Abstract
CBFA2(AML1) has emerged as a gene critical in hematopoiesis; its protein product forms the DNA-binding subunit of the heterodimeric core-binding factor (CBF) that binds to the transcriptional regulatory regions of genes, some of which are active specifically in hematopoiesis. CBFA2 forms a fusion gene with ETO andMDS1/EVI1 in translocations in myeloid leukemia and withETV6(TEL) in the t(12;21) common in childhood pre-B acute lymphoblastic leukemia. We have analyzed samples from 30 leukemia patients who had chromosome rearrangements involving 21q22 by using fluorescence in situ hybridization (FISH). Our analysis showed that 7 of them involved CBFA2 and new translocation partners. Two patients had a t(17;21)(q11.2;q22), whereas the other 5 had translocations involving 1p36, 5q13, 12q24, 14q22, or 15q22. Five of these novel breakpoints in CBFA2 occurred in intron 6; this same intron is involved in the t(3;21). One breakpoint mapped to the t(8;21) breakpoint region in intron 5, and 1 mapped 5′ to that region. All 7 CBFA2 rearrangements resulted from balanced translocations. All 7 patients had myeloid disorders (acute myeloid leukemia or myelodysplastic syndrome); 2 were de novo and 5 had treatment histories that included topoisomerase II targeting agents. The association of therapy-related disorders with translocations involving CBFA2 was significant by Fisher’s exact test (P < .003). These results provide further evidence that this region of CBFA2 is susceptible to breakage in cells exposed to topoisomerase II inhibitors.
© 1998 by The American Society of Hematology.
RECURRING CHROMOSOME translocations often are specifically associated with particular subtypes of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). The 8;21 translocation, t(8;21)(q22;q22), is one of the most common rearrangements found in AML and is associated with French-American-British (FAB) subtype M2 (AML-M2), often with a unique t(8;21) phenotype.1,2 The translocation fuses the 5′ part of CBFA2(AML1) on chromosome 21 and nearly the entire geneETO(MTG8) on chromosome 8.3,4 The fusion gene is oriented such that transcription proceeds from the telomeric (5′) end toward the centromere (reviewed in Nucifora and Rowley5).
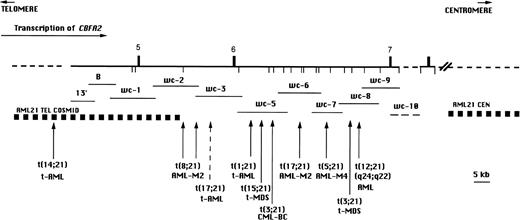
CBFA2 also contributes 5′ sequences to fusion genes produced by the 3;21 translocation.6 The t(3;21) is associated with therapy-related AML and MDS (t-AML/t-MDS)7and with chronic myelogenous leukemia in blast crisis (CML-BC),8 but it is rarely found in AML de novo.9 The t-AML patients with the t(3;21) often were treated with topoisomerase II-targeting agents for their primary malignancies.10-12 The t(3;21) fuses the 5′ region ofCBFA2 with up to three different genes (EAP,MDS1, and EVI1) located over a region of 400 to 750 kb through remarkable alternative splicing.13 The t(3;21) breakpoints are located about 60 kb downstream of the t(8;21) breakpoint region, such that an additional CBFA2 exon (exon 6) is present in the t(3;21) fusion message (Fig 1).6
BamHI restriction map of CBFA2 on chromosome 21 in the region containing the breakpoints of the t(8;21), t(3;21) and newly identified translocations involving CBFA2(vertical arrows). The breakpoint region for the t(12;21)(p13;q22) associated with childhood B-cell ALL is located 5′ of the map area. The numbered vertical bars above the line represent exons and the vertical lines below the line indicate BamHI sites. The direction of transcription is indicated by a horizontal arrow. The recombinant phage walking clones 13′ through wc-10 are indicated by horizontal lines below the map. wc-10 was not precisely mapped and is indicated by a dashed line. Genomic regions indicated by a dashed line have not been precisely mapped.
BamHI restriction map of CBFA2 on chromosome 21 in the region containing the breakpoints of the t(8;21), t(3;21) and newly identified translocations involving CBFA2(vertical arrows). The breakpoint region for the t(12;21)(p13;q22) associated with childhood B-cell ALL is located 5′ of the map area. The numbered vertical bars above the line represent exons and the vertical lines below the line indicate BamHI sites. The direction of transcription is indicated by a horizontal arrow. The recombinant phage walking clones 13′ through wc-10 are indicated by horizontal lines below the map. wc-10 was not precisely mapped and is indicated by a dashed line. Genomic regions indicated by a dashed line have not been precisely mapped.
The t(12;21)(p13;q22) was identified by fluorescence in situ hybridization (FISH) analysis with whole chromosome painting probes for chromosome 12 in 3 of 8 patients with acute lymphoblastic leukemia (ALL); the breakpoint on 21q was localized by using chromosome 21-specific painting probes and yeast artificial chromosomes (YACs) for that region.14 A fusion gene forTEL/AML1(ETV6/CBFA2), from 12p13 was cloned.15-17In contrast to the fusion genes produced in the t(8;21) and t(3;21), the promoter and 5′ sequences in this fusion are provided byETV6, and the 3′ sequences include almost the entireCBFA2 gene, including the runt homology domain. Moreover, all of these patients had B-precursor ALL (CD10+, CD19+). The t(12;21) has been shown to occur in about 25% of childhood B-cell ALL and appears to be associated with a good prognosis.18 19
CBFA2(AML1) shows homology to the Drosophilasegmentation gene runt, which has a DNA-binding region as well as a domain for protein-protein interaction. The AML1/ETOfusion gene in the t(8;21)4 as well as the fusion genes formed by the t(3;21) (AML1/EAP, AML1/EVI1, andAML1/MDS) include the runt domain.6 Further downstream, CBFA2 has a transactivation domain. CBFA2 forms a heterodimer with the subunit CBFB, which confers a greater DNA-binding activity. Remarkably, CBFB is rearranged by the inv(16) associated with AML-M4Eo, producing a fusion geneCBFB/MYH11.20 Significantly, CBFA2 has a large number of splice variants in both the 5′ and 3′ regions, and at least two promoters (two 5′UTRs) have been identified.21 Mouse homologs have also been identified; mice with homozygous mutations in CBFA2 are deficient in hematopoiesis and die during embryonic development.22 23
We asked whether translocations involving 21q22 and other chromosome partners would show breaks within the CBFA2 gene, in a manner similar to the large number of partner chromosomes that produce fusion genes with 5′ sequences derived from MLL on chromosome 11 band q23 (reviewed in Rowley24). A preliminary survey identified 2 patients with translocation breakpoints localized within the same intron of CBFA2(AML1), which is involved in the majority of t(3;21) patients.25 We show here that these balanced translocations involving CBFA2 occur predominantly in therapy-related disorders and define a genomic region that appears to be susceptible to exposure to cytotoxic drugs, especially those that target DNA topoisomerase II.
MATERIALS AND METHODS
Patient material and cytogenetic analysis.
We examined samples from 30 patients (27 bone marrow and 3 peripheral blood) with rearrangements of chromosome band 21q22, not including t(3;21) or t(8;21). Most patients were identified at the University of Chicago Hematology/Oncology Cytogenetics Laboratory between 1970 and 1996; 1 sample was contributed from the University Hospital (Copenhagen, Denmark). Most samples were obtained at diagnosis of leukemia (N = 25), 4 were obtained at the time of relapse, and 1 after initiation of treatment. Diagnosis was based on morphologic and cytochemical studies of peripheral blood smears and bone marrow aspirate and biopsy specimens based on the FAB classification. Twenty-four patients had myeloid disorders (AML de novo, primary MDS, t-AML, and t-MDS) and 6 had ALL. Cytogenetic analysis with a trypsin-Giemsa banding technique was performed on cells from bone marrow and/or peripheral blood according to standard procedures. Chromosomal abnormalities are described according to the International System of Human Cytogenetic Nomenclature (1995).
FISH analysis.
The procedure used for FISH has been described.26 Probes were labeled with biotin or digoxigenin and detected by fluorescein isothiocyanate (FITC)-conjugated avidin or rhodamine-conjugated antidigoxigenin antibodies. Metaphase chromosomes were identified by 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) staining. Fluorescence images were captured using a cooled charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) and Nu200 software. Separate images of DAPI-stained cells and the fluorescence signal were merged using NIH Image 1.60 software (NIH, Bethesda, MD).
Probes.
For CBFA2(AML1), the contiguous phage λ walking clones (probe 13′ and wc-1 through wc-10) and cosmid contigs that flank theCBFA2 breakpoint (AML21 centromeric and AML21 telomeric probes; Oncor, Inc, Gaithersburg, MD) have been described (Fig1).4,6,27 A chromosome 21 painting probe (WCP 21q SpectrumOrange; Vysis, Downers Grove, IL) was used for confirmation of derivatives. For chromosome 17, two cosmids containing sequences fromRARA were used: COS34A, which is split by the t(15;17) associated with acute promyelocytic leukemia, and PC7-611, which is centromeric to the breakpoint (Richard S. Lemons, MD, University of Utah, Salt Lake City, UT).27 The NF1YAC was a gift from Douglas A. Marchuk, PhD (Duke University Medical Center, Durham, NC). For chromosome 5, YAC A (contains D5S112 sequences) and YAC C (contains D5S401) were used; both are distal to the SMA locus in 5q13 (Lalitha Nagarajan, PhD, MD Anderson Cancer Center, Houston, TX).
RESULTS
Breakpoints on 21q22.
Seven patients were shown to have reciprocal translocations involving CBFA2 when the cosmid contigs that flank the t(8;21) breakpoint were used. The locations of the breakpoints, clinical information, and complete karyotypes of these patients are listed in Table 1. With trypsin-Giemsa banding, 2 patients had apparently identical translocations t(17;21) and either de novo or t-AML (patients no. 1 and 4, Table 1; Fig 2A and B). A cryptic translocation, t(1;21)(p36;q22), was detected in patient no. 7 (Fig 3F), who, by standard trypsin-Giemsa banding, appeared to have an interstitial deletion, del(21)(q21q22.1) (patient no. 70 reported in Pedersen-Bjergaard and Philip12). In patient no. 6, with the t(14;21), both derivative chromosomes were labeled with the telomeric cosmid contig (Fig 3E). The der(21)t(14;21), but not the der(14), was labeled by probe 13′, which was the most telomeric (5′) walking clone used in this study, indicating that this patient’s breakpoint maps 5′ of exon 5 of CBFA2 (Fig 1). The telomeric cosmid contig is 5′ to the t(8;21) breakpoint and 3′ to the t(12;21) breakpoint in 5 of 7 patients.27 Therefore, the breakpoint in the t(14;21) is between that in most t(12;21) ALL patients and the t(8;21) breakpoint region.
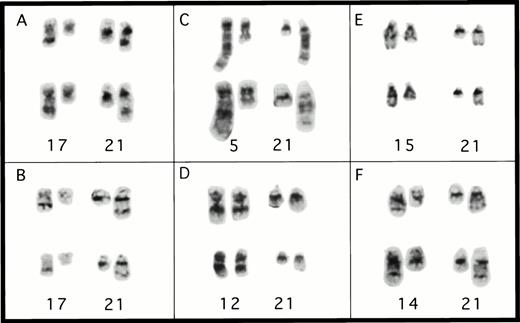
Partial karyotypes from patients with rearrangements ofCBFA2. (A) Patient no. 1, t(17;21)(q11.2;q22); (B) patient no. 4, t(17;21)(q11.2;q22); (C) patient no. 2, t(5;21)(q13;q22); (D) patient no. 3, t(12;21)(q24;q22); (E) patient no. 5, t(15;21)(q22;q22); and (F) patient no. 6, t(14;21)(q22;q22). The abnormal homolog is on the right in every example.
Partial karyotypes from patients with rearrangements ofCBFA2. (A) Patient no. 1, t(17;21)(q11.2;q22); (B) patient no. 4, t(17;21)(q11.2;q22); (C) patient no. 2, t(5;21)(q13;q22); (D) patient no. 3, t(12;21)(q24;q22); (E) patient no. 5, t(15;21)(q22;q22); and (F) patient no. 6, t(14;21)(q22;q22). The abnormal homolog is on the right in every example.
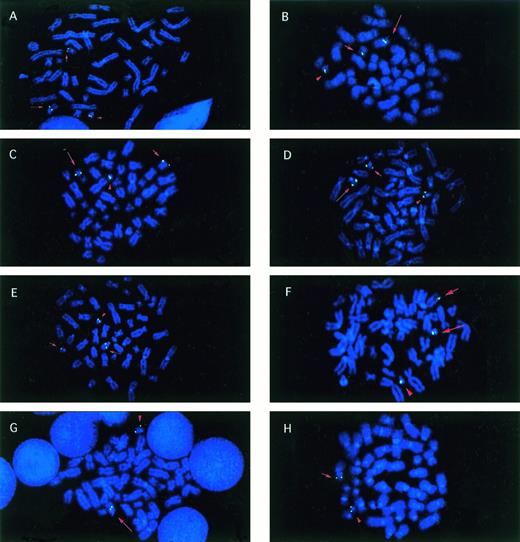
Metaphase cells from patients with balanced reciprocal translocations involving CBFA2. Signals (green) fromCBFA2 probes that were split by the translocation are indicated on the der(21) (long arrow), the partner derivative (short arrow), and the normal 21 (arrowhead) chromosomes in all examples, except as noted. (A) Patient no. 1 with the t(17;21)(q11.2;q22) hybridized with the biotinylated clone wc-6. (B) Patient no. 2, t(5;21) with walking clone 7. (C) Patient no. 3, t(12;21) with wc-8. (D) Patient no. 5, t(15;21) with wc-5. (E) Patient no. 6, t(14;21) with AML21-telomeric cosmid contig. (F) Patient no. 7, t(1;21)(p36;q22) with wc-5. (G) Patient no. 4, t(17;21) with AML21-centromeric contig. The der(21) (long arrow) and normal 21 (arrowhead) were labeled. (H) Patient no. 4, t(17;21) with AML21-telomeric contig. The der(17) (short arrow) and normal 21 (arrowhead) were labeled.
Metaphase cells from patients with balanced reciprocal translocations involving CBFA2. Signals (green) fromCBFA2 probes that were split by the translocation are indicated on the der(21) (long arrow), the partner derivative (short arrow), and the normal 21 (arrowhead) chromosomes in all examples, except as noted. (A) Patient no. 1 with the t(17;21)(q11.2;q22) hybridized with the biotinylated clone wc-6. (B) Patient no. 2, t(5;21) with walking clone 7. (C) Patient no. 3, t(12;21) with wc-8. (D) Patient no. 5, t(15;21) with wc-5. (E) Patient no. 6, t(14;21) with AML21-telomeric cosmid contig. (F) Patient no. 7, t(1;21)(p36;q22) with wc-5. (G) Patient no. 4, t(17;21) with AML21-centromeric contig. The der(21) (long arrow) and normal 21 (arrowhead) were labeled. (H) Patient no. 4, t(17;21) with AML21-telomeric contig. The der(17) (short arrow) and normal 21 (arrowhead) were labeled.
Only 2 of the 30 patients had 21q22 breakpoints proximal toCBFA2; 1 of these 2 had a t(10;21)(q22;q22) and the other had a complex karyotype with a gain of a der(21)t(15;21). The other rearrangements of 21q22 were distal (telomeric) to CBFA2(N = 14) or proved to be complex rearrangements (N = 3) or unbalanced translocations leading to loss of chromosomal material (N = 4), rather than balanced reciprocal translocations.
Localization of breakpoints on partner chromosomes.
Two patients [patient no. 1, t(17;21), and patient no. 2, t(5;21)] had sufficient material to investigate the location of the breakpoints of the partner chromosomes. For patient no. 1, two cosmid probes specific for the RARA locus localized to the der(21)t(17;21). A YAC probe containing sequences for NF1 localized to the der(17). Thus, the breakpoint on chromosome 17 is between the loci forNF1 and RARA.
For patient no. 2, two YAC probes that are distal to the SMAlocus in 5q13 were shown to flank the breakpoint: YAC A, which includes D5S112, was on the der(5) and YAC C, which includes D5S401, was on the der(21). This spans a distance of about 10 Mb (L. Nagarajan, personal communication, July 1998).
Association with previous chemotherapy.
Of the 7 patients with balanced translocations involving CBFA2, 5 had a history of previous chemotherapy, whereas 2 did not. Four of the 5 had presented with t-AML or t-MDS, and the fifth patient was found to have a t(12;21)(q24;q22) after chemotherapy for AML evolving from primary MDS. In contrast, of the 23 patients with 21q22 rearrangements but determined not to have translocations ofCBFA2, only 2 had t-AML/t-MDS. The karyotypes and FISH results for these 2 patients are listed in Table 2, as are the treatment histories for all of the t-AML/t-MDS patients. The association of a history of chemotherapy with CBFA2translocations was significant by a Fisher’s exact test (P < .003, N = 30).
The fifth patient with a translocation in CBFA2 and a history of chemotherapy (patient no. 3) presented a diagnostic problem at the time when the t(12;21)(q24;q22) was found. Initially, this 66-year-old man had been diagnosed with primary MDS (refractory anemia with excess blasts in transformation) progressing toward AML-M6 (Table 1). A normal karyotype was noted, and interphase analysis of 500 cells from this pretreatment sample did not indicate a CBFA2 translocation. The patient received daunorubicin and cytarabine and achieved a complete remission but relapsed after 11 months. He then had residual disease for 18 months, at which time Auer rods were noted in a peripheral blood sample. A bone marrow sample was analyzed, and the t(12;21) was noted in 18 of 22 cells. Hence, the t(12;21) represents a change from the pretreatment sample, but it is not clear whether it was associated with a therapy-related myeloid leukemia or progression of the initial disease.
DISCUSSION
We have shown that CBFA2 is involved in reciprocal translocations between 21q22 and chromosome bands 17q11.2, 5q13, 1p36, 12q24, 15q22, and 14q22 in myeloid disorders (AML, MDS, t-AML, and t-MDS). The breakpoints for 5 of the 7 CBFA2 translocations were localized to intron 6, the same intron that contains the breakpoint in most patients with the t(3;21).6 This is one intron downstream from the location of most of the breakpoints in the t(8;21).28 By analogy with the t(8;21) and t(3;21), the fusion transcript should include the 5′ region through exon 6. It will be of interest to compare motifs of these partner genes with those of ETO and EAP, MDS, and EVI1 to see whether they share similar characteristics.
Thus, at present, we know of 9 translocation partners forCBFA2. Six are described here; 1 of these was found to be a potential recurring abnormality [the t(17;21)]. In 8 translocations, including the t(8;21) and t(3;21), CBFA2 provides the 5′ part of the fusion gene and the leukemias are myeloid, whereas in the t(12;21)(p13;q22), CBFA2 is 3′ and the phenotype is ALL.
The finding of 2 AML patients (1 with AML-M2 [patient no. 1] and 1 t-AML [patient no. 4]) with identical cytogenetic breakpoints t(17;21)(q11.2;q22) suggests a rare recurring abnormality, despite the different diagnoses. It is well established that recurring translocations involving 11q23 occur after treatment with topoisomerase II inhibitors,10,11,29,30 and MLL was demonstrated to be affected in such cases.31 Thus, by analogy toMLL, it is not surprising to find that CBFA2 is translocated in both de novo and therapy-related myeloid disorders with a balanced translocation of 21q22, even when the partner chromosome is the same. In fact, the t(3;21) that involves CBFA2 and is usually associated with prior therapy has also been observed in at least one de novo case.9
The latency period from the initial treatment for the primary neoplasia to development of t-AML/t-MDS in these 4 patients had a mean of 59 months (range, 52 to 63 months). The interval from initial exposure to topoisomerase II inhibitors to development of t-AML/t-MDS was, on average, 53 months (range, 36 to 63 months), which is longer than for most MLL translocations. In one such study, the time from initial treatment to t-AML/t-MDS was an average of 23 months (range, 9 to 35 months; N = 9); for the time from first topoisomerase II exposure, the average was 20 months (range, 9 to 35 months).31 Furthermore, t-MDS was present before t-AML in 2 patients reported here (patients no. 5 and 6), whereasMLL-rearranged t-AML typically presents abruptly, without a preleukemic phase.
The association of CBFA2 rearrangements with prior therapy supports the conclusions of Stanulla et al,32 who found site-specific CBFA2(AML1) cleavage inducible in vivo by topoisomerase II inhibitors for several cell types. In their study,CBFA2 sites were less specific (in that fainter bands suggestive of additional cleavage sites were found) and less sensitive (in that fewer cell lines and fewer cells had significant breakage at the main cleavage site) than in MLL. The major site-specific cleavage site in CBFA2 mapped to intron 5, slightly 5′ of the breaks for most of the patients reported here. In this study, we found 2 of 4 therapy-related breakpoints in intron 6, 1 was 5′ of exon 5 and only 1 breakpoint was in intron 5 (Fig 1 and Table 1). Unfortunately, there was no material available to determine the precise location of this patient’s breakpoint. However, the 2 breakpoints in intron 6 are located in wc-5, within 10 to 20 kb of the in vivo cleavage site (Fig 1). This same walking clone contained the breakpoint of a t(3;21) from a CML blast crisis patient.6 It will be important to examine this region for the presence of a topoisomerase II consensus cleavage site. However, characterizaton of the functions of the various CBFA2 fusion proteins will be critical to understanding the process of transformation in de novo and therapy-related leukemia.
NOTE ADDED IN PROOF
Another CBFA2 translocation has been described, the t(16;21)(q24;q22), a rare translocation found in patients with MDS (N=1) or t-MDS/t-AML (N=3). The CBFA2 breakpoint occurs between exons 5 and 6. As in the t(8;21) and t(3;21), CBFA2contributes 5′ sequences to a fusion gene. The partner gene,MTG16, is highly homologous toMTG8(ETO).33
ACKNOWLEDGMENT
For expert technical assistance, we thank the staff of the Gene Mapping Laboratory. For the contribution of patient material used in the survey, we thank Jens Pedersen-Bjergaard, MD, PhD, Avery Sandberg, MD, DSc, Rod Morgan, MS, and Andrew Carroll, PhD. We thank the following physicians for providing patient treatment histories: John E. Godwin, MD, Patrick J. Stiff, MD, Jim Cohn, MD, and Michael J. Thirman, MD.
Supported in part by National Institutes of Health Grants No. CA40046 (J.D.R., R.A.L., and M.M.L.) and CA42557 (J.D.R.) and by the G. Harold and Leila Y. Mathers Foundation (J.D.R.). G.N. is a Scholar of the Leukemia Society of America.
Address reprint requests to Diane Roulston, PhD, Section of Hematology/Oncology, The University of Chicago Pritzker School of Medicine, 5841 S Maryland Ave (MC2115), Chicago, IL 60637.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.