Abstract
In the Rh blood system, RhAG (Rh-associated glycoprotein, or Rh50) is thought to be involved in Rh30 (D, CE) expression by forming a protein complex on the red cell surface. To obtain further insight into the Rh complex, we chose nonerythroid COS-1 cells instead of proerythroblast-like K562 cells, which produce endogenous Rh proteins as cell host, for the expression of both RhAG and RhD. The RhAG cDNA was subcloned into a retroviral vector, and a stable COS-1 cell line was then established via retroviral transduction. Surface expression of RhAG on the COS-1 cells was monitored by flow cytometry using mouse monoclonal anti-RhAG(2D10). Under these conditions, we detected significant expression of RhAG on the cell surface, compared to stable COS-1 cells transduced with the vector alone. To confirm the results, we isolated RhAG by immunoprecipitation from the lysate of the COS-1 cells, which were metabolically labeled with [35S]-methionine. A strong band of the 32 kd on SDS-PAGE was obtained, corresponding to the results obtained from other cultured cells (K562 cell and others), which always produce partially glycosylated RhAG with a molecular weight of 32 kd. Thus, RhAG was expressed without Rh30 and other Rh-related glycoproteins (LW, glycophorin B) in nonerythroid cells. Using the same strategy, however, we could not express RhD epitopes on COS-1 cells even in the presence of RhAG cDNA, suggesting that other factors might be required for the surface expression of RhD antigen. (Blood. 2000;95:336-341)
The elucidation of the RhD antigenicity is of clinical importance because of its involvement in transfusion medicine and hemolytic disease of the newborn.1-3 Molecular analysis of partial Ds in DIV, DV, DVI, Dc- and DFR cells as well as the commonly occurring antigens (C/c, E/e, and D) and the identification of these antigen-carrying membrane proteins have been reported.4-11 Furthermore, a 30-epitope model was recently established for D antigen using a number of partial D phenotypes and human monoclonal anti-D antisera.12However, the nature of RhD antigenicity is still unknown.
Expression of recombinant RhD cDNA in eukaryotic cells would greatly contribute to our understanding of RhD antigenicity and biological function of this protein if it exists. Although RhD cDNA and RhAG cDNA can be transiently expressed in COS-1 cell lysates, surface expression of these proteins was not successful.13-15 Iwamoto et al tried expression of RhD or RhcE antigen on both nonerythroid cell, 293, and erythroid cell, human erythroleukemia (HEL) together with RhAG. However, they could not detect RhD or RhcE antigen on them and suggested that a second coexpressing factor will be needed to express Rh antigens.16 Because Rh antigenicity is believed to be erythroid cell–specific and -conformational, the correct environment may be important for the proper folding and expression of the Rh epitopes. Smythe et al reported the surface expression of RhD, Rhc, and RhE antigens in K562 cells transfected with RhD or RhcE cDNA, respectively.17 Also, Liu et al demonstrated that D epitopes were expressed by retroviral vectors containing mutant RhD cDNA.18 Recently, Zhu et al showed clearly endogenous expression of D antigen on K562 cells with human monoclonal anti-D and reported the expression of D antigen epitopes in K562 cells using RhD fusion proteins.19 Because K562 cells resemble proerythroblasts and express D antigen on their cell surfaces endogenously, it is likely that they produce the correct environment for the expression.20
The presence of RHAG-like sequences in nematode and the sponge is reported.3,21 The human RhAG protein is known to share a certain degree of homology (20% to 27% identical) with the Mep/Amp family of NH4+ transporter, permeases that have been identified from bacteria to yeast and plant (but not animals).22 In fact, Caenorhabditis elegans is found to possess at least 4 ammonium transporter genes and 2RHAG-like sequences. Interestingly, Cherif-Zahar et al proposed that mutant alleles of RHAG are likely candidates for suppressors of the RH locus, accounting for most cases of Rh deficiency (Rhnull, regulator type).23 It has been suggested that the deficiency of Rh30 proteins on red cells from individuals in Rhnull amorph type results in homozygosity for a silent allele at the RH locus and is associated with the lack or reduced expression of Rh-related glycoproteins. Huang et al and Cherif-Zahar et al demonstrated the molecular defects in the RHCE gene from Rhnull amorph type.24,25 Therefore, at least these 2 proteins (Rh30 and RhAG) seem to be closely interacting with each other. Moore and Green also immunoprecipitated RhAG glycoprotein with human anti-D and anti-c antibodies.26Moreover, Hartel-Schenk et al coprecipitated Rh components as a 170-kd complex by density ultracentrifugation of [3H]-palmitate–labeled Rh30 proteins.27
Rh expression in nonerythroid cells like the COS-1 cell is useful for reconstitution of Rh complex because K562 cell possesses Rh30 and Rh-related glycoproteins (Rh50, LW, CD47, and glyco- phorin B) endogeneously. To shed light onto the physiologic role of RhAG and functional role of RhAG in Rh30 expression, we produced a stable COS-1 cell line with RhAG cDNA using a retroviral vector. We report in this paper surface expression of RhAG glycoprotein in stable COS-1 cells transfected with the retroviral vector containing RhAG cDNA.
Materials and methods
Materials
COS-1 cells were obtained from American Type Culture Collection; culture media and G418 were purchased from Life Technologies Inc; pLNCX vector, PT67 cells, and CalPhos Maximizer Transfection Kit were obtained from Clontech Laboratories Inc; [3H]-mannose was purchased from ICN and L-[35S]-methionines from Amersham for metabolical labeling grade and from Dupont NEM for translation grade. Transcription and translation-coupled reticulocyte lysate system and canine pancreatic microsomal membranes were obtained from Promega, restriction enzymes (Xba I, Hind III, Hpa I) from New England BioLabs, goat F(ab')2 anti-human immunoglobulin G (IgG) conjugated with phycoerythrin and goat F(ab')2anti-mouse IgG conjugated with phycoerythrin from Biosource International, and polybrene from Sigma. Purified mouse monoclonal anti-RhAG(2D10) was kindly given by Dr Albert von dem Borne (Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, Amsterdam, Netherlands) and human monoclonal anti-D (LOR15C9) by Dr A. Blancher (Hopital Purpan, Toulouse, France).
Methods
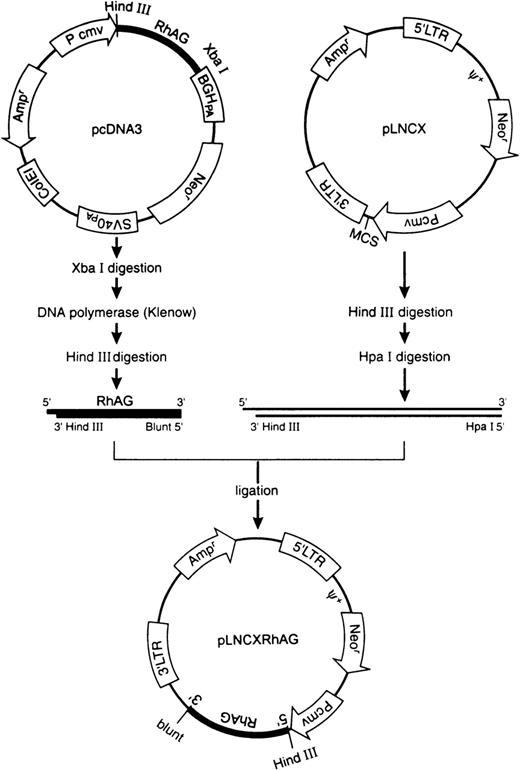
Construction of retroviral expression vectors.
The pcDNA3RhAG vector previously constructed was cut with Xba I restriction enzyme and filled in with Klenow DNA polymerase. Then, RhAG cDNA was released from the vector using Hind III restriction enzyme and purified. The retroviral expression vector pLNCX was digested with Hind III followed by Hpa I and purified. The RhAG cDNA released was ligated with the Hind III/Hpa I-cleaved pLNCX, and the ligates were transformed into DH5α cells (Figure 1). After confirming the orientation and the sequence by dideoxy- chain termination method, the plasma vector was referred to as pLNCXRhAG. The pcDNA3RhD vector was also cut with Bam H I/Xba I, and the released RhD cDNA was filled in with T4 DNA polymerase and purified. The pLNCX vector was cut with Hpa I, dephosphorylated with calf intenstine phosphatase, and then purified. This vector was ligated with the purified RhD cDNA, and the newly produced vector was referred to as pLNCXRhD. Packaging cell line (PT67 cell) was transfected with the constructed plasmid vector (10 μg/100 mm plate, pLNCX, or pLNCXRhAG, or pLNCXRhD) using CalPhos Maximizer Transfection Kit and cultured for 48 hours; the produced viruses then were collected from the culture medium of the transfected PT67 cells. The virus-containing medium was used for transduction of actively dividing COS-1 cells. The medium was filtered through 0.45-μm cellulose acetate filter and diluted to 10% and then added to COS-1 cells (5 × 105 cells/100 mm plate). The cells were incubated for 24 hours after addition of 4 μg/mL polybrene. They were then re-placed into fresh regular medium and cultured for 24 hours. By antibiotic selection with G418 (0.5 mg/mL), the stable COS-1 cells with RhAG or RhD cDNA were produced. Construction of pR-RhD was described previously.19
Expression of RhAG in vitro.
Expression of RhAG in vitro was performed with a transcription and translation-coupled reticulocyte lysate system (TNT-T7) following the method described previously and using pLNCXRhAG as expression vector.14 The reaction mixtures were incubated at 30°C for 90 minutes, diluted 1:10 in lysis buffer (1% Triton X-100 + 1 mM TPCK + 1 mM phenylmethylsulfonyl fluoride + 100 u/mL aprotinin in 20 mM tris, pH 7.4) and incubated with 2D10 overnight at 4°C. The immune complexes were eluted from protein A-Sepharose beads by boiling for 5 minutes in 1% sodium dodecyl sulfate (SDS) with 8 mol/L urea and 5% β-mercaptoethanol in 0.16 mol/L tris, pH 6.8, and run on SDS-PAGE (SDS-polyacrylamide gel electrophoresis) (12%).
Expression of RhAG and RhD polypeptides in stably transduced COS-1 cells.
Expression of RhAG and RhD proteins in stable COS-1 cells (5 × 106 cells/60 mm plate) transduced with the viruses containing pLNCX, or pLNCXRhAG, or pLNCXRhD was performed as follows. Logarithmically growing stable COS-1 cells were labeled with 200 μCi/mL [35S]-methionine in 1mL methionine-free medium for 2 hours at 37°C. As described above, the cells were washed 3 times with ice-cold phosphate buffered saline (PBS), lysed with lysis buffer, and centrifuged. The supernatants were immunoprecipitated with rabbit polyclonal anti-Rh or 2D10, and the immune complexes were eluted from protein A-Sepharose (anti-Rh) or protein G-Sepharose(2D10) and run on SDS-PAGE (12%) under the reducing conditions.
[3H]-mannose incorpration into RhAG glycoprotein in stable COS-1 cells transduced with the viruses containing pLNCXRhAG.
Logarithmically growing COS-1 cells (5 × 106 cells) were incubated with [3H]-mannose (135 μCi/mL) in glucose-free RPMI 1640 for 5 hours at 37°C. After washing 3 times with PBS, the cells were lysed as described above. Supernatant (500 μL) from the lysate was incubated with 5 μL of 1:10 diluted 2D10 overnight at 4°C. The immune complexes extracted by protein G-Sepharose were run on SDS-PAGE (12%) under the reducing conditions, followed by fluorography.
Fluorescence-activated cell sorting (FACS).
Stable COS-1 cells (5 × 105 cells) were incubated with 0.1 mL of 1:100 diluted 2D10 (1 mg/mL) for 30 minutes at 37°C and washed 3 times in PBS with 0.5% bovine serum albumin. The cells were reincubated with phycoerythrin-conjugated goat F(ab')2 anti-mouse IgG for 30 minutes at room temperature, washed 3 times, and resuspended in PBS. The cells were analyzed by FACS IV (Becton Dickinson, Sunnyville, CA). Data were presented as fluorescence intensity versus cell frequency.
Results
Expression of RhAG glycoprotein in vitro
Previously, we were able to detect RhAG polypeptide by immunoprecipitation in COS-1 cells transiently transfected with pSVLRhAG vector using purified mouse monoclonal anti-RhAG(2D10), but surface expression of the RhAG antigen on the cells was not successful when examined by flow cytometry.14 Now, as shown in Figure1, we constructed a retroviral vector (pLNCXRhAG) carrying the RhAG cDNA. After confirming the authenticity of the vector by checking the orientation and the DNA sequence, the vector was used for expression of the RhAG glycoprotein in a transcription and translation-coupled reticulocyte lysate system. [35S]-methionine-labeled RhAG glycoprotein was immunoprecipitated with 2D10. RhAG polypeptides immunoprecipitated from reticulocyte lysate without microsomal membranes resided in the 30-kd region of a 12% acrylamide SDS-PAGE, as shown in Figure 2. Addition of microsomal membranes to this reticulocyte system caused partial glycosylation of the protein, as we expected, producing 32-kd protein.
Expression of RhAG glycoprotein in a transcription and translation-coupled reticulocyte lysate system.
Autoradiogram of [35S]-methionine-labeled proteins separated by 12% SDS-PAGE (for details, see Materials and Methods). Lane 1, pLNCXRhAG vector; lane 2, pLNCXRhAG vector with microsomal membranes. Arrow indicates nonglycosylated RhAG; arrow head, glycosylated RhAG.
Expression of RhAG glycoprotein in a transcription and translation-coupled reticulocyte lysate system.
Autoradiogram of [35S]-methionine-labeled proteins separated by 12% SDS-PAGE (for details, see Materials and Methods). Lane 1, pLNCXRhAG vector; lane 2, pLNCXRhAG vector with microsomal membranes. Arrow indicates nonglycosylated RhAG; arrow head, glycosylated RhAG.
Expression of RhAG and RhD proteins in stable COS-1 cells
COS-1 cells were transduced with the viruses containing pLNCX, or pLNCXRhAG, or pLNCXRhD; the stable COS-1 cell lines were then established by antibiotic selection with G418. Expression of RhAG glycoprotein in stable COS-1 cells transduced with the viruses containing pLNCXRhAG was demonstrated by immunoprecipitating the [35S]-methionine labeled proteins in the supernatants of the cell lysates with 2D10, and the immunoprecipitates were analyzed by SDS-PAGE. The stable COS-1 cells exhibited a 32-kd band, which signifies expression of RhAG glycoprotein (Figure3A). This partially glycosylated protein was similar to those produced in a microsomal in vitro translation system (Figure 2) and in K562 cells endogenously, as reported previously.14 To confirm whether the 32-kd band is a glycoprotein, RhAG was immunoprecipitated from the lysate of the stable COS-1 cells metabolically labeled with [3H]-mannose using 2D10, and the immune complex was analyzed by SDS-PAGE (Figure4). Again, [3H]-mannose–labeled RhAG migrated to the 32-kd region of the gel as single band, confirming that the band at 32 kd is RhAG glycoprotein. Therefore, bands at a higher molecular weight region (>40 kd) seen in Figure 3A are not RhAG-related proteins. These results suggest that the stable COS-1 cells in tissue culture cannot glycosylate the RhAG to the same extent (40 to 100 kd in size) as is found in human red cells. Expression of RhD protein was also accomplished, as evident from the presence of the immunoprecipitated 32-kd proteins in the lysates of COS-1 cells transduced with pLNCXRhD using rabbit polyclonal anti-Rh, which specifically recognizes Rh30 proteins but not in the lysates of COS-1 cells transduced with the pLNCX vector alone (Figure 3B).
Autoradiogram of immunoprecipitates obtained from stable COS-1 cells transduced with the viruses containing RhAG or RhD RNA using 2D10 or rabbit polyclonal anti-Rh.
Logarithmically growing COS-1 cells were incubated with [35S]-methionine for 2 hours, and the cell lysates were subjected to immunoprecipitation with 2D10 or rabbit polyclonal anti-Rh; the immunoprecipitates were then separated by 12% SDS-PAGE under reducing conditions (for details, see Materials and Methods). (A) Lane 1, stable COS-1 cells transduced with the viruses containing vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhAG. Arrow shows RhAG glycoprotein. (B) Lane 1, stable COS-1 cells transduced with the viruses containing vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhD. Arrowhead shows RhD polypeptides.
Autoradiogram of immunoprecipitates obtained from stable COS-1 cells transduced with the viruses containing RhAG or RhD RNA using 2D10 or rabbit polyclonal anti-Rh.
Logarithmically growing COS-1 cells were incubated with [35S]-methionine for 2 hours, and the cell lysates were subjected to immunoprecipitation with 2D10 or rabbit polyclonal anti-Rh; the immunoprecipitates were then separated by 12% SDS-PAGE under reducing conditions (for details, see Materials and Methods). (A) Lane 1, stable COS-1 cells transduced with the viruses containing vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhAG. Arrow shows RhAG glycoprotein. (B) Lane 1, stable COS-1 cells transduced with the viruses containing vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhD. Arrowhead shows RhD polypeptides.
Autoradiogram of immunoprecipitates obtained from stable COS-1 cells transduced with the viruses containing pLNCXRhAG using 2D10.
Logarithmically growing COS-1 cells were incubated with [3H]-mannose for 5 hours, and the cell lysates were subjected to immunoprecipitation with 2D10; the immunoprecipitates were then separated by SDS-PAGE under reducing conditions (for details, see Materials and Methods). Lane 1, stable COS-1 cells transduced with the viruses containing pLNCX vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhAG. Arrow shows RhAG glycoprotein.
Autoradiogram of immunoprecipitates obtained from stable COS-1 cells transduced with the viruses containing pLNCXRhAG using 2D10.
Logarithmically growing COS-1 cells were incubated with [3H]-mannose for 5 hours, and the cell lysates were subjected to immunoprecipitation with 2D10; the immunoprecipitates were then separated by SDS-PAGE under reducing conditions (for details, see Materials and Methods). Lane 1, stable COS-1 cells transduced with the viruses containing pLNCX vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhAG. Arrow shows RhAG glycoprotein.
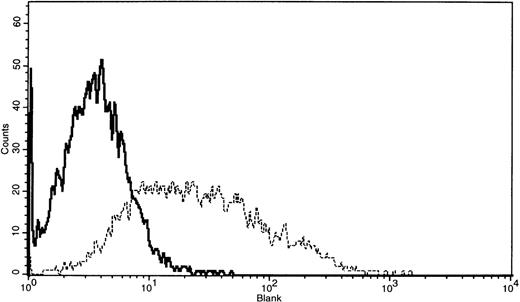
Surface expression of RhAG glycoprotein on the stable COS-1 cells transduced with the retroviruses containing pLNCXRhAG
Surface expression of RhAG glycoprotein on stable COS-1 cells transduced with the viruses containing pLNCXRhAG was determined by flow cytometry using 2D10. As shown in Figure 5, the cells sensitized with this antibody followed by incubation with phycoerythrin-conjugated mouse anti-IgG, exhibited a significant fluorescence intensity when compared with the control stable COS-1 cells transduced with the viruses containing pLNCX vector alone. Because G418-resistant COS-1 cells were not cloned, the broad range of fluorescence was observed. This is probably due to different transport efficiency of RhAG to plasma membrane by each cell. After one-time cell sorting, we achieved sharper and higher distribution of the fluorescence (data not shown). This indicates, for the first time, expression of RhAG glycoprotein on the membranes of the cultured nonerythroid cells without any Rh30 proteins and Rh-related glycoproteins. Surface expression of RhD protein on the stable COS-1 cells transduced with the viruses containing pLNCXRhD was also determined by flow cytometry using human monoclonal anti-D (LOR15C9), which shows broad D specificity and also recognizes denatured D polypeptide. However, surface expression of RhD protein on the cells was not recognized (data not shown). Other human monoclonal anti-D (LOR1, SAL-20, LOR-12E2) also did not bind to the cells (data not shown). Furthermore, a new stable COS-1 cell line obtained by transfecting the stable COS-1 cells carrying RhAG cDNA with RhD cDNA-containing plasmid vector (pR-RhD) was established. However, this cell line could not express any D epitopes, suggesting that the presence of both RhD protein and RhAG glycoprotein is not enough to produce D antigen on COS-1 cells and that other factors might be involved in expression of RhD antigen. However, as described in “Materials and Methods,” all the stable cell lines were established after 2-week growth in antibiotics rather than grown from individual colonies. Therefore, the level of protein expression is likely to vary among cells. In the case of the COS-1 cells coexpressing RhAG and RhD, it may be worthwhile—using cell sorting in flow cytometry analysis—to examine the possibility that only a very few transducted cells are expressing RhD antigens.
FACS profile of stable COS-1 cells transduced with the viruses containing pLNCXRhAG using 2D10.
COS-1 cells were incubated with 2D10 (10 μg/mL) containing 0.5% bovine serum albumin in PBS. After washing, the cells were stained with phycoerythrin-conjugated goat F(ab')2 anti-mouse IgG and then analyzed by flow cytometry (for details, see Materials and Methods). The solid line represents control cells (COS-1 cells transduced with the viruses containing vector alone), and the dotted line, COS-1 cells transduced with the viruses containing pLNCXRhAG.
FACS profile of stable COS-1 cells transduced with the viruses containing pLNCXRhAG using 2D10.
COS-1 cells were incubated with 2D10 (10 μg/mL) containing 0.5% bovine serum albumin in PBS. After washing, the cells were stained with phycoerythrin-conjugated goat F(ab')2 anti-mouse IgG and then analyzed by flow cytometry (for details, see Materials and Methods). The solid line represents control cells (COS-1 cells transduced with the viruses containing vector alone), and the dotted line, COS-1 cells transduced with the viruses containing pLNCXRhAG.
Discussion
Rh30 and RhAG proteins are thought to be the same protein family because of a significant amount of homology.28-32 Rh30 expression was successful using the retroviral gene transfer technology to generate stable K562 clones expressing the Rh blood group antigens (D, G, c, E).17 Recently, expression of the RhD antigen in K562 cells has been reported by a number of groups.17,19,33It is worth noting that wild-type K562 cells produce a low level of RhD antigens on the cell surface, and appropriate conditions are critical for the detection by flow cytometry analysis.19 The presence of endogenous Rh proteins in K562 cells supports the notion that K562 cells contain the proper cellular mechanism and necessary components for the surface expression of RhD.20 On the other hand, the use of K562 cells in the study of de novo synthesis and potential function of the Rh complex may thus be limited. Thus, we have been implored to express Rh30 and RhAG in COS-1 cells. This paper mainly describes expression of RhAG on COS-1 cells.
We constructed RhD- and RhAG-expressing vectors using a retroviral vector (pLNCX) as shown in Figure 1 and produced stable COS-1 cells with RhD cDNA or RhAG cDNA. RhAG was expressed on the cells when determined by flow cytometry (Figure 5). However, RhD was not detected on the cells, although RhD protein was immunoprecipitated by rabbit polyclonal anti-Rh or human monoclonal anti-D (LOR15C9) from the lysates of the stable COS-1 cells transfected with RhD cDNA (Figure 3). Therefore, RhAG was simply expressed, but RhD was not expressed on the cell surface regardless of the identical experimental conditions, such as the same expression vector (pLNCX) and the same host cells (COS-1 cells). This means that expression of RhAG does not require Rh30 and other Rh-related glycoproteins. However, in humans, CD47 has a very wide tissue distribution and is expressed as different splicing isoforms. Therefore, the presence of a protein homologous to CD47 in COS-1 cells (green monkey kidney cells) may not be neglected. These results support that the expression of RhAG glycoprotein in red cells does not require the coexpression of Rh30 polypeptides, because RhnullU+ erythrocytes have RhAG glycoprotein but not Rh30.34 Moreover, recent studies indicated that K562 cells carried about 60 × 103 to 90 × 103 copies of RhAG/cell.25Although the number of RhD protein expressed on K562 cells is not known but is probably extremely low, it is likely that most of the RhAG proteins on intact K562 cells are not complexed with the protein. The studies with transduced COS-1 cells confirm these observations. However, maximum expression of RhAG may require the presence of Rh30 and other Rh-related glycoproteins. We have shown previously that endogenous RhAG of K562 cells is underglycosylated.14 This is most likely explained by the very low level of Rh30 protein expressed at the cell surface of these cells, which correlates well with the studies of Ridgwell et al34showing underglycosylation of RhAG in RhnullU+ red cells lacking Rh30. The expression level of RhAG glycoprotein on some Rhnull cells, however, is always very low.23,25Therefore, the extent of RhAG glycosylation should depend on the transit time in the Golgi and would be modulated by the presence or absence of the Rh30 protein. However, transduction of the RhAG-expressing COS-1 cells with RhD vector did not alter glycosylation of RhAG (data not shown). Recently, it was also reported that Rhnull amorph–type individuals bearing normal RHAGgene and mutant RHCe gene have RhAG glycoprotein but not RhCe protein on red cells.24,25 On the other hand, expression of RhD protein seems to be dependent on the presence of RhAG glycoprotein, because Rhnull regulator–type individuals bearing normalRh30 gene and mutant RHAG gene do not carry both Rh30 and RhAG proteins on their red cells. However, surface expression of RhD was not recognized even in stable COS-1 cells carrying both RhD and RhAG cDNAs. COS-1 cells transduced with RhD vector grew normally, suggesting that Rh30 proteins are not toxic to the cells (data not shown). For the RhD expression, another Rh-related glycoprotein (glycophorin B, LW, CD47) may be required, or unknown factors including cytoskeletal protein(s) of red cells, which is thought to interact with Rh30 proteins, may be involved. However, it is clear that neither LW nor glycophorin B are necessary, because variant phenotypes (LW-negative, and S-s-) express normally the Rh antigens, notwithstanding that LW-negative and S-s- red cells are deficient in LW glycoprotein and glycophorin B, respectively. Band 3 might be one of the factors, because Beckmann et al reported that band 3 transduced in K562 cells enhances the cell surface reactivity of Rh antigens.35
Mallinson et al suggested that 2D10 epitope is chymotrypsin-sensitive and N-glycan–dependent, because the epitope is destroyed by endoglycosidase F.36 However, the partially glycosylated RhAG protein was immunoprecipitated by 2D10, indicating that 2D10 epitope may not be carbohydrate-dependent and may have been destroyed by protease contaminants in their glycosidase preparations. RhAG glycoproteins obtained from in vitro expression (Figure 2), intact K562 cells,14 stable COS-1 cells transduced with RhAG cDNA (Figures 3 and 4), and 2-phase liquid culture—which was recently established by us to produce erythroid cells from the progenitor cells of fresh peripheral blood37—are always immunoprecipitated as a 32-kd protein. Only red cells possess highly heterogeneous RhAG glycoproteins (40 to 100 kd in size) and the maximum copies of Rh30. Higher glycosylation of RhAG may be necessary for the maximum transport of both RhAG and Rh30 to plasma membranes. Stable COS-1 cells expressing RhAG glycoprotein were established and will be useful for studying physiologic functions such as ion transport (eg, NH4+, Ca++).
Acknowledgments
We are grateful to Dr Albert von dem Borne (Amsterdam, Netherlands) for the generous gift of mouse monoclonal anti-RhAG(2D10) and Dr Antoine Blancher for human monoclonal anti-D (LOR15C9). We thank Ruth Croson-Lowery and Edward Beharry at the New York Blood Center for their excellent technical assistances in flow cytometric analysis. We would like to thank Andrea Molinaro of the Laboratory of MicroChemistry for her outstanding work in DNA sequencing. We also thank Tellervo Huima for her excellent photography.
Reprints:Kimita Suyama, The New York Blood Center, 310 East 67th St, New York, NY 10021; e-mail: ksuyama@nybc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 2. Expression of RhAG glycoprotein in a transcription and translation-coupled reticulocyte lysate system. / Autoradiogram of [35S]-methionine-labeled proteins separated by 12% SDS-PAGE (for details, see Materials and Methods). Lane 1, pLNCXRhAG vector; lane 2, pLNCXRhAG vector with microsomal membranes. Arrow indicates nonglycosylated RhAG; arrow head, glycosylated RhAG.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/1/10.1182_blood.v95.1.336/5/m_bloo00146002w.jpeg?Expires=1763533866&Signature=MsdGErP4L-~uqnElSK7UvoNyi5dAeE7e7-aY1B-KKdX1EYCHMoOaB5oGo97JGMEcN~9QFRXmfJrp5T-3VuvKtbseICEK4IRThQDH~CRlIfLnPUhRqLu5QzwBqyT6mDYMlZpAd4msxVTlyglmtAR7nQFt3j71ForUrXPFYv2HkSnpi0yDykDDgkU~nkAQ9ukWKBsUK~G44ZfJXRsg27aPDrwkiapHEOxfbvZ0XA-QDJAERW0OIXjEmdg0ZlPUFTcXZ5-vxZSZNvoA-4S-tkbPqqbP7omQayrTEh3Ic8h7l3~UWgFRQ8Qvih2EjLyqbgrAvasPFO9qrXkEkZrRxAOR1Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Autoradiogram of immunoprecipitates obtained from stable COS-1 cells transduced with the viruses containing RhAG or RhD RNA using 2D10 or rabbit polyclonal anti-Rh. / Logarithmically growing COS-1 cells were incubated with [35S]-methionine for 2 hours, and the cell lysates were subjected to immunoprecipitation with 2D10 or rabbit polyclonal anti-Rh; the immunoprecipitates were then separated by 12% SDS-PAGE under reducing conditions (for details, see Materials and Methods). (A) Lane 1, stable COS-1 cells transduced with the viruses containing vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhAG. Arrow shows RhAG glycoprotein. (B) Lane 1, stable COS-1 cells transduced with the viruses containing vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhD. Arrowhead shows RhD polypeptides.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/1/10.1182_blood.v95.1.336/5/m_bloo00146003w.jpeg?Expires=1763533866&Signature=ckmlNkyw8yqoYu~3r1umKIjZFA6AvvUU6gIXkxKxIeI8M9bf2vIBCFbRL7EhG31g4SCZkSU6gqXOtdIw6y-SNZSzC5Tdfe7SoifwyfkRj~4iQ4rwOc~Z7LWt9fO22Uj1SQKJzSU6qlWsPEbUpNzNrsoEhFN6h2bNq1L2fbKtuCs6Q2qEjNDAKg9BTHEkmQp2xDdHJFi8CMdVvT8x8UpvdBBFSnIve3wf3NytpqQhFKkAmPayl6sYz8Bf4LvxslqEp2noEVcDzT9Yu2CSwFAH5-FTEKUN2G4CDDzsiJ2zPBP-usNMhxKEQG7UcLY3xp2RozcdENviERbh~d1GTGuX4A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Autoradiogram of immunoprecipitates obtained from stable COS-1 cells transduced with the viruses containing pLNCXRhAG using 2D10. / Logarithmically growing COS-1 cells were incubated with [3H]-mannose for 5 hours, and the cell lysates were subjected to immunoprecipitation with 2D10; the immunoprecipitates were then separated by SDS-PAGE under reducing conditions (for details, see Materials and Methods). Lane 1, stable COS-1 cells transduced with the viruses containing pLNCX vector alone; lane 2, stable COS-1 cells transduced with the viruses containing pLNCXRhAG. Arrow shows RhAG glycoprotein.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/1/10.1182_blood.v95.1.336/5/m_bloo00146004w.jpeg?Expires=1763533866&Signature=E64-Z1V~cIs2vkjRuxYv0GQpet90n2Ea-gIphcpH5i~FZqstuYEm4dqAUqj4V8U0XMjrRbiYjCZLVC7HGdo5FvEQbCZM0lcZ4ITeGzgy9T~ioGnD00ZZYWV2S-gwTlStuu5H2JTe-FGVkRn~4av2okRT5-VHlay1ZhaJqSLTon8AGNndE1L-sKz6M1JTTT6BD9a57HCItisqxSyvxsPhbRhlwwVWsEurkSLLmnjSnyaAo86iWoCQpW4DTWbITgNkyq9yQ2p14hHIzlic6skkbqMMzIEIDsoWZ5b22v7FkJJRhqhhfQLAsY-OIgLTwu0HLnihE6CA2hOGllfR~SEVkw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)