VITAMIN K, AN ESSENTIAL vitamin, is a cofactor for a single known enzymatic reaction: the conversion of glutamic acid to γ-carboxyglutamic acid in vitamin K-dependent proteins during their biosynthesis. Since the discovery of vitamin K and its association with blood coagulation,1 many milestones have been passed on the road to understanding the biological role of vitamin K. Important early landmarks include the discovery of vitamin K antagonists and their introduction as pharmacologic agents for anticoagulation2; the discovery of γ-carboxyglutamic acid in blood clotting proteins3,4; the identification of γ-carboxyglutamic acid as a metal binding amino acid that confers metal binding properties on proteins, a requirement for protein-membrane interaction5,6; the detection of an enzymatic activity (ie, the vitamin K-dependent γ-glutamyl-carboxylase) that catalyzes the incorporation of CO2 into glutamic acid5; the identification of an intervening sequence (the propeptide) between the signal peptide and mature vitamin K-dependent protein7; and the discovery of the requirement for8 and the sufficiency of9 the γ-carboxylation recognition site within the propeptide in directing synthesis of γ-carboxyglutamic acid on the adjacent Gla domain on the precursor protein. More recent advances include the purification and cloning of the vitamin K-dependent carboxylase (carboxylase)10,11; the determination of the three-dimensional structure of the Gla domain of prothrombin and observation of an internal carboxylate-Ca2+network12; the proposal of a mechanism of vitamin K-mediated base enhancement of carboxylase action13; and the regulation of enzymatic vitamin K epoxidase activity by glutamate containing substrate.14 Because carboxylase activity is found in essentially all mammalian tissues15 and because γ-carboxyglutamic acid has been observed in both vertebrates and invertebrates, this amino acid must play an important biological role in protein function. The biosynthesis of γ-carboxyglutamic acid is the topic of this review.
γ-CARBOXYGLUTAMIC ACID: CHEMISTRY OF BIOSYNTHESIS
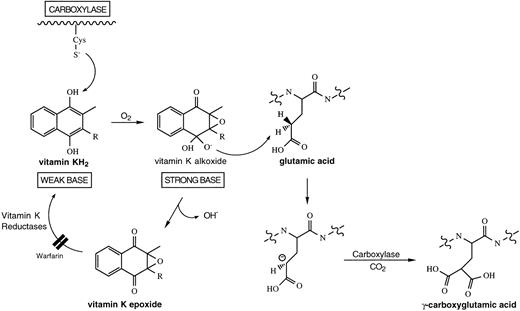
The requirement for vitamin K as an enzyme cofactor is unique to the vitamin K-dependent carboxylase and the biosynthesis of γ-carboxyglutamic acid. In this reaction, the γ-proton on glutamic acid is abstracted, followed by the addition of CO2; the intermediate in this reaction is a γ-glutamyl carbanion16(Fig 1). The mechanism by which vitamin K participates as a cofactor with the γ-carboxylase remains a puzzle. The most attractive hypothesis is that an active oxygenated species of vitamin K abstracts a hydrogen from the γ-carbon of glutamic acid, with subsequent collapse of the activated vitamin K species to vitamin K epoxide. Carbon dioxide is subsequently added to the γ-carbon of glutamic acid. Based on a nonenzymatic model,13 a “base strength amplification mechanism” has been proposed to explain the conversion of vitamin KH2 into an oxygenated intermediate of sufficient basicity to abstract a hydrogen from the γ-carbon of a glutamic acid.17 The proposed intracellular intermediate is short-lived. “Footprints” of the formation of such an unstable intermediate by the mechanism proposed have been discovered by the identification of a stable, isotopically labeled degradation product of vitamin K dioxide during enzymatic vitamin K-dependent carboxylation in vitro.18 19
Biosynthetic pathway for vitamin K-dependent production of γ-carboxyglutamic acid. It has been hypothesized that a free cysteine residue in the carboxylase converts vitamin KH2into a “strong base” of sufficient basicity to abstract a hydrogen from the γ-carbon of glutamic acid.17Subsequently, CO2 is added to the γ-carbon of glutamic acid to form γ-carboxyglutamic acid. The activated vitamin K species collapses into vitamin K epoxide and is recycled back to vitamin KH2, following the action of two vitamin K reductases, one of which is sensitive to warfarin.25
Biosynthetic pathway for vitamin K-dependent production of γ-carboxyglutamic acid. It has been hypothesized that a free cysteine residue in the carboxylase converts vitamin KH2into a “strong base” of sufficient basicity to abstract a hydrogen from the γ-carbon of glutamic acid.17Subsequently, CO2 is added to the γ-carbon of glutamic acid to form γ-carboxyglutamic acid. The activated vitamin K species collapses into vitamin K epoxide and is recycled back to vitamin KH2, following the action of two vitamin K reductases, one of which is sensitive to warfarin.25
Thus, the vitamin K-dependent carboxylase is also a vitamin K epoxidase, and, under normal conditions, for each molecule of γ-carboxyglutamic acid generated, a molecule of vitamin K epoxide is generated. The coupling of γ-carboxyglutamic acid and vitamin K epoxide production was first noted with partially purified carboxylase20 and has been confirmed with purified carboxylase.21 In addition to its role as a recognition sequence,8,9 binding of the propeptide of a vitamin K-dependent protein substrate to the carboxylase stimulates γ-glutamyl carboxylation.22 Because oxidation of reduced vitamin K precedes reaction at the γ-C-H on the substrate glutamate residue and γ-carboxylation cannot occur without formation of vitamin K epoxide, this observation suggests that vitamin K epoxidation by the carboxylase may be stimulated by propeptide. Kinetic studies have determined that the propeptide increase in carboxylase catalytic efficiency is in selective lowering of an energy barrier preceding the γ-glutamyl carbanion intermediate either by accelerating formation of the reactive vitamin K intermediate or proton abstraction from a substrate glutamyl residue.23
The short-lived highly reactive vitamin K intermediate is potentially toxic, and it would be undesirable for it to be generated intracellularly in the absence of substrate glutamate residues. Indeed, it has been recently demonstrated that the carboxylase has no epoxidase activity until the enzyme system is activated.14Glutamate-containing substrates bound to the vitamin K-dependent carboxylase convert its vitamin K epoxidase function from an inactive to an active state. Therefore, epoxidase activity is turned-off, and no highly reactive vitamin K intermediate is generated, unless a carboxylase substrate is bound to the enzyme.14
One intriguing question regarding the mechanism of the vitamin K-dependent carboxylase is whether the enzyme is processive or distributive. If processive, then once a vitamin K-dependent protein substrate has bound the enzyme all target Glu residues are converted to Gla residues before the protein substrate dissociates from the enzyme. If distributive, the protein substrate associates with and dissociates from the carboxylase multiple times before carboxylation is complete. A second interesting question is whether there is a preferred order of carboxylation of the target Glu residues in a vitamin K-dependent protein substrate. Morris et al24 have reported that, when purified carboxylase carboxylates a peptide analog of the propeptide and Gla domain of factor IX, the products of the reaction are skewed toward highly carboxylated forms. These investigators suggest that only two mechanisms can support these results: either the carboxylase is processive or it is distributive with the requirement that partially carboxylated forms of the factor IX peptide analog are more efficiently carboxylated than the uncarboxylated peptide. Although the currently available data suggest that the carboxylase is processive, conclusive experiments remain to be performed. No information is currently available regarding the order of carboxylation of Glu residues within a vitamin K-dependent protein substrate.
RECYCLING OF THE CARBOXYLASE COFACTOR VITAMIN K
In its naturally occurring state, vitamin K is at the quinone oxidation state and must be reduced to the hydroquinone form, the active cofactor for the vitamin K-dependent carboxylase. The enzyme responsible for this conversion is known as the vitamin K epoxide reductase, so named because it also reduces the vitamin K epoxide formed during the carboxylation reaction.25 The vitamin K epoxide reductase works at low concentrations of vitamin K epoxide and vitamin K quinone and is likely the physiologically important enzyme for recycling vitamin K.26 This is the enzyme that is inhibited by warfarin, resulting in insufficient vitamin K hydroquinone to support full carboxylation of the vitamin K-dependent proteins of blood coagulation, thus its role as a pharmacologically useful anticoagulant. It is believed that the enzymatic activity of the vitamin K epoxide reductase resides in a membrane-bound, multiprotein enzyme complex in the endoplasmic reticulum. Recently, two components of this complex have been identified: microsomal epoxide hydrolase27 and a member of the glutathione S-transferase super gene family.28
A second enzyme, DT-diapharase, an NAD(P)H dehydrogenase, reduces the quinone form of vitamin K but not vitamin K epoxide.25Furthermore, this enzyme requires high concentrations of vitamin K26 and likely does not play a role in vitamin K recycling at physiologic tissue concentrations of vitamin K. This enzyme may play an important role when vitamin K, in the quinone form, is used to overcome warfarin intoxication.
HOW ARE PROTEINS SELECTED FOR γ-CARBOXYLATION? ROLE OF THE PROPEPTIDE AND THE γ-CARBOXYLATION RECOGNITION SEQUENCE
The propeptides of the vitamin K-dependent proteins contain a γ-carboxylation recognition site that is required for γ-glutamyl carboxylation.8 In one known example, matrix Gla protein, the γ-carboxylation recognition site resides within the mature protein sequence.29 The amino acids of the γ-carboxylation recognition site bind directly to the vitamin K-dependent carboxylase.30 Although no obvious consensus sequence prevails in the carboxylation recognition sites of vitamin K-dependent proteins (Fig 2), this site is best defined by a Z-F-Z-X-X-X-X-A motif, where Z is an aliphatic hydrophobic residue (Ile, Val, Leu), F is phenylalanine, A is alanine, and X is any amino acid. Recent studies have emphasized that phenylalanine at residue −16 is preferred in carboxylase substrates, but leucine, valine, and lysine at this position also support carboxylation.31 Indeed, gas6, a recently described Gla-domain containing protein,32 has a leucine at residue −16 and is carboxylated.33
The γ-carboxylation recognition sites of known vitamin K-dependent proteins. The propeptides of the vitamin K-dependent blood coagulation proteins contain a γ-carboxylation recognition site that directs carboxylation.8 A phenylalanine at position −16 and an alanine at position −10 are well conserved within the propeptides of carboxylase substrates, as are aliphatic hydrophobic residues (isoleucine, leucine, and valine) at position −17 and −15. Recent data suggest that, whereas phenylalanine at −16 is preferred, leucine, valine, and lysine at this position can also support carboxylation.31
The γ-carboxylation recognition sites of known vitamin K-dependent proteins. The propeptides of the vitamin K-dependent blood coagulation proteins contain a γ-carboxylation recognition site that directs carboxylation.8 A phenylalanine at position −16 and an alanine at position −10 are well conserved within the propeptides of carboxylase substrates, as are aliphatic hydrophobic residues (isoleucine, leucine, and valine) at position −17 and −15. Recent data suggest that, whereas phenylalanine at −16 is preferred, leucine, valine, and lysine at this position can also support carboxylation.31
Disruption of this carboxylation recognition site in factor IX yields a mature factor IX protein that either lacks or is deficient in γ-carboxyglutamic acid.8 Thus, this site is required for γ-carboxylation. It is most likely that this region of the blood coagulation protein precursor docks with the membrane-bound carboxylase, bringing the active site of the carboxylase in close proximity to the substrate glutamate residues on the precursor form of the vitamin K-dependent proteins. There is no evidence that the amino acid sequence or three-dimensional structural elements in the immediate vicinity of the glutamate residues contribute significantly to the recognition of the protein substrate by the carboxylase. Indeed, a double point mutant of prothrombin, in which serine substituted for cysteines 17 and 22 disrupted a conserved disulfide loop, was fully carboxylated when expressed in CHO cells.9 In addition, a prothrombin propeptide/thrombin chimera was constructed by deleting the Gla, aromatic amino acid stack, and kringle domains of prothrombin. This construct has the signal peptide and γ-carboxylation recognition site-containing propeptide juxtaposed to a glutamate-rich C-terminal region of prothrombin. Seven or eight of the eight glutamic acids within the first 40 residues of the NH2-terminus adjacent to the propeptide underwent complete carboxylation when this protein was expressed in CHO cells.9 These results indicate that the prothrombin γ-carboxylation recognition site on the propeptide is sufficient to direct carboxylation of adjacent glutamic acid residues in the propeptide/thrombin chimera by the vitamin K-dependent carboxylase without regard for the sequence context of the glutamic acid substrate or structures defined by disulfide bonds. Thus, we hypothesize that any protein will undergo γ-carboxylation if it meets the following criteria: (1) the protein includes a γ-carboxylation recognition site that interacts with the γ-glutamyl carboxylase; (2) the protein is trafficked through the rough endoplasmic reticulum during protein biosynthesis; (3) the cell has the carboxylase enzyme associated with the rough endoplasmic reticulum; (4) there are glutamic acid residues within 40 residues of the γ-carboxylation recognition site; and (5) intracellular vitamin K is present.
Others have proposed that a second recognition site might reside within the mature sequence of the nascent vitamin K-dependent protein.29 A Gla domain consensus sequence, E16XXXE20XC22, common to the vitamin K-dependent blood and bone proteins, was disrupted by site-specific mutagenesis.34 This was associated with the expression of incompletely carboxylated recombinant protein C in a mammalian expression system. Furthermore, recombinant protein C in which glutamic acid residues in the Gla domain were systematically mutated to aspartic acid residues was incompletely carboxylated.35 These results were interpreted to suggest that this consensus sequence is important for carboxylation. In contrast, we prepared 10 mutants of prothrombin in which each of the glutamic acid residues in the Gla domain were modified to aspartic acid.36 Expression of these mutant prothrombins generated fully carboxylated protein. These results most likely relate to differences in the heterologous mammalian expression systems used or efficiency of carboxylation of individual vitamin K-dependent proteins and not to a specific role for glutamic acid in ligand binding energy.
Naturally occurring propeptide mutants of factor IX have been described in humans. The mutation of alanine −10 to threonine is associated with marked sensitivity of patients to warfarin anticoagulation and severe inhibition of carboxylation, well in excess of that observed with the other vitamin K-dependent proteins.37Similarly, mutation of alanine −10 to either threonine or to valine is associated with marked sensitivity of patients to phenprocoumon.38 In all cases, the phenotype and factor IX levels of patients with these mutations at alanine −10 were normal, unless they were exposed to a vitamin K antagonist. Alanine −10 has been shown to be a part of the carboxylation recognition site by mutational analysis and using small peptide substrates for carboxylation. Mutation is associated with diminished γ-carboxylation both during expression of recombinant protein in CHO cells8,39 and during in vitro37γ-carboxylation. However, there is no evidence of defective carboxylation of factor IX in men with these mutations when they are not challenged with warfarin. This paradoxical effect has not been adequately explained to date. With a reduced affinity for the carboxylase, the mutant profactor IX may be at a kinetic disadvantage when the concentration of the other carboxylase substrate, reduced vitamin K, is limited by inhibition of the recycling enzymes and other vitamin K-dependent proteins can compete for enzyme (Fig 1).
VITAMIN K-DEPENDENT CARBOXYLASE: THE ENZYMATIC MACHINERY FOR THE SYNTHESIS OF γ-CARBOXYGLUTAMIC ACID
Vitamin K-dependent carboxylase activity was first demonstrated in liver in 1975, shortly after the discovery of γ-carboxyglutamic acid.5 Because the enzyme is membrane-associated and unstable, purification of the enzyme was difficult and refractory to standard techniques. However, considerable insight about this enzyme system was gained from the analysis of solubilized and partially purified carboxylase.25 With the discovery that the propeptide of the vitamin K-dependent proteins directs γ-carboxylation,8 the use of the propeptide for affinity chromatography led first to significant purification30 and finally to the isolation of the carboxylase.10 Molecular cloning of the human and bovine vitamin K-dependent carboxylase11,40 predicted a single chain protein of 758 amino acids. The protein, with a molecular weight of about 94,000, is dominated by hydrophobic amino acids, particularly toward the N-terminal third of the molecule, but the presence of transmembrane spanning regions is uncertain. The human carboxylase gene is localized on chromosome 2 at 2p12.41 The gene is 13 kb in length and contains 15 exons.42 In humans in the tissues examined, two transcripts were observed. In rats, the gene is 16.3 kb in length, with similar intron/exon boundaries as in the human gene. Only one transcript was observed in the rat.43
The predicted sequences of human, bovine, and rat carboxylase have been reported, and these share a high degree of homology: 88% between human and rat carboxylase and 94% between human and bovine carboxylase.43 The carboxylase has minimal sequence homology with other proteins, with the closest being NADH-ubiquinone oxidoreductase chain 2, cytochrome C, and soybean lipoxygenase. Price and Williamson44 observed that carboxylase shares homology over 24 residues (495-518) with the region of matrix Gla protein that bears the γ-carboxylation recognition site. These investigators suggested that this sequence might play a regulatory role, blocking the active site of the enzyme and preventing nonspecific protein carboxylation. The propeptide of a vitamin K-dependent protein could displace this internal γ-carboxylation recognition site to gain access to the enzyme active site. Berkner and Pudota45 have recently reported that the carboxylase can itself be carboxylated, with 3 moles of Gla per mole of enzyme. It appears that this modification can occur in the absence of an exogenous peptide containing a γ-carboxylation recognition site, suggesting that the regulatory model proposed by Price and Williamson44 does not apply to carboxylation of the carboxylase. The functional significance of this modification of the carboxylase is currently unknown.
Although the protein sequence of the carboxylase is known, the general organization of functional regions of the protein is appreciated only at low resolution. The known functional properties of this enzyme include a carboxylase active site, an epoxidase active site, a propeptide binding site that docks substrate, a propeptide binding site that stimulates carboxylase and epoxidase activity, and a vitamin K binding site. Using substrate- and propeptide-based affinity labeling reagents, the vitamin K-dependent carboxylase active site and propeptide binding site were located, at least in part, within the hydrophobic N-terminal third of the protein between residues 1 and 225.46-49 A preliminary study based on the yeast two-hybrid system concludes that the propeptide binds to the N-terminal third of the enzyme.50 However, one recent report suggests that the propeptide binding site on the carboxylase is located on the more hydrophilic C-terminal two thirds of the enzyme between residues 438 and 758.51 Another study reports that the active site of the enzyme is located in a central region of the carboxylase between residues 350 and 508, with both an N-terminal and C-terminal tryptic peptide labeled with a propeptide-substrate based affinity label.52 Several sets of mutagenesis experiments bear on this question as well. Point mutations at the charged residues 234/235, 406/408, and 513/515 result in carboxylase species with reduced affinity for propeptide.49 Truncations of bovine carboxylase from the C-terminus at amino acids 712 or 676 result in carboxylase species that bind to propeptide and glutamate containing substrates equivalently to the wild-type enzyme, suggesting that the extreme C-terminal region is not involved in propeptide binding.53 It is not clear what experimental variations have led to various affinity-labeling experiments giving differing results. The data given above, taken in toto, suggest that the propeptide and perhaps the glutamate binding site of the carboxylase include regions in the N-terminal third and central third of the enzyme. Alternatively, there may be more than one propeptide binding site.22 Without a structural model on which to base functional studies, it is not presently possible to reconcile these results.
INTRACELLULAR SITE OF γ-CARBOXYLATION DURING PROTEIN SYNTHESIS
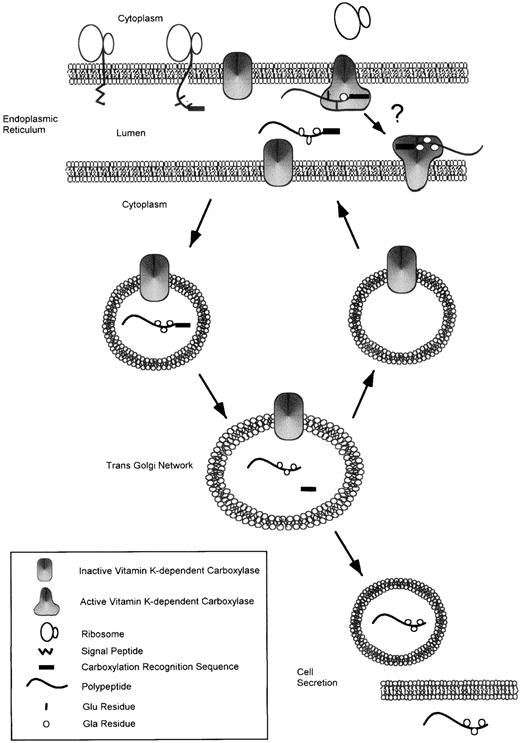
Subcellular fractionation methods have previously demonstrated that carboxylase activity resides in both the endoplasmic reticulum54 and the Golgi apparatus.55 Studies of the glycoforms of prothrombin56 and protein C57 during biosynthesis have indicated that core glycosylation but not formation of complex carbohydrate proceeds carboxylation. These data suggest that carboxylation is complete in the endoplasmic reticulum. In both cases, removal of the propeptide occurs late in the processing pathway. We have used antibodies to directly localize carboxylase and the various processed forms of prothrombin during biosynthesis. Using anticarboxylase antibodies, we have confirmed the intracellular localization of the carboxylase in both the endoplasmic reticulum and the Golgi. The intracellular sites of carboxylation and subsequent propeptide cleavage along the pathway of prothrombin biosynthesis were monitored for recombinant prothrombin synthesis in Chinese hamster ovary cells expressing prothrombin.58 Antibodies specific to processing intermediates (uncarboxylated prothrombin or uncarboxylated proprothrombin, carboxylated proprothrombin, or carboxylated prothrombin) were used for immunocytolocalization. These studies showed that uncarboxylated proprothrombin undergoes γ-carboxylation to carboxylated proprothrombin in the endoplasmic reticulum. The carboxylated proprothrombin leaves the endoplasmic reticulum intact and is further processed in the Golgi apparatus to remove the propeptide. Furin has been implicated in the cleavage of the propeptide from factor IX.59 However, there is a large family of related proconvertases that are involved in cleavage of propeptides that contain Arg or Lys at the −2 position and Arg at −1.60 The actual proconvertase that removes the propeptides of the vitamin K-dependent proteins is not known. Although carboxylase is detected in the Golgi complex by immunostaining, carboxylation appears to be complete in the endoplasmic reticulum. The carboxylase as a membrane-bound protein may be carried to the Golgi by bulk flow and may get recycled to the endoplasmic reticulum (Fig 3).
Pathway for synthesis of vitamin K-dependent proteins. Vitamin K-dependent proteins are synthesized in the endoplasmic reticulum as precursor proteins containing a signal peptide and a γ-carboxylation recognition site usually found within a propeptide of the proprotein. After cleavage of the signal peptide, the proprotein binds to the endoplasmic reticulum membrane-associated vitamin K-dependent carboxylase via its γ-carboxylation recognition site. Recent data suggest that binding of a glutamic acid-containing substrate to the carboxylase converts it from an inactive to an active state.14 The carboxylase catalyzes the conversion of glutamic acid residues to γ-carboxyglutamic acid residues. As is indicated by an arrow with a question mark connecting two active carboxylase molecules, it is unclear whether carboxylation of proproteins occurs by a processive mechanism. The carboxylated proprotein is trafficked through the cell to the trans Golgi network, where the propeptide is cleaved by an unknown proconvertase. The inactive form of the carboxylase may be carried to the Golgi and then recycled back to the ER. The fully modified, mature protein is then secreted from the cell.
Pathway for synthesis of vitamin K-dependent proteins. Vitamin K-dependent proteins are synthesized in the endoplasmic reticulum as precursor proteins containing a signal peptide and a γ-carboxylation recognition site usually found within a propeptide of the proprotein. After cleavage of the signal peptide, the proprotein binds to the endoplasmic reticulum membrane-associated vitamin K-dependent carboxylase via its γ-carboxylation recognition site. Recent data suggest that binding of a glutamic acid-containing substrate to the carboxylase converts it from an inactive to an active state.14 The carboxylase catalyzes the conversion of glutamic acid residues to γ-carboxyglutamic acid residues. As is indicated by an arrow with a question mark connecting two active carboxylase molecules, it is unclear whether carboxylation of proproteins occurs by a processive mechanism. The carboxylated proprotein is trafficked through the cell to the trans Golgi network, where the propeptide is cleaved by an unknown proconvertase. The inactive form of the carboxylase may be carried to the Golgi and then recycled back to the ER. The fully modified, mature protein is then secreted from the cell.
Prothrombin and the other γ-carboxylated extracellular vitamin K-dependent proteins bind to acidic membranes in the presence of calcium ions. The question arises as to what prevents the fully γ-carboxylated precursor vitamin K-dependent proteins, such as proprothrombin, from binding to endoplasmic reticular membranes during transit through the biosynthetic pathway and getting “hung up?” The calcium concentration in the endoplasmic reticulum is high, in contrast to the cytoplasm, and is sufficient to support protein-membrane interaction. It is known that chemical modification of the N-terminus of prothrombin inhibits membrane binding.61Similarly, fully carboxylated profactor IX does not bind to membranes in the presence of calcium ions, whereas factor IX does.59It would appear that the propeptide attached to factor IX prevents proper folding of the Gla domain, the expression of the phospholipid binding site, and the interaction of profactor IX with membranes.
THE FUNCTION OF γ-CARBOXYGLUTAMIC ACID
Why is γ-carboxyglutamic acid so broadly used in biology? The answers are not in yet, but knowledge of the function of γ-carboxyglutamic acid is expanding. γ-Carboxyglutamic acid has been extensively studied in three protein families. These include the vitamin K-dependent blood coagulation (factor IX, factor VII, factor X, and prothrombin) and regulatory (protein C and protein S) proteins, proteins of mineralized tissue (bone Gla protein, matrix Gla protein), and neurotoxins in the venom of cone snails. There is high likelihood that many other vitamin K-dependent proteins exist outside of these three families. Several proteins that contain Gla domains homologous to the Gla domains of blood coagulation but of unknown function have recently been identified. These include Gas6 and two proline-rich γ-carboxyglutamic acid-containing proteins (Table 1).
An extensive review of the literature supporting our current knowledge of the role of γ-carboxyglutamic acid is beyond the scope of this review. A brief discussion is included to provide the reader with an indication of the consequences of this posttranslational modification. γ-Carboxyglutamic acid distinguishes itself from aspartic acid and glutamic acid by containing two carboxyl groups in its side chain. The bivalent nature of γ-carboxyglutamic acid is similar to Igs or fibrinogen, for example. In these cases, a structural framework is formed via the linking of one ligand to another through a common bivalent, symmetrical molecule. As is described below, such is the case for the formation of the calcium-carboxylate network that stabilizes the Gla domains and, in the vitamin K-dependent proteins of blood coagulation, allows expression of the phospholipid binding site (Fig 4).
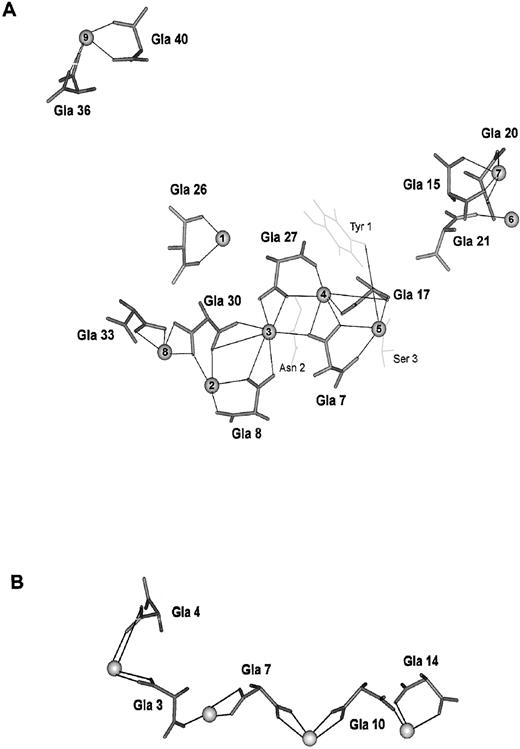
The calcium-carboxylate networks of the factor IX Gla-domain and conantokin G. One role of γ-carboxyglutamic acid is to stabilize a protein structure by formation of an extended calcium-carboxylate network. This property of γ-carboxyglutamic acid is used to stabilize the factor IX Gla-domain and conantokin G. (A) The calcium ligands for the Gla-domain of factor IX determined by NMR and molecular dynamics simulation64,65 are shown. In addition to the γ-carboxyglutamic acid residues, Tyr 1 and Ser 3 provide ligands to calcium 5 and Asn 2 provides a ligand to calcium 3. Tyr 1, Ser 3, and Asn 2 are shown in light grey. Calcium ions 3 through 5 are buried within the protein and not exposed to water. (B) The calcium ligands for conantokin G determined by NMR and molecular dynamics simulation78 are shown. Each calcium ion is coordinated by three or four carboxylate oxygens contributed by two γ-carboxyglutamic acid residues. The γ-carboxyglutamic acid residues and the bound calcium ions are solvent exposed.
The calcium-carboxylate networks of the factor IX Gla-domain and conantokin G. One role of γ-carboxyglutamic acid is to stabilize a protein structure by formation of an extended calcium-carboxylate network. This property of γ-carboxyglutamic acid is used to stabilize the factor IX Gla-domain and conantokin G. (A) The calcium ligands for the Gla-domain of factor IX determined by NMR and molecular dynamics simulation64,65 are shown. In addition to the γ-carboxyglutamic acid residues, Tyr 1 and Ser 3 provide ligands to calcium 5 and Asn 2 provides a ligand to calcium 3. Tyr 1, Ser 3, and Asn 2 are shown in light grey. Calcium ions 3 through 5 are buried within the protein and not exposed to water. (B) The calcium ligands for conantokin G determined by NMR and molecular dynamics simulation78 are shown. Each calcium ion is coordinated by three or four carboxylate oxygens contributed by two γ-carboxyglutamic acid residues. The γ-carboxyglutamic acid residues and the bound calcium ions are solvent exposed.
The Plasma Proteins of Blood Coagulation
The vitamin K-dependent blood coagulation and regulatory proteins contain 10 to 12 γ-carboxyglutamic acid residues in the Gla domain, located within the first 40 residues of the N-terminus of the mature proteins. The Gla domain in association with the adjacent aromatic amino acid stack domain functions as a membrane binding component of these proteins. γ-Carboxyglutamic acid is a metal binding amino acid that confers metal binding properties on the vitamin K-dependent proteins. With the addition of calcium ions, these proteins undergo a structural transition that leads to the exposure of a phospholipid binding site. In most cases in which it has been investigated, neither aspartic acid nor glutamic acid will substitute for the function of γ-carboxyglutamic acid, emphasizing the importance of both carboxyl groups. The inability of glutamic acid to substitute for γ-carboxyglutamic acid is perhaps best illustrated by the usefulness of the anticoagulant drug sodium warfarin. As described above and shown in Fig 1, sodium warfarin prevents the recycling of vitamin K from the oxidized to the reduced form, leading to decreased γ-glutamyl carboxylation due to insufficient active cofactor. The abnormal forms of the vitamin K-dependent proteins are undercarboxylated, having glutamic acid residues at some or all of the positions that are usually carboxylated, and are for the most part biologically inert. Systematic mutation of individual carboxylatable glutamic acid residues in recombinant prothrombin36 or protein C35,62 63has yielded molecules with modest to severe functional defects, again indicating the importance of the bifunctional γ-carboxyglutamic acid side chain.
Prothrombin.
The X-ray crystal structure of prothrombin fragment 1 first showed that the Gla domain is highly structured, and many of the γ-carboxyglutamic acid residues in this domain coordinate internal calcium ions that are not exposed to solvent.12 Most of the γ-carboxyglutamic acid side chains point inward to a linear array of internal calcium ions. Several of these calcium ions are completely sequestered inside the core of the Gla domain, ie, not exposed to solvent.
Factor IX.
The factor IX Gla domain, studied by 2D NMR spectroscopy in a peptide consisting of the Gla-aromatic amino acid stack domain, is characterized by a similar fold of the polypeptide backbone.64 The location of the calcium binding sites in the internal core structure is nearly identical in prothrombin and factor IX.65
Factor VII.
The crystal structure of the factor VII-tissue factor complex showed the structure of the Gla domain of factor VII in the presence of calcium ions.66 Its structure is nearly identical to its homolog in prothrombin.
The phospholipid membrane binding site of the vitamin K-dependent proteins is expressed on the surface of the Gla domain. X-ray structure showed the curious exposure of three hydrophobic residues on the protein surface, suggesting a potential role for these residues in membrane interaction.12 Site-specific mutagenesis of homologous residues in protein C interfered with the membrane binding properties of this protein, but mutation of other hydrophobic residues did not perturb membrane binding significantly.67Sunnerhagen et al68 compared the NMR-determined solution structure of the calcium-free Gla-EGF domain pair from factor X with a model of the calcium-bound factor X Gla domain based on presumed structural homology with prothrombin and concluded that calcium-induced exposure of hydrophobic amino acids in the Gla domain is critical for membrane binding. Direct comparison of the NMR determined structures of the calcium-stabilized form of factor IX, which binds to membranes, and the magnesium-stabilized form of factor IX, which does not bind to membranes, implicated the N-terminal 11 amino acid residues that form a loop in the Gla domain.69 Based on correlation of membrane binding properties of the vitamin K-dependent proteins of plasma with homology considerations, McDonald et al70 has recently proposed an alternative membrane contact site that implicates residues 11, 33, and 34 in this process.
Gla-Containing Proteins of Mineralized Tissue
Whereas the role of γ-carboxyglutamic acid is well defined in the plasma proteins, this role remains uncertain in proteins/peptides outside of this family. There have been no successful studies to date on the structure of γ-carboxyglutamic acid-containing bone proteins, including osteocalcin (bone Gla protein) and matrix Gla protein. Although these proteins bind to calcium ions, the specific structural role of γ-carboxyglutamic acid has not yet been defined. Osteocalcin function remains uncertain. However, increased bone formation, including higher bone mass and bones of improved functional quality, were observed in osteocalcin-deficient mice.71 Spontaneous and ultimately fatal calcification of arteries and cartilage was observed in mice lacking matrix Gla protein,72 suggesting that one of its functions is to control and limit extraosseous calcification.
Other Mammalian Vitamin K-Dependent Proteins
Gas6 is a vitamin K-dependent protein with marked sequence homology in the Gla domain to the vitamin K-dependent blood coagulation and regulatory proteins and in particular with protein S.32Gas6 is released from and potentiates the growth of vascular smooth muscle cells. Gas6 is a calcium-dependent ligand for the receptor tyrosine kinases Axl and Sky/Rse.73,74 The domain structure includes a Gla domain, loop domain, EGF domain, and two tandem “globular” (or G) domains. This protein contains γ-carboxyglutamic acid, and Gas6 synthesized in the presence of warfarin and thus lacking γ-carboxyglutamic acid demonstrates no thrombin-inducible growth potentiating activity or receptor binding ability.33 In contrast, Gas6 lacking the entire Gla domain is a functional growth factor, indicating that the tandem G domains are sufficient to activate Rse phosphorylation.75 This suggests that the Gla domain may be a negative regulator of the structure of a growth factor domain located elsewhere on the molecule.
PRGP1 and PRGP2, two proteins named for theirProline-Rich Gla Protein characteristic, were identified by searching an expressed sequence tag database with a protein query sequence based on a consensus sequence derived from the analysis of the Gla domain of the vitamin K-dependent blood clotting and regulatory proteins.76 PRGP1, with a predicted molecular mass of 23 kD, is composed of 198 amino acids after cleavage of the putative propeptide. PRPG2, with a predicted molecular mass of 17 kD, has 153 amino acids after the cleavage of the putative signal peptide and propeptide. Both proteins are characterized by Gla domains preceded by prototypic propeptides, a transmembrane domain and a cytoplasmic domain rich in proline and containing the motifs PPXY and PXXP. These proteins are expressed in a variety of extrahepatic tissues, in contrast to the blood clotting proteins. The function of these newly discovered proteins is unknown.
As discussed above, the carboxylase itself has sequence homology with the region of matrix Gla protein containing the γ-carboxylation recognition site.44 Under some conditions, the carboxylase has been reported to be carboxylated,45 although the role of γ-carboxyglutamic acid in the enzyme remains to be determined.
Conantokins
Despite an extensive search for γ-carboxyglutamic acid across most phyla, the distribution of this amino acid had been thought to be limited to vertebrates—until a conotoxin from the venomous cone snail was isolated and sequenced.77 Conotoxins are small, paralyzing neuroactive peptides that are injected into prey after a cone snail harpoons its victim. Conantokin G, the sleeper peptide, is 17 residues long and contains five γ-carboxyglutamic acid residues. In the conantokin G:Ca2+ complex, the Gla side chains are oriented externally, and Gla 3, Gla 7, Gla 10, and Gla 14 are aligned in a linear array.78 This structural motif is clearly distinct from that of the vitamin K-dependent blood coagulation proteins. However, a carboxylate:Ca2+ network is also present. A similar structure and carboxylate:metal ion network has been described in the conantokin G:Mg2+ complex.79 A related conotoxin, conantokin T, has a similar helical structure in the presence of magnesium ions80 and calcium ions.81 A propeptide that serves a similar role as those of vertebrate vitamin K-dependent proteins in identifying them as substrates for the γ-carboxylase has recently been discovered for conantokin G.82 The propeptide sequence does not contain the γ-CRS of vertebrate vitamin K-dependent proteins, and the γ-CRS within the conantokin G propeptide has not been identified. A large number of neuroactive Gla-containing conotoxins have recently been identified.
SUMMARY
The sole known biological role of vitamin K is as a cofactor for the vitamin K-dependent γ-glutamyl carboxylase in the synthesis of γ-carboxyglutamic acid. This enzyme has been purified to homogeneity and both its cDNA sequence and gene structure determined. The mechanism of action of vitamin K in the reaction catalyzed by this enzyme remains hypothetical.
Protein precursors destined to undergo posttranslational γ-carboxylation contain a γ-carboxylation recognition site, often within the propeptide of a precursor protein, that binds to the γ-carboxylase. This recognition element assures conversion of glutamic acid to γ-carboxyglutamic acid on carboxylase substrates. γ-Carboxyglutamic acid is a calcium-binding amino acid and is required for the function of vitamin K-dependent proteins. The blood clotting and regulatory proteins require γ-carboxyglutamic acid for Ca2+-induced interaction with membrane surfaces.
A common feature of vitamin K-dependent proteins is the formation of a network of protein carboxylate ligands bound to Ca2+. This network, first described in prothrombin, is defined by γ-carboxyglutamic acid and is associated with the stabilization of a unique conformer and the exposure of solvent-accessible hydrophobic amino acids that are available for membrane or protein interaction.
ACKNOWLEDGMENT
The authors thank members of this laboratory, past and present, who have contributed so much to the development of our understanding of γ-carboxyglutamic acid synthesis and function.
The work performed in this laboratory was supported by grants from the National Institutes of Health (Grants No. HL18834, HL38216, and HL42443). B.A.B. was supported by a Judith Graham Pool Postgraduate Research Fellowship from the National Hemophilia Foundation.
REFERENCES
Author notes
Address reprint requests to Bruce Furie, MD, BIDMC Cancer Center, KS 158, Beth Israel Deaconess Medical Center, 330 Brookline Ave, Boston, MA 02215.