Abstract
The Ets variant gene 6 (ETV6/TEL) gene is rearranged in the majority of patients with 12p13 translocations fused to a number of different partners. We present here a case of acute myeloid leukemia M4 with eosinophilia (AML-M4Eo) positive for the CBFb/MYH11 rearrangement and carrying a t(1;12)(q25;p13) that involves the ETV6 gene at 12p13. By 3′rapid amplification of cDNA ends-polymerase chain reaction (3′RACE-PCR), a novel fusion transcript was identified between the ETV6 and the Abelson-related gene (ARG) at 1q25, resulting in a chimeric protein consisting of the HLH oligomerization domain of ETV6 and the SH2, SH3, and protein tyrosine kinase (PTK) domains of ARG. The reciprocal transcript ARG-ETV6 was also detected in the patient RNA by reverse transcriptase-polymerase chain reaction (RT-PCR), although at a lower expression level. The ARG gene encodes for a nonreceptor tyrosine kinase characterized by high homology with c-Abl in the TK, SH2, and SH3 domains. This is the first report on ARGinvolvement in a human malignancy.
THE SHORT ARM OF chromosome 12 (12p) is a region commonly involved in a wide variety of hematological malignancies in both children and adult patients.1 TheETV6/TEL gene was cloned by virtue of the translocation t(5;12)(q33;p13) in the leukemic cells of a patient with chronic myelomonocytic leukemia (CMML).2 More recently,ETV6 has been shown to be rearranged in the majority of patients with 12p13 translocations3,4 and fused to a number of different partners (reviewed in Rubnitz et al5; see also other studies6-9). Two possible mechanisms of transformation have been proposed for ETV6/TEL fusion genes. First, aberrant transcription factors can result from joining the 3′ region of ETV6 (Ets-family DNA binding domain) to the 5′ region of the partner gene (reviewed in Rubnitz et al5). Second, constitutive kinase phosphorylation, by dimerization of protein tyrosine kinases, can result from joining of the 5′ region (HLH domain) of ETV6 to the 3′ region of the partner gene (reviewed in Rubnitz et al5). In fact, the ETV6 gene has been previously reported to form fusion transcripts with several tyrosine kinase genes.2,6-10 In particular, ETV6 was found to be fused to Abl in some rare acute lymphocytic leukemia (ALL),11 acute myeloid leukemia (AML),6 and chronic myeloid leukemia (CML)12 cases.
We report here the identification of the Abelson-related geneARG as a fusion partner of ETV6 in a t(1;12)(q25;p13) found in the leukemic cells of an adult patient with AML M4 with eosinophilia and inv(16). This is the first demonstration ofARG involvement in a translocation in a human malignancy.
MATERIALS AND METHODS
Patient.
The patient was a 54-year-old woman presenting the following laboratory features: red blood cell count, 3.54 × 109/L; white blood cell count, 116 × 109/L (with 6% lymphocytes and 94% blasts); hemoglobin, 11.5 g/dL; and platelets, 36 × 109/L in peripheral blood. Diagnosis of AML was performed by morphology: 45% myeloblasts, 22% monoblasts, 30% eosinophils, and 3% plasma cells were detected. Thus, the patient was classified as AML-M4 with eosinophilia, according to the French-American-British (FAB) classification. The presence of type A inv(16), undetectable by conventional cytogenetics, was demonstrated by reverse transcriptase-polymerase chain reaction (RT-PCR)13 (data not shown). At the end of the induction therapy, the patient achieved complete hematological remission (CR). Six months later, she suffered from bone marrow (BM) relapse, with 60% blast invasion of the BM. CR was obtained after 2 courses of salvage therapy, but 4 months later, the patient suffered a second BM relapse and central nervous system involvement and died 4 months later from progression of disease.
Fluorescence in situ hybridization (FISH).
Probes used for FISH were as follows: (1) YAC 964c10, containing the entire ETV6 gene (CEPH, Paris, France); and (2) cosmids from the ETV6 gene14: 2G8 (exons 3 and 4) and 148B6 (exon 8; kindly provided by Dr P. Marynen, Centre for Human Genetics, University of Leuven, Leuven, Belgium). FISH analysis was performed on BM metaphases from archival methanol:acetic acid-fixed chromosome suspension stored at −20°C as previously described.6
PCR.
First-strand cDNA was reverse transcribed from 1 μg of total RNA with Superscript II reverse transcriptase (GIBCO-BRL, Life Technologies, Milano, Italy), according to standard procedures, using the already described primer R2N6.7 Nested 3′rapid amplification of cDNA ends-polymerase chain reaction (3′-RACE-PCR) was performed using primers specific for ETV6 exon 5 in combination with primers R2N6R1 and R2N6R2,7 respectively. In particular, the ETV6 primers designed for t(12;21) detection were used.13 Colonies with recombinant plasmids containing the PCR products were screened by hybridization using standard protocols. Two oligonucleotide probes specific for both ETV6 exon 5 and exon 6 were used. Clones positive for the exon 5 probe and negative for the exon 6 probe were selected. These colonies were further screened by RT-PCR using the ETV6 forward primer T-9 (5′-TGAAGAGCACGCCATGCCCATTG-3′) and the R2N6R2 as a reverse primer.
To confirm the presence of the ETV6-ARG fusion product, RT-PCR was performed on patient RNA using standard procedures, with the TA-4 forward primer (5′-ATCGGGAAGACCTGGCTTACA-3′) onETV6 exon 5 and the TA-5 reverse primer (5′-TGCCTGGGGTTCAACATCAC-3′) on ARG. To detect the reciprocal ARG-ETV6 transcript, first PCR was performed using AT-10 forward primer (5′-GGAGCCGAGGAGGAATGT-3′), specific for ARG, in combination with AT-11 reverse primer (5′-TGATTTCATCTGGGGTTTTCA-3′) on ETV6 exon 6. Nested PCR was performed using AT–8 forward primer (5′-GATGGGGCAGCAGGTG-3′) on ARG in combination with AT–9 reverse primer (5′-CAGGGCTCTGGACATTTTCTC-3′) onETV6 exon 6.
RESULTS AND DISCUSSION
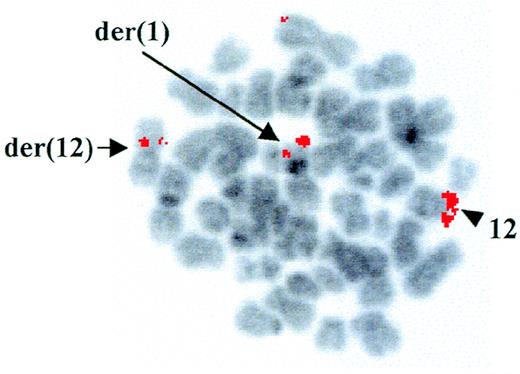
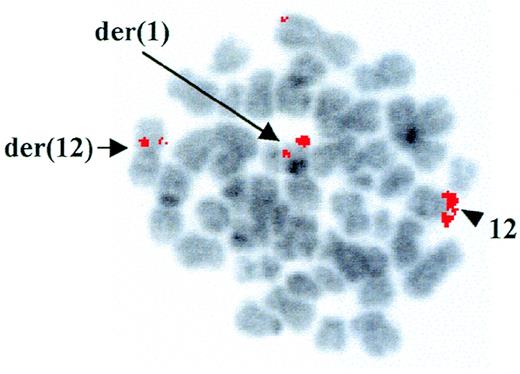
Cytogenetic analysis at diagnosis of a patient with AML-M4Eo showed the presence of a t(1;12)(q25;p13) as the sole chromosomal abnormality associated with the disease. However, using RT-PCR, we demonstrated positivity for type A inv(16), which is frequently detected in AML-M4Eo patients.15 FISH with the YAC 964c10, containing the entireETV6 gene,14 showed a fluorescent signal on both the der(12) and the der(1), indicating that the breakpoint on chromosome 12 was contained within this clone (Fig 1). Further FISH analyses with cosmids from the ETV6 gene confirmed that the breakpoint was withinETV6, between cosmid 2g8 (exons 3 and 4) and 148b6 (exon 8) (data not shown).
FISH analysis of the ETV6 rearrangement. FISH with the ETV6-containing YAC 964c10 to leukemic metaphases. Arrows indicate fluorescent signals corresponding to YAC 964c10 on the der(1) and the der(12); the arrowhead shows the normal chromosome 12 homologue. The G-banding on the chromosomes was obtained by inverting the DAPI-counterstained image.
FISH analysis of the ETV6 rearrangement. FISH with the ETV6-containing YAC 964c10 to leukemic metaphases. Arrows indicate fluorescent signals corresponding to YAC 964c10 on the der(1) and the der(12); the arrowhead shows the normal chromosome 12 homologue. The G-banding on the chromosomes was obtained by inverting the DAPI-counterstained image.
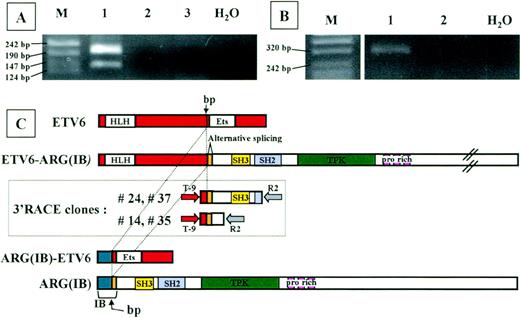
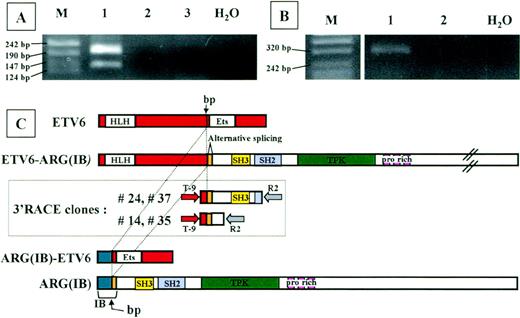
To identify the fusion partner of ETV6, we performed 3′-RACE analysis on RNA from the patient, using nested primers located in ETV6 exon 5. The whole 3′RACE-PCR product was cloned in pMOS vector, and 200 bacterial colonies were screened using both an ETV6-exon 6 primer and an ETV6-exon 5 primer (internal to the one used in nested PCR) as probes. Using double screening, we could exclude the presence of the unrearrangedETV6 allele (21 positive colonies with exon 6 probe) and consider only colonies ETV6-exon 6 negative andETV6-exon 5 positive (n = 40). To exclude PCR artefacts, these colonies were further screened by PCR using T-9 forward primer and R2 reverse primer; 29 of 40 colonies were positive. Thirteen of 29 plasmids were sequenced. Seven clones represented the normalETV6 allele, 2 sequences corresponded to ETV6 intron 5, and the remaining 4 clones were completely sequenced in both directions. Sequence analysis of the 4 clones detected an unknown sequence fused to the ETV6 exon 5 sequence. This sequence was proven to be the Abl-related gene ARG cDNA by BLAST database searching (GenBank accession no. M35296). Two clones (no. 24 and 37) extended from nt 362 of ARG to nt 466; the other 2 (no. 14 and 35) corresponded to ARG cDNA through to nt 815 (Fig 2C).
Analysis of the chimeric proteins. RT-PCR analysis ofETV6-ARG (A) and ARG-ETV6 (B) fusion products. Total RNA from the patient (lane 1) and 2 different healthy donor white blood cells (lanes 2 and 3) was used for RT-PCR with primers located in the relevant exons of ETV6 and ARG(see Materials and Methods). Marker sizes are indicated on the left. (A) RT-PCR analysis of ETV6-ARG. Two specific bands of 194 and 131 bp, respectively, were detected. The different PCR products are the result of an alternative splicing event in the ARGgene, as shown in (C). (B) RT-PCR analysis of ARG-ETV6. One specific band of 343 bp was detected. (C) Schematic representation of ETV6 (top) and ARG(IB) (bottom) cDNAs, 3′RACE clones (boxed), and predicted ETV6-ARG andARG-ETV6 fusion proteins. The vertical arrow indicates the breakpoint. The alternatively spliced hypothetical ARG exon is indicated. HLH, helix-loop-helix domain; ETS, ETS-family DNA binding domain; SH3, SH3 domain; PTK, protein-tyrosine kinase domain; pro rich, proline-rich domains. Full-length cDNAs from the ARG andETV6 genes are shown at the top and bottom, respectively. The 2 different types of 3′RACE-PCR clones are shown in the box. The nested PCR primers are indicated as arrows at the extremities of the RACE clones.
Analysis of the chimeric proteins. RT-PCR analysis ofETV6-ARG (A) and ARG-ETV6 (B) fusion products. Total RNA from the patient (lane 1) and 2 different healthy donor white blood cells (lanes 2 and 3) was used for RT-PCR with primers located in the relevant exons of ETV6 and ARG(see Materials and Methods). Marker sizes are indicated on the left. (A) RT-PCR analysis of ETV6-ARG. Two specific bands of 194 and 131 bp, respectively, were detected. The different PCR products are the result of an alternative splicing event in the ARGgene, as shown in (C). (B) RT-PCR analysis of ARG-ETV6. One specific band of 343 bp was detected. (C) Schematic representation of ETV6 (top) and ARG(IB) (bottom) cDNAs, 3′RACE clones (boxed), and predicted ETV6-ARG andARG-ETV6 fusion proteins. The vertical arrow indicates the breakpoint. The alternatively spliced hypothetical ARG exon is indicated. HLH, helix-loop-helix domain; ETS, ETS-family DNA binding domain; SH3, SH3 domain; PTK, protein-tyrosine kinase domain; pro rich, proline-rich domains. Full-length cDNAs from the ARG andETV6 genes are shown at the top and bottom, respectively. The 2 different types of 3′RACE-PCR clones are shown in the box. The nested PCR primers are indicated as arrows at the extremities of the RACE clones.
Sequence analysis showed a deduced 1466 amino acid open reading frame for the full-length ETV6-ARG fusion, starting at theETV6 first ATG and ending at the ARG TAG stop codon. The predicted size of the protein encoded by this transcript is 161 kD.
RT-PCR analysis on patient material was performed to confirm the fusion transcript between the ETV6 and ARG mRNA. Two fragments were unexpectedly detected using forward primer on ETV6 exon 5 (TA-4) and reverse primer on ARG (TA-5) (Fig 2A). Sequence analysis showed that the 2 bands represent 2 alternatively splicedETV6-ARG transcripts. The larger product results from the junction of exon 5 of ETV6 (nt 1033) to nt 362 of ARG, exactly as the 3′RACE clones do. The smaller product spliced exon 5 of ETV6 to nt 425 of ARG. Interestingly, a CAG triplet is present at the end of ETV6 exon 5 (nt 1031-1033) and also at the nt 422-424 of ARG, corresponding to the very beginning of the region common to ARG(IA) and(IB).16 17 The sequence of the lower band in the RT-PCR experiment on the patient showed the absence of 1 of the 2 repeated CAG triplets expected as a result of the mRNA splicing. Both splicing forms result in an open reading frame linking the HLH oligomerization domain of ETV6 to the complete SH3, SH2, and PTK domain of ARG. PROSITE analysis of the fusion proteins showed that no additional domains were added by virtue of the new generated region at the junction site. Moreover, the only difference between the alternatively spliced forms consists of an additional Casein Kinase II phosphorylation site in the larger version. Indeed, the absence of a CAG triplet in the spliced form does not generate any difference in the recognized domains.
Interestingly, the alternative spliced portion of ARG (nt.362-422), which is responsible for the lower RT-PCR band, does not have any homology with Abl.17 Thus, we can expect that this small portion represents a single exon of ARG.
The arg protein is a nonreceptor tyrosine kinase characterized by high homology with c-abl in the TK, SH2, and SH3 domains (95%).16,17 Moreover, the products of the humanARG gene and human, mouse, Drosophila, and nematode Ablgenes are characterized by high homology, indicating that they can have an important function conserved during the evolution. By contrast, the variation in the N-terminal and C-terminal domains of arg and c-abl may account for their different functions.17 In particular, arg is cytoplasmic,18 but c-abl has both nuclear and cytoplasmic localization.19 The abl and argproducts have also different transforming activity mediated by the distinct arg and abl C-terminal domains (CTD).20 So far, there is little information on the role of Arg in the cell; moreover, the role of Arg in the hematological system is still unknown.
Receptor and nonreceptor tyrosine kinases are known to play an important role in cell growth and differentiation. Amplifications, mutations, or recombinations involving these genes can result in increased activation of their kinase domain, which leads to cellular transformation. In both the ETV6-PDGFRb and ETV6-Ablfusion transcripts,21-23 it has been demonstrated that the HLH domain contained in ETV6 confers the oncogenic activity to the chimeric PTK proteins by forming HLH domain-dependent homo-oligomers that result in ligand-independent tyrosine kinase activation.21-23ETV6-Abl, like BCR-Abl[which is consistently observed in t(9;22)-positive CML or ALL], leads to increased tyrosine kinase activity contained within the abl part and relocalization of abl from the nucleus to the actin cytoskeleton of the cytoplasm.22 Moreover, BCR-Abland ETV6-Abl seem to activate similar signal transduction pathways23 and show similar transforming activity.22,23 In addition, it has been demonstrated that the ETV6-Abl expression in factor-dependent murine hematopoietic precursor cells transforms these cells, converting them to factor independence for both survival and growth.24
Because of the tight homology between ARG and c-Abl and the very similar activity of both etv6-abl and bcr-abl chimeric proteins,22 23 the homodimerization property of the HLH domain can also be proposed as an activation mechanism of theetv6-arg chimeric protein kinase. However, further experiments are needed to confirm this hypothesis.
In contrast to the t(5;12), t(12;15), and t(9;12), in which the reciprocal transcripts ETV6-PDGFR, ETV6-TRKC, ETV6-ABL, andETV6-JAK2 were not detected,2 7-12 we could demonstrate the presence of reciprocal ARG-ETV6 transcript in the t(1;12) (Fig 2B). However, the presence of the latter was detected only by nested PCR, indicating that this transcript was expressed at a low level in the patient leukemic cells. The reciprocal full-lengthARG-ETV6 transcript accounts for a 168 amino acid open reading frame, starting at the ARG first ATG and ending at theETV6 TGA stop codon. This transcript consists of the fusion between the NH2 region of ARG to the ETV6 exon 6, which retains the Ets-DNA binding domain of ETV6 and results in a hypothetical 20-kD protein. The presence of this protein and its role in the leukemic cells have still to be fully investigated. However, it is possible to speculate that the aberrant DNA-binding protein would interact with the physiological ETV6 target genes, although under the regulation of the Arg promoter.
In conclusion, this is the first demonstration of the involvement of the Abelson-related gene ARG in a translocation in a human hematologic malignancy. In future experiments, the analysis of the chimeric proteins that arise from this translocation should elucidate both the physiological and the aberrant role of ARG in hematopoietic cells.
Supported by Fondazione M. Tettamanti, Associazione Italiana per la Ricerca sul Cancro (AIRC), MURST 40%. S.T. and L.K. are supported by the Leukaemia Research Fund, UK, and the Medical Research Council, respectively.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Giovanni Cazzaniga, PhD, Clinica Pediatrica Università di Milano-Bicocca, Centro Ricerca Tettamanti, Nuovo Ospedale San Gerardo, Via Donizetti, 106, 20052 Monza, Italy; e-mail: fondazione.tettamanti@galactica.it.