKI-DLCL-1 by PFGE.
High molecular weight DNA from peripheral blood lymphocytes (lane 1) and SKI-DLCL-1 cells (lane 2) was digested with NotI and resolved through PFGE with a window of resolution of 50 kb to 1500 kb (A-E) or 25 to 500 kb (F, G). The identical blot was sequentially hybridized with a series of probes, as indicated in A to E. A second blot was used (F, G) for hybridization by the indicated probes. The samples are as follows: lane 1, PBL; lane 2, ascitic fluid; lane 3, SKI-DLCL-1. Complete removal of the different probes after stripping was confirmed by exposure to film. ZNR, zone of no resolution.
We prepared a cosmid library derived from an MboI partial digestion. The library was screened for Cγ-positive clones. We expected to obtain clones derived from all the Cγ loci, both on the normal and on the translocated chromosomes. Thirteen clones were obtained that formed 3 restriction patterns. Two of them, S7D and S5F, represented overlapping cosmids on the basis of shared restriction fragments. This mini-contig hybridized with probes for Cα, Sγ, and Cγ (Figure 3); therefore, these cosmids were candidates to span the break between Cγ2 and Cγ4 derived from der(14). Two additional clones, S5A and S7A, represented a second mini-contig based on the restriction pattern and hybridized to both the Cγ and the Sγ probes. The lack of Cα hybridization and the difference in restriction map from the S7D/S5F contig made them candidates to be derived from the reciprocal der(1) chromosome.
Megabase map of the t(1;14)(q21;q32) and the location of the cosmid and P1 clones.
Clones S7D (Cγ+, Cα−) and S5F (Cγ+, Cα+) span the breakpoint on der(14). The restriction maps overlap, creating a mini-contig. Clones S5A and S7A (Cγ+) also comprise a mini-contig, and they span the breakpoint on the der(1) chromosome. The sizes of the translocated bands detected by PFGE are 140 kb for the derivative chromosome 14 and 600 kb for the derivative chromosome 1. PI-PAC clones 35(A12) and 35(A12) span a 100-kb region centered around the MUC1 gene.
Megabase map of the t(1;14)(q21;q32) and the location of the cosmid and P1 clones.
Clones S7D (Cγ+, Cα−) and S5F (Cγ+, Cα+) span the breakpoint on der(14). The restriction maps overlap, creating a mini-contig. Clones S5A and S7A (Cγ+) also comprise a mini-contig, and they span the breakpoint on the der(1) chromosome. The sizes of the translocated bands detected by PFGE are 140 kb for the derivative chromosome 14 and 600 kb for the derivative chromosome 1. PI-PAC clones 35(A12) and 35(A12) span a 100-kb region centered around the MUC1 gene.
Clone S7D, a candidate to span the breakpoint on the der(14) chromosome, was screened by FISH on normal cells. Clone S7D identified signals at 14q32 and 1q21 (Figure 4A), confirming that it must span the breakpoint on chromosome 1. The cosmid was then subcloned as a BamHI sublibrary. Clones representative of each of the BamHI fragments were selected from the libraries and were screened for homology to a switch gamma (Sγ) probe. Clone B109 hybridized with Sγ and therefore likely carried the translocation breakpoint. Clones B101, B22, and B61 failed to show hybridization with Cγ and were presumed to be derived from chromosome 1 (Figure 5).
Characterization of genomic clones by FISH.
(A) FISH analysis of normal cells using cosmid clone S7D, a candidate to span the t(1;14) breakpoint. Biotinylated probes for the chromosome 14 centromere-specific probe and the heterochromatic region of chromosome 1 were cohybridized with digoxigenin dUTP-labeled clone S7D. The centromeric and heterochromatic regions were identified with phycoerythrin-SA (red) and the breakpoint probe with FITC-antidigoxigenin (green). The result confirmed that clone S7D split the breakpoint. (B) FISH analysis of SKI-DLCL-1 cells using P1 PAC clone 35(A12) derived from the normal chromosome 1. The P1 PAC clone 35(A12) probe was labeled with digoxigenin dUTP and detected by FITC-antidigoxigenin. The finding of signals on the normal chromosome 1 and on the der(1) and der(14) chromosomes confirms that P1 spans the region on chromosome 1 involved with the breakpoint.
Characterization of genomic clones by FISH.
(A) FISH analysis of normal cells using cosmid clone S7D, a candidate to span the t(1;14) breakpoint. Biotinylated probes for the chromosome 14 centromere-specific probe and the heterochromatic region of chromosome 1 were cohybridized with digoxigenin dUTP-labeled clone S7D. The centromeric and heterochromatic regions were identified with phycoerythrin-SA (red) and the breakpoint probe with FITC-antidigoxigenin (green). The result confirmed that clone S7D split the breakpoint. (B) FISH analysis of SKI-DLCL-1 cells using P1 PAC clone 35(A12) derived from the normal chromosome 1. The P1 PAC clone 35(A12) probe was labeled with digoxigenin dUTP and detected by FITC-antidigoxigenin. The finding of signals on the normal chromosome 1 and on the der(1) and der(14) chromosomes confirms that P1 spans the region on chromosome 1 involved with the breakpoint.
Restriction map of cosmid S7D.
Cosmid S7D was subcloned as a BamHI sublibrary. Sequence of the ends of clone B61 was found to be colinear with YAC MNG1 and was homologous to the thrombospondin 3 gene. MUC1 gene extends from clone B22 to clone B109, which spans the breakpoint. Two boxes indicate the position of the fragments used as probes (RsaI 3.5-kb andBstxI 1.1-kb fragment). Restriction sites are indicated: N,NotI; B, BamHI; E, EcoRI; M,MluI; Bss, BssHII.
Restriction map of cosmid S7D.
Cosmid S7D was subcloned as a BamHI sublibrary. Sequence of the ends of clone B61 was found to be colinear with YAC MNG1 and was homologous to the thrombospondin 3 gene. MUC1 gene extends from clone B22 to clone B109, which spans the breakpoint. Two boxes indicate the position of the fragments used as probes (RsaI 3.5-kb andBstxI 1.1-kb fragment). Restriction sites are indicated: N,NotI; B, BamHI; E, EcoRI; M,MluI; Bss, BssHII.
Localization of chromosome 1–derived sequences
The BamHI fragments subcloned from cosmid S7D were sequenced from the ends, using primers homologous to the flanking T3 and T7 promoters. These sequences were screened by BLAST searching.16 Clone B61 was found to be colinear with a previously sequenced region from YAC MNG1.17 The sequence of the YAC in this region spans a 75,000-bp region extending from the CLK2 kinase at the centromeric end, and it spans a cluster of genes near the Gaucher disease locus ending in the thrombospondin 3 gene. B61 was colinear with a 6-kb BamH1 fragment derived from a region corresponding to thrombospondin 3. Thus, we were able to establish that the cloned material was in fact derived from chromosome 1, and we had additional information regarding the physical map in this region. The T7 end of clone B22 corresponded to the 3′ end of the thrombospondin cDNA, consistent with this fragment's downstream location from the YAC MNG1. The T3 end of this clone was homologous to the MUC1 gene, which is known to lie immediately downstream of thrombospondin 3.18 Clone B101 proved to be colinear with the MUC1 gene. The T3 end of clone B109 was colinear with exon 7 of MUC1 and a portion of the 3′ untranslated region of the MUC1 gene. The T7 terminus was homologous to Cγ. Thus, the chromosomal breakpoint occurred within the B109 clone, as expected. A 1.1-kbBstXI fragment derived from the T3 end of B109 was used as a probe on a Southern blot of DNA extracted from SKI-DLCL-1 and PBL DNA from a healthy subject and was digested with BamHI and EcoRI. As seen in Figure 6, the probe identified a 9-kb BamHI fragment in the tumor that corresponded to B109. In addition, a second 5.1-kb BamHI fragment was identified in normal DNA and in the tumor. This corresponded to the nontranslocated chromosome 1 allele. In theEcoRI digestion, the germline fragment measured 5.6 kb, and the translocated fragment measured 15 kb.
Southern blot analysis confirming that B109 spanned the t(1;14) breakpoint.
DNA from PBL (lane 1) and SKI-DLCL-1 (lane 2) was digested withBamHI or EcoRI, and the blot was successively hybridized with a Cγ probe and the 1.1-kb BstXI fragment from clone B109. The BstXI probe identified a nongermline-rearranged band for each enzyme (9-kb band forBamHI and 15-kb band for EcoRI). The nongermline bands comigrated with nongermline bands detected with a Cγ probe.
Southern blot analysis confirming that B109 spanned the t(1;14) breakpoint.
DNA from PBL (lane 1) and SKI-DLCL-1 (lane 2) was digested withBamHI or EcoRI, and the blot was successively hybridized with a Cγ probe and the 1.1-kb BstXI fragment from clone B109. The BstXI probe identified a nongermline-rearranged band for each enzyme (9-kb band forBamHI and 15-kb band for EcoRI). The nongermline bands comigrated with nongermline bands detected with a Cγ probe.
To identify precisely the chromosomal breakpoint, the B109 fragment was sequenced. To obtain the sequence of the normal chromosome 1 allele, a P1 artificial chromosome (PAC) library was screened with a 3.5-kbRsaI fragment derived from clone B101, identifying 2 clones—PAC 35(A12) and PAC 280(09) (Figure 3). PAC 35(A12) was used as a FISH probe on the SKI-DLCL-1 cell line (Figure 4B). In addition to the normal chromosome 1, the der(1) and the der(14) chromosomes were identified, confirming that this P1 spans the chromosomal breakpoint. These PAC clones overlap, and each has a 5.1-kb BamHI fragment that hybridizes to the 1.1-kb BstXI probe derived from B109 (data not shown). The 5.1-kb BamHI fragment was subcloned as clone B5 and sequenced. Because clone B5 spanned the breakpoint on chromosome 1, it would be expected to identify clones derived from both the der(1) and the der(14) chromosomes. It was used as a probe to screen the Cγ-positive cosmids and hybridized to clones S5A and S7A, which confirmed that they were derived from the der(1) chromosome as expected. The breakpoint was contained within a 5-kb BamHI fragment. This region, adjacent to the breakpoint, was sequenced. The sequences of the der(1), der(14), normal chromosome 1, and switch gamma 4 sequences were aligned (Figure 7). The break occurred 0.86 kb downstream of the final MUC1 exon on chromosome 1 and within the Sγ4 region on chromosome 14. The normal chromosome 1, der(1), and der(14) all share the sequence CATT at the breakpoint. The der(1) allele has a 7-bp deletion of chromosome 14 sequences just 5′ of the CATT, and the der(14) has a 5-bp deletion just 3′ of the CATT. This is consistent with a staggered double-strand break with 3′ to 5′ exonuclease removal of some bases from the free ends. The differences between the Sγ4 sequence and the der(1) and der(14) sequences likely are allelic variations between the cell line and the switch region sequence reported in GenBank.
Sequence of the der(1) and der(14) breakpoints aligned with Sγ4 and the normal sequence of chromosome 1.
Clone B109 was derived from cosmid S7D (Figure 3) and corresponds to the der(14) chromosome. Clone B10 is derived from cosmid S5A (Figure 3) and corresponds to the der(1) chromosome. P48-B5 is derived from the normal chromosome 1 P1. The switch gamma 4 sequence was obtained from GenBank. It is uncertain whether the minor differences between the switch gamma 4 sequence and the B10 sequence represent polymorphism, sequencing errors, or differences arising during the translocation.
Sequence of the der(1) and der(14) breakpoints aligned with Sγ4 and the normal sequence of chromosome 1.
Clone B109 was derived from cosmid S7D (Figure 3) and corresponds to the der(14) chromosome. Clone B10 is derived from cosmid S5A (Figure 3) and corresponds to the der(1) chromosome. P48-B5 is derived from the normal chromosome 1 P1. The switch gamma 4 sequence was obtained from GenBank. It is uncertain whether the minor differences between the switch gamma 4 sequence and the B10 sequence represent polymorphism, sequencing errors, or differences arising during the translocation.
The productive VDJ allele is translocated to chromosome 1
The PFGE mapping of the IGH locus in SKI-DLCL-1 (see above) demonstrated that 1 of the JH loci was deleted and that the productive allele was subtended on a 600-kb NotI fragment. A fragment derived from clone B5, corresponding to the der(1) chromosome, was used as a hybridization probe on the blot of PBL and SKI-DLCL-1 DNA digested with NotI (Figure 2E). A 140-kb doublet band was seen in the cell line, and a single 140-kb band was seen in PBL (the doublet is best seen in the alternative window of resolution shown in Figure2F). The band seen in both the PBL and the cell line must correspond to the germline fragment. The second band at 140 kb in the cell line comigrated with the band detected with the Cα and Cγ probes (Figures 2C, 2D, and 2E) thus represented a rearranged fragment. There was an additional 600-kb band present only in the cell line. The fragment comigrated with the band detected with the JH and Cμ probes (Figures 2A and 2B). Because the cell line expresses an sIgM and there is only 1 JH allele, this 600-kbNotI fragment represents the productive allele. Thus, in this cell line, the productive IGH locus is on the der(1) chromosome and the normal 14 must have a deletion involving a portion of the IGH locus.
MUC1 is highly expressed in this cell line
Given that MUC1 was adjacent to the chromosomal breakpoint, a 3.5-kbRsaI fragment was derived from clone B101 and used to evaluate the steady-state level of transcription of this gene. MUC1 mRNA was highly expressed in the SKI-DLCL-1 cell line, though it was not expressed in a number of other lymphoma cell lines (Figure8A). Two transcripts were detected: a 4.4-kb transcript was strongly expressed, and a 6-kb transcript was weakly expressed. It is likely that these represented alternative splice variants.19 Expression of protein was demonstrated by Western blot hybridization (Figure 8B). The HMGF2 antibody recognizes the repeat region of MUC1. The level of expression is significantly greater than in the breast carcinoma cell line MCF7. In addition, it was expressed in the original ascites fluid and in the SKI-DLCL-1 cell line (lanes 3 and 4). An identical pattern was observed with the HMGF1 antibody that recognizes the glycosylated form of MUC1. Cell surface expression was confirmed by immunofluorescence using an MUC1-specific monoclonal antibody that demonstrated a strong expression of MUC1 (Figure 8C).
Expression of the MUC1 gene in SKI-DLCL-1.
(A) Expression of mRNA was evaluated by Northern blot hybridization. One microgram of poly-A+ RNA was electrophoresed on a 1% formaldehyde agarose gel. The blot was hybridized successively with an MUC1 probe and a β-actin probe. Length of exposure on film is 1 hour for both probes. Lanes 1 to 6 represent different cell lines: lane 1, SKI-DLCL-1; lane 2, OCI-Ly8; lane 3, Daudi; lane 4, FL-318; lane 5, Raji; lane 6, Molt-4. (B) Protein expression was evaluated by Western blot hybridization using the MUC1-specific monoclonal antibodies HMFG1 (glycosylation sensitive) and HMFG2 (specific for the repeat motif). Lane 1, Raji; lane 2, MCF-7; lane 3, primary ascites; lane 4, SKI-DLCL-1. (C) Immunofluorescence was used to assess cell-surface expression. Staining of SKI-DLCL-1 cells and MCF-7 cells with HMFG2 antibody demonstrated a strong expression of MUC1. OCI-LY8 cells were used as a negative control in this experiment.
Expression of the MUC1 gene in SKI-DLCL-1.
(A) Expression of mRNA was evaluated by Northern blot hybridization. One microgram of poly-A+ RNA was electrophoresed on a 1% formaldehyde agarose gel. The blot was hybridized successively with an MUC1 probe and a β-actin probe. Length of exposure on film is 1 hour for both probes. Lanes 1 to 6 represent different cell lines: lane 1, SKI-DLCL-1; lane 2, OCI-Ly8; lane 3, Daudi; lane 4, FL-318; lane 5, Raji; lane 6, Molt-4. (B) Protein expression was evaluated by Western blot hybridization using the MUC1-specific monoclonal antibodies HMFG1 (glycosylation sensitive) and HMFG2 (specific for the repeat motif). Lane 1, Raji; lane 2, MCF-7; lane 3, primary ascites; lane 4, SKI-DLCL-1. (C) Immunofluorescence was used to assess cell-surface expression. Staining of SKI-DLCL-1 cells and MCF-7 cells with HMFG2 antibody demonstrated a strong expression of MUC1. OCI-LY8 cells were used as a negative control in this experiment.
MUC1 alone is overexpressed among a cluster of genes within 90 kb of the breakpoint
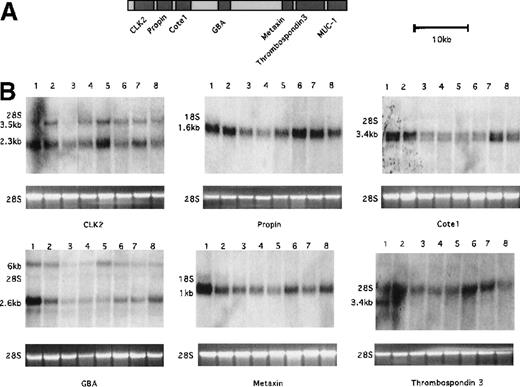
The MUC1 gene lies in a region of chromosome 1q21 that has an unusually high density of genes18 (Figure9A). Approximately 80 kb upstream of MUC1 is the CLK2 gene. Between CLK2 and MUC1 there are 5 additional genes—propin, COTE1, GBA, metaxin, and thrombospondin 3, which ends less than 1 kb upstream of MUC1.17 To determine whether the translocation resulted in the dysregulation of any other loci, we examined the expression of the genes upstream of MUC1 in the SKI-DLCL-1 cell line and compared this with a number of other cell lines. The results are shown in Figure 9B. Expression of the upstream genes is not significantly augmented in the SKI-DLCL-1 cell line in comparison with the other cell lines. Thus, the translocation resulted in the dysregulated expression only of the MUC1 gene. The basis for this narrow effect is under continued investigation.
Expression of loci within 90 kb of the t(1;14) translocation breakpoint.
(A) Physical map of the cluster of genes identified near the Gaucher disease locus based on sequence data (map based on previously published data in Adolph et al18). (B) Expression of the cluster of genes upstream of the breakpoint by Northern blot analysis included the following loci: CLK2, propin, cote1, GBA, metaxin, and thrombospondin 3. Total RNA (10 μg) was electrophoresed on a 1% formaldehyde agarose gel. 28S was used as a control for RNA loading. The size of the expected transcripts is indicated. Lanes 1 to 8 represent different cell lines: lane 1, Hela; lane 2, UC726/G; lane 3, SKI-DLCL-1; lane 4, Daudi; lane 5, OCI-LY8; lane 6, FL318; lane 7, Raji; lane 8, Molt-4.
Expression of loci within 90 kb of the t(1;14) translocation breakpoint.
(A) Physical map of the cluster of genes identified near the Gaucher disease locus based on sequence data (map based on previously published data in Adolph et al18). (B) Expression of the cluster of genes upstream of the breakpoint by Northern blot analysis included the following loci: CLK2, propin, cote1, GBA, metaxin, and thrombospondin 3. Total RNA (10 μg) was electrophoresed on a 1% formaldehyde agarose gel. 28S was used as a control for RNA loading. The size of the expected transcripts is indicated. Lanes 1 to 8 represent different cell lines: lane 1, Hela; lane 2, UC726/G; lane 3, SKI-DLCL-1; lane 4, Daudi; lane 5, OCI-LY8; lane 6, FL318; lane 7, Raji; lane 8, Molt-4.
Discussion
We described the cloning and characterization of the chromosomal breakpoint of a reciprocal translocation involving chromosomes 1q21 and 14q32. The breakpoint is not crossed by probes for either the MUM2 or the BCL9 gene (data not shown).3 5 The MUC1 gene, immediately adjacent to the breakpoint, is markedly dysregulated. Expression of 6 other genes in proximity to the breakpoint are not dysregulated as a consequence of the translocation.
The overexpression of mucins by adenocarcinoma tumors have been associated with high metastatic potential and poor prognosis.20-22 Expression of MUC1 is associated with immunosuppression because it inhibits T-cell proliferation.23 MUC1 expression has been described in multiple myeloma (MM).24,25 Of note, 1q21 abnormalities are seen in multiple myeloma, and some may occur near the breakpoint described in this article.26 This is a subject for further investigation.
The consequence of the expression of the MUC1 gene in this case is uncertain. However, it is notable that the patient had lymphoma in the form of abdominal ascites. It is possible that the expression of mucin in this case contributed to the extranodal pattern of growth of this lymphoma. MUC1 is an antiadhesive protein that interrupts cell–cell and cell–matrix interactions.27,28 There is a report of expression of EMA (which is equivalent to MUC1) in body cavity lymphoma infected by human herpesvirus 8 (HHV8)29 and of anaplastic large-cell lymphoma that frequently is accompanied by extranodal, particularly cutaneous, disease.30-32 EMA is also expressed in lymphocyte-predominant Hodgkin disease (75%), plasmacytoma (75%), and T-cell lymphoma (50%).33 The tumor cells in this case were tested and were found to be HHV8 negative (data not shown). It is possible that a consequence of HHV8 expression is the dysregulation of the MUC1 gene. This is under investigation.
The level of expression of MUC1 in the original tumor material and in the derived cell line is remarkable and exceeds that of the MCF7 cell line. As a consequence of the translocation, the MUC1 gene is brought into proximity with the Cγ4 and Cα2 loci. Downstream of Cα2 is an enhancer element homologous to the well-characterized murine and rat 3′IgH-E(hs1,2).34 In the mouse, this enhancer is active late in B-cell differentiation,35 and, though the activity of the human homolog has not been fully defined, it is active in mature B cell lines in transient transfection assays.34It is likely that the overexpression of the gene is driven by the juxtaposition of the Eα enhancer to the MUC1 gene. The human 3′IgH enhancer has an octamer binding motif and an ETS/AP-1 motif. Regulatory elements have been identified within the MUC1 promoter, and the regulation is complex. It remains to be determined whether and how the Eα enhancer alters the expression from the MUC1 promoter.
Interestingly, the translocation appears to result in the dysregulation of only MUC1 in a cluster of 7 genes within 80 to 90 kb of the translocation breakpoint. In other translocations involving the IGH or IGL loci, the gene dysregulated can be remote from the breakpoint. In the t(14;18) translocation characteristic of follicular lymphoma,12 the breaks occur in 2 major cluster regions on chromosome 18. The MBR is within the 3′ untranslated region of exon 3 of BCL2, and the MCR is approximately 30 kb downstream of BCL2. The BCL2 gene has an unusually large second intron that measures approximately 270 kb in length; thus the breakpoint is 270 kb to 300 kb away from the BCL2 promoter region.36 Because the translocation alters the rate of transcription of the BCL2 gene and does not affect the message half-life,36 it is clear that the translocation affects transcription at a significant distance. Similarly, the dysregulation of cyclin D1, which is a consequence of the t(1;14) translocation, can result from chromosome breaks as far as 150 kb downstream of the gene.37,38 In some patients with Burkitt lymphoma with the variant t(2;8) chromosomal translocation, the c-myc locus is up to 280 kb away from the breakpoint but is nonetheless dysregulated.39-41 In the present case, the consequence of the translocation is the dysregulation only of the gene immediately 5′ to the breakpoint. The possibility remains that this t(1;14)(q21;q32) translocation results in the dysregulation of other loci more than 90 kb upstream of MUC1 or of loci on the der(1) allele, which translocates the Eμ enhancer to chromosome 1.
Acknowledgments
We thank Dr Philip Livingston for the gift of the HMFG1 and HMFG2 monoclonal antibodies, Dr Lloyd Old and Marc Fediricci for performing the MUC1 immunofluorescence, and Dr Stephen Nimer for helpful discussions. We also thank Letha Menon for secretarial assistance in the preparation of this manuscript.
Supported in part by the Lymphoma Research Fund and by National Institutes of Health grant R01-CA-61429 (A.D.Z.).
F.G. and A.G. contributed equally to this study.
Reprints:Andrew D. Zelenetz, Memorial Sloan–Kettering Cancer Center, 1275 York Avenue, Box 330, New York, NY 10021; e-mail:a-zelenetz@ski.mskcc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.