Abstract
Mucin-like molecules represent an emerging family of cell surface glycoproteins expressed by cells of the hematopoietic system. We report the isolation of a cDNA clone that encodes a novel transmembrane isoform of the mucin-like glycoprotein MGC-24, expressed by both hematopoietic progenitor cells and elements of the bone marrow (BM) stroma. This molecule was clustered as CD164 at the recent workshop on human leukocyte differentiation antigens. CD164 was identified using a retroviral expression cloning strategy and two novel monoclonal antibody (MoAb) reagents, 103B2/9E10 and 105.A5. Both antibodies detected CD164/MGC-24v protein expression by BM stroma and subpopulations of the CD34+ cells, which include the majority of clonogenic myeloid (colony-forming unit–granulocyte-macrophage [CFU-GM]) and erythroid (blast-forming unit-erythroid [BFU-E]) progenitors and the hierarchically more primitive precursors (pre-CFU). Biochemical and functional characterization of CD164 showed that this protein represents a homodimeric molecule of approximately 160 kD. Functional studies demonstrate a role for CD164 in the adhesion of hematopoietic progenitor cells to BM stromal cells in vitro. Moreover, antibody ligation of CD164 on primitive hematopoietic progenitor cells characterized by the cell surface phenotype CD34BRIGHTCD38− results in the decreased recruitment of these cells into cell cycle, suggesting that CD164 represents a potent signaling molecule with the capacity to suppress hematopoietic cell proliferation.
© 1998 by The American Society of Hematology.
A WIDE VARIETY OF cell surface molecules participate in the regulation of hematopoiesis. Of these, cell adhesion molecules (CAMs) play a major role in mediating interactions between primitive hematopoietic progenitor cells (HPCs) and various components of the bone marrow (BM) stroma. Based on domain structure and function, these CAMs can be classified into 5 main groups, including the Ig, integrin, cadherin, selectin, and mucin-like molecules families.1 2
The mucin-like molecules represent an emerging family of glycoproteins expressed by tissues of the hematopoietic system.3-6 Within this family are the L-selectin ligands GlyCAM-1 and CD34,6,7 PSGL-18 and MAdCAM-1,9,10a counter receptor on high endothelial venules (HEV) in mucosal lymph nodes for L-selectin, and integrin α4β7.11 Other members include the ubiquitously expressed CD43 (leukosialin, sialophorin),12-15 CD45RA,16,17CD68,18 and tactile (CD96).19 At least four of these mucin-like molecules are expressed at high level on primitive human HPCs, including CD34,20,21 CD43,22,23CD45RA,24 and PSGL-1 (CD162).25,26 Although exhibiting limited homology at the cDNA level, mucin-like molecules all share the common characteristic of being highly glycosylated polypeptides, containing predominantly O-linked carbohydrate side chains linked to serine and threonine residues.3,27,28Their nonglobular, thread-like structure resembles that of classical mucins expressed at the surface of and in the mucosal secretions of epithelial cells. Carbohydrate chains in mucins are predominantly attached by an α1,3 linkage between N-acetyl galactosamine and the oxygen atom of serine or threonine (O-glycosidic bond), although some oligosaccharides are attached by a linkage between the nitrogen of asparagine and N-acetylgalactosamine (N-glycosidic bond).27 28
The dense array of O-linked side chains in mucin-like molecules conveys at least two important structural implications that may influence function.3 The first is the extended structure, making many of the mucin-like molecules long enough to protrude beyond the polysaccharide glycocalyx that surrounds the cell. The second being the optimal exposure and high multiplicity of the terminal sugars. By virtue of their negative charge and extended configuration, mucin-like glycoproteins may act as a repulsive barrier around the cell; however, when an opposing cell has specific receptors for the mucin, adhesion surmounts repulsion. This is well exemplified by the molecular partners for the members of the selectin family, including the P-/E-/L-selectin ligand, PSGL-1,8,29,30 and the L-selectin ligands, GlyCAM-1,7 murine CD34,31,32 and MAdCAM-1,9 10 which all mediate a rapid proadhesive tethering of leukocytes to endothelia under conditions of flow.
The studies presented here describe the isolation of a cDNA clone that encodes a transmembrane mucin-like glycoprotein expressed by both HPCs and elements of the BM stroma. Recently clustered as CD164,33 the cDNA clones were identified using two novel monoclonal antibodies (MoAbs), 103B2/9E10 and 105.A5, and a previously described retroviral expression cloning strategy.34 35 Flow cytometric analyses using both MoAbs showed that the CD164 protein was expressed by subpopulations of CD34+ cells. These include the majority of clonogenic myeloid (colony-forming unit–granulocyte-macrophage [CFU-GM]) and erythroid (blast-forming unit-erythroid [BFU-E]) progenitors and the hierarchically more primitive precursors (pre-CFU). Biochemical and functional characterization of CD164 indicate that this protein exists as a homodimeric molecule of approximately 160 kD that participates in the adhesion of CD34+ cells to BM stroma and functions as a signaling molecule, inhibiting the recruitment of primitive HPCs into cell cycle.
MATERIALS AND METHODS
Generation and Characterization of the CD164 MoAbs, 105.A5 and 103B2/9E10
CD164-specific MoAbs (MGC-24v MoAbs), 9E10/103B2 (MGC-24v.1) and 105.A5 (MGC-24v.2), were generated after immunization of BALB/c mice with the megakaryocytic cell line MOLM-136 according to previously described methods.37 In brief, BALB/c mice were injected intraperitoneally 3 to 4 times with 107 MOLM cells in 300 μL phosphate-buffered saline (PBS) at weekly intervals. Ten days after the final intraperitoneal immunization, mice were boosted with 5 × 105 cells administered intrasplenically. Four days later, the spleen cells were fused with the SP2/0 myeloma cell line using polyethylene glycol (PEG; Sigma Chemical Co, St Louis, MO) using a modification of the method first described by Köhler and Milstein.38 The resulting hybridomas were grown in RPMI 1640 containing 10% (vol/vol) fetal calf serum (FCS; PA Biologicals, Sydney, Australia) supplemented with hypoxanthine-aminopterin-thymidine (HAT; Sigma). Culture supernatants were screened on the immunizing cell line and positive hybridomas were cloned twice by limiting dilution. MoAbs 105.A5 and 103B2/9E10 were isotyped as IgM and IgG3, respectively, by means of a mouse MoAb isotyping enzyme-linked immunosorbent assay (ELISA; Boehringer Mannheim, Mannheim, Germany), as recommended by the manufacturer.
Preparation of BM Mononuclear Cells (BMMNCs)
Normal BM was aspirated into preservative-free, sodium heparin-containing tubes (1,000 U/mL; Fisons Pharmaceuticals, Homebush, New South Wales, Australia) from the sternum and posterior iliac crest of healthy young volunteers after informed consent had been obtained. The use of normal BM cells for these studies was approved by the Human Ethics Committee of the Royal Adelaide Hospital. Low-density BMMNCs were collected after centrifugation at 400g over Ficoll-Hypaque (Lymphoprep, 1.077 g/dL; Nycomed Pharma AS, Oslo, Norway) for 30 minutes at room temperature. MNCs were obtained by selecting the interface cells, which were subsequently washed three times by centrifugation at 4°C in HHF (Hank’s balanced salt solution [HBSS; Life Technologies, Gaithersburg, MD] supplemented with 20 mmol/L HEPES, pH 7.35, and 5% [vol/vol] FCS).
Isolation of CD34+ Cells From Normal Human BM
BMMNCs obtained as described above were incubated in blocking buffer (HHF supplemented with 2% normal human serum) for 30 minutes on ice as described above. Labeling was performed with the anti-CD34 HPCA-2-phycoerythrin (PE) (Becton Dickinson, Mountain View, CA) as previously described.39 Cell sorting was performed using a FACStarPLUS cell sorter and the threshold for selection of CD34+ cells was based on the level of staining obtained with an isotype-matched control IgG1-PE antibody (Coulter, Hialeah, FL). CD34+ cells within the lymphocyte/blast region40 were sorted into Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 50 Kunitz units/mL DNase I (Sigma) and 20% FCS. Purity of the separated CD34+ cells was assessed by analysis of an aliquot of sorted cells and was routinely greater than 98%.
Alternatively, BMMNCs obtained as described above were washed twice with ice-cold HHF buffer before the addition of anti-CD34 (561) Dynabeads (Dynal, Oslo, Sweden) at a 1:1 ratio of beads:cells. This suspension was incubated at 4°C on a rotary mixer for 60 minutes. Cells rosetted by the CD34 Dynabeads were purified by multiple rounds of washing and capture using an a cobalt-samarium magnet (MPC-1; Dynal). The CD34+ cells were then recovered by incubating the cell-bead complexes in DETACHaBEAD reagent (Dynal) according to the manufacturer’s recommendation. The released CD34+ cells were washed several times in HHF buffer and a portion was labeled with HPCA-2-PE (as described above) to assess purity. In all experiments performed, this procedure yielded CD34+ populations that were greater than 95% pure. CD34 cells were resuspended in 1× IMDM supplemented with 10% FCS for use in all subsequent assays.
Multiparameter Flow Cytometric Analysis and Sorting of BMMNCs
Multiple-color immunophenotypic analysis was performed to examine the expression of activation/differentiation antigens by CD34+CD164+ cells. Before immunostaining, ficoll-separated BMMNCs were incubated on ice for 10 minutes with HHF-5% (vol/vol) normal human serum (NHS) to block Fc receptors. The cells were labeled for 30 minutes at 4°C with biotinylated MoAb 103B2/9E10, anti-CD34-fluorescein isothiocyanate (FITC), or anti-CD34-PE (clone 8G12), and either anti-HLA-DR (clone L243), anti-CD33-PE (clone P67.6), anti-CD38-PE (clone HB-7), anti-CD71-PE (clone L01.1), anti-CD90-PE (clone 9E10), or anti-CD117-PE (clone 95C3). After washing twice in cold HHF, specifically bound biotinylated 103B2/9E10 was detected by incubation with streptavidin-allophycocyanin (SAV-APC) for 15 minutes at 4°C. Anti-CD34-FITC, anti-CD34-PE, anti-HLA-DR-PE, anti-CD33-PE, anti-CD38-PE, and anti-CD71-FITC were from Becton Dickinson (Heidelberg, Germany). Anti-CD90-PE was purchased from PharMingen (Hamburg, Germany), and anti-CD117-PE was purchased from Immunotech (Hamburg, Germany). Flow cytometric analysis was performed using a Profile II flow cytometer (Coulter). Twenty thousand events were collected per sample as list mode data and analyzed using Coulter ELITE software. Alternately, after cell labeling, subpopulations of cells were selectively isolated using either a FACStarPLUS or FACSVantage cell sorter (Becton Dickinson, Sunnyvale, CA) fitted with a 250-mW argon laser enabling the simultaneous detection of FITC and PE at emission wavelengths of 530 and 570 nm, respectively. The APC fluorescence was excited with a mixed-gas ion laser (Spectrum 70; Coherent) at 647 nm and emission was detected at 670 nm. Appropriate instrument alignment, optimization of the second laser, and compensation of FITC versus PE signals was accomplished using a mixture of Calibrite and APC beads (Becton Dickinson). One hundred thousand events were collected per sample and analyzed using the LYSIS II software (Becton Dickinson) or sorted directly into IMDM (Trace Biosystems, Castle Hill, Australia) supplemented with 10% FCS.
HPC Assays
Clonogenic assays of HPCs.
Various populations of cells obtained by fluorescence-activated cell sorting (FACS) and cells derived from pre-CFU assay (see below) were assayed for their content of granulocyte-macrophage colony forming cells (CFU-GM), primitive erythroid progenitors (BFU-E), committed erythroid colony-forming cells (CFU-E), and multipotential colony-forming cell (colony-forming unit granulocyte, erythroid, monocyte, megakaryocyte [CFU-GEMM] or CFU-Mix), as previously described.41 Cultures were established in triplicate by plating 1 × 103 CD34+ cells or subpopulations thereof per 35-mm dish in 1 mL of IMDM supplemented with 0.9% methylcellulose (Dow Chemicals, Lake Jackson, TX), 30% FCS, 1% deionized bovine serum albumin (BSA; Sigma; Batch 2153), 3 mmol/L L-glutamine, and 5 × 10−5 2-mercaptoethanol. Colony growth was stimulated by the addition of a combination of the following recombinant human (rHu) hematopoietic growth factors: interleukin-1β (IL-1β), IL-3, IL-6, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), stem cell factor (SCF) (all used at 10 ng/mL; generously provided by Amgen Inc, Thousand Oaks, CA), and 4 U of erythropoietin (Eprex; Janssen Cilag, Auckland, New Zealand). All colonies were scored according to standard criteria after 14 days of incubation in a fully humidified incubator containing 5% CO2 in air.
Pre-progenitor cell (pre-CFU) culture.
This is a stroma-free, cytokine-dependent suspension culture system initially described by Iscove et al42 and modified by Haylock et al39 that measures the de novo generation of CFU-GM as an index of precursors (pre-CFU) of CFU-GM. Immunolabeled BMMNCs were sorted into cell fractions using the FACStarPLUS cell sorter and resuspended into pre-CFU medium (IMDM supplemented with 30% FCS, 1% deionized BSA, 3 mmol/L L-glutamine, and 5 × 10−5 mol/L β-mercaptoethanol) at a concentration of 1 × 103cells/mL. Triplicate 1 mL suspension cultures were established in 24-well plates in pre-CFU medium supplemented with each of the following human hematopoietic growth factors (HGFs) at a final concentration of 10 ng/mL: rHu IL-1β, IL-3, IL-6, G-CSF, GM-CSF, and SCF. Clonogenic assays were performed in triplicate to determine the number of CFU-GM in the input population of cells used to initiate the pre-CFU cultures. The cultures were incubated at 37°C in 5% CO2 for 28 days. At days 7, 14, 21, and 28, the contents of each well were removed and washed in IMDM, and cell counts were performed to determine cell production over the previous week. One tenth of the harvested cells were assayed for their content of CFU-GM (as discussed above), and a further tenth were set up in pre-CFU culture with fresh growth medium supplemented with six HGFs. The remainder of the cells were used for immunophenotypic analysis or for the preparation of cytospins to assess cell morphology.
Human BM Stromal Cell (HBMSC) Cultures
Stromal cultures were established essentially as described by Simmons et al.43 BMMNCs were prepared by buoyant density gradient centrifugation as described above. After washing three times in HHF, the BMMNCs were resuspended in 10 mL of α-minimal essential medium (GIBCO, Melbourne, Australia) supplemented with folic acid (0.01 mg/mL), myo-inositol (0.4 mg/mL; Sigma), 50 mmol/L 2-mercaptoethanol, 1 mmol/L hydrocortisone sodium succinate (Sigma), 12.5% FCS, and 12.5% horse serum (CSL, Melbourne, Australia) and cultured in a 25-cm2 flask (Becton Dickinson Labware, Franklin Lakes, NJ). Upon development of confluent stromal layer, the cells were detached using 0.05% (wt/vol) trypsin-EDTA in PBS (GIBCO) and replated in the same medium at 2 × 105 cells/mL in 2 × 75 cm2 tissue culture flasks (Becton Dickinson Labware).
In Situ Immunofluorescence Staining of Cultured HBMSCs
Cultures of HBMSCs cultures were trypsinized as described above and seeded at 2 × 104 cells per well of an 8-chamber slide (Nunc, Inc, Naperville, IL). Before immunostaining, cultures were washed three times with ice-cold HHF and then fixed in acetone/methanol 1:1 at −20°C for 15 minutes. After washing three times in PBS, the cells were blocked in 5% normal goat serum (NGS) for 1 hour at room temperature. The blocking buffer was removed and saturating levels of the anti-CD164 MoAbs 103B2/9E10, 105.A5, or isotype-matched nonbinding controls were added for 60 minutes at room temperature. The slides were washed three times in PBS + 0.05% (vol/vol) Triton X-100 (Sigma). To show primary antibody reactivity, cells were incubated with a 1/50 dilution of FITC-conjugated goat antimouse F(ab)2antisera (Silenus, Hawthorn, Victoria, Australia) for 60 minutes at room temperature. The cells were then washed as described above and mounted in aqueous mountant (Uvinert; BDH, Poole, UK). The labeled specimens were examined using an Olympus BH2-RFCA fluorescence microscope (Olympus, Tokyo, Japan).
Adhesion of CD34+ Cells to BM Stromal Cells
CD34+ cells prepared with 561-Dynabeads (see above) were washed twice, resuspended in 500 μL cell adhesion medium (RPMI-2% FCS), and labeled with sodium chromate as previously described.44 Briefly, 50 to 100 μCi of Na251CrO4 (New England Nuclear, Cambridge, MA) was added to the cells and incubated for 1 hour at 37°C. After radio-labeling, cells were washed three times with adhesion medium; resuspended to 1 × 105cells/mL in either adhesion medium or 20 μg/mL of nonbinding, isotype-matched control MoAbs, 105.A5 or 103B2/9E10 (all diluted in adhesion medium); and incubated at 4°C for 30 minutes. In a separate group, MoAbs P4C2, which binds to VLA-4 (CD49d/CD29) blocking adhesive function (generously provided by Dr E. Wayner, University of Minnesota Medical School, St Paul, MN), and PHM2, a function-blocking MoAb directed to VLA-5 (CD49e/CD29; a gift from Prof R.A. Aitkins, Monash Medical Centre, Melbourne, Australia), were included as adhesion-blocking controls. Triplicate 100-μL aliquots of CD34+ cells were added directly (without washing) into 96-well plates seeded 24 hours with 2 × 104 HBMSCs. The entire procedure was performed on ice. Plates were centrifuged at 1,000 rpm for 5 minutes at 4°C to sediment cells onto the stroma. Plates were quickly warmed for 2 minutes to 37°C using a heating block before transfer to a humidified incubator at 37°C for 30 minutes. Assay medium was removed by aspiration and wells were washed three times by the addition of 150 μL of the adhesion assay medium and flicking off. After the last wash, cell adhesion was examined using an inverted-phase contrast microscope before lysing in 150 μL 1% sodium dodecyl sulfate (SDS), 1% NaOH solution. Lysates were counted after 10 minutes using a gamma counter. Data are presented as the percentage of control adhesion obtained in the presence of a cocktail of IgG3 and IgM nonbinding control MoAbs.
The Effect of CD164 Ligation on Recruitment and Proliferation of HPCs
Terasaki plates (Nunc InterMed A/S, Kamstrup, Denmark) were coated overnight at 4°C with 0.5 μL of the following IgG3-UNLB isotype MoAbs, 103.B2 (CD164), P4C2 (CD29/CD49d), and (Southern Biotech IgG3 negative control; catalogue no. 105-01) diluted to a final concentration of 20 μg/mL in PBS. After three rounds of washing in PBS to remove unbound antibody, wells were then treated with IMDM supplemented with 2% BSA for 2 hours at 37°C to block excess protein binding sites. After blocking, the IMDM/BSA were then replaced with 10 μL complete serum-deprived medium (SDM) composed of IMDM, BSA, low-density lipoprotein (20 μg/mL; kindly provided by Prof P. Barter, Lipid Research Laboratory, Hanson Centre for Cancer Research, Adelaide, Australia), recombinant human insulin (Novo Nordisk, Copenhagen, Denmark), and 2-ME (5 × 10−5 mol/L; Sigma) supplemented with purified recombinant human IL-3, IL-6, G-CSF, and SCF (all generously supplied by AMGEN Inc, Thousand Oaks, CA) at concentrations of 10, 10, 100, and 100 ng/mL, respectively. BMMNCs prepared and labeled with CD34 and CD38 antibodies as previously described were then subjected to FACS using a FACStarPLUS cell sorter equipped with an automatic cell deposition unit (ACDU) to deposit single CD34+CD38− cells into each well of the antibody-coated Terasaki plates. Two plates (120 wells) coated with each of the three antibodies were seeded with single cells in each experiment. The plates were then incubated at 37°C in a fully humidified atmosphere containing 5% CO2 in air for the duration of the experiment. The accuracy of single-cell deposition was assessed by visual inspection of all plates using an inverted phase contrast microscope 2 to 3 hours after deposition. Thereafter, the plates were examined on days 3, 7, 10, and 14 to determine the proportion of single CD34+CD38− cells that were induced to divide under each antibody condition. A positive response in this assay is defined as greater than 1 division during the 14-day time course of the experiment. For cells recruited into division, their subsequent proliferative history was monitored by counting the number of cells per well at each successive time point.
Protein Analysis
Immunoprecipitation and PAGE.
Biotinylated NP40-lysates of cells were prepared as described by Cole et al.45 Goat antimouse Ig-coupled Sepharose (AH-Sepharose 4B; Pharmacia, Piscataway, NJ) was washed twice in 1% (vol/vol) NP40-50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1 mmol/L EDTA (TSE) before the addition of 400 μL of hybridoma supernatant. This mixture was then incubated at 4°C for a minimum of 6 hours, with rotation. The resulting prearmed Sepharose was washed twice in 1% NP40 TSE, as described above, and excess supernatant was aspirated to give 12.5 μL of 100% Sepharose. To these, 1.0-mL aliquots of the appropriate NP40 cell lysate were added. The samples were incubated overnight at 4°C with rotation. The immunoprecipitates were then washed twice in 1% (vol/vol) NP40-TSE, once in 0.1% (vol/vol) NP40-TSE, and once in TSE, pH 8.0. The supernatant was then removed and samples stored at −20°C or used immediately for electrophoresis. Each immunoprecipitate represented the material from 1 × 107 cells.
Samples were boiled for 3 minutes in 25 μL reducing sample buffer (62.5 mmol/L Tris, 3% [wt/vol] SDS, 10% [vol/vol] glycerol, and 5% [vol/vol] 2-mercaptoethanol) and analyzed by 10% (wt/vol) SDS-polyacrylamide gel electrophoresis.46 After electrophoresis, proteins were transferred to Hybond-C (Amersham Int, Amersham, Bucks, UK) at 80 mA using a semidry blotting apparatus (Hoefer Scientific Instruments, San Francisco, CA). The filter was blocked by overnight incubation in PBS/3% (wt/vol) BSA at room temperature, washed four times in PBS/0.5% (vol/vol) Tween-20, and subsequently incubated with streptavidin-biotin-HRPO complex (Amersham). The filter was washed four times in PBS/0.5% (vol/vol) Tween-20 and immunoreactive proteins were visualized by enhanced chemiluminescence (ECL; Amersham) as recommended by the manufacturer.
Western blotting.
NP40-lysates of cells were prepared as previously described.45 Lysates were diluted in reducing (inclusion of β-mercaptoethanol) and nonreducing sample buffer and electrophoresed on a 10% SDS-polyacrylamide gel. Proteins were then transferred to Hybond-C membrane as described above. After blocking by overnight incubation at 4°C with 5% skim milk powder-0.05% Tween-20 in PBS, filter strips were incubated with either MoAb 103B2/9E10 or 105.A5 antibodies or isotype-matched controls (all at 10 μg/mL) for 1 hour at room temperature. Filter strips were subsequently incubated with goat antimouse conjugated to horseradish peroxidase (HRPO; Immunotech, Marseille, France). Immunoreactive proteins were visualized by ECL, as recommended by the manufacturer (Amersham).
Expression Cloning of CD164 cDNA
The cDNA encoding the cell surface antigen identified by the MoAbs 103B2/9E10 and 105.A5 were isolated from an HBMSC cDNA library in the retroviral vector, pRUFneo, as recently described.34 Briefly, cDNA synthesised from mRNA from HBMSC cultures was cloned into the retroviral vector pRUFneo. Plasmid DNA from the library was used to transfect a viral packaging line (PA317). Virus-containing supernatant from these cells was used to infect the packing cell line Ψ2, which in turn was used to infect the murine factor-dependent cell line FDC-P1. Infected cells were selected for G418 resistance, and cells expressing genes encoding the 103B2/9E10 and 105.A5 antigens were selected using MoAb and expanded into clonal populations using multiple rounds of immuno-magnetic bead (Dynabead) selection followed by FACS sorting. Genomic DNA prepared from FDC-P1 cells was used in a polymerase chain reaction (PCR) using retroviral specific primers to recover proviral cDNA inserts.
Partial Sequencing of PCR-Rescued cDNA Clones and Computer Analysis
As described previously,34 cDNA clones generated by PCR were gel-purified and subcloned into the pGEM T vector (Promega, Madison, WI), as recommended by the manufacturer. Double-stranded DNA was prepared by standard alkaline lysis mini-prep method47and 1 to 2 μg was used per sequencing reaction. Reactions were prepared using the PRISM Ready Reaction Cycle sequencing kit (Applied Biosystem, Foster City, CA), as recommended by the manufacturer. Reactions analyzing both cDNA strands were run on a Applied Biosystems 373 automated sequence analyzer and 500 to 600 bp of 5′ and 3′ sequence data was routinely obtained per clone. Sequence data were then analyzed by accessing the Genbank and European Molecular Biology Laboratory (EMBL) data bases at the National Centre for Biotechnological Information (NCBI).
Recloning of 103B2/9E10 cDNA Clone Into pRUFneo and Validation of Surface Antigen Expression
After PCR recovery of proviral cDNA inserts from genomic DNA, uniqueBamHI and Xho I restriction sites present in the 5′ and 3′ flanking regions, respectively, were used to reclone the cDNA into the MCS of the retroviral vector pRUFneo.Escherichia coli DH10B cells were transformed as described above and plasmid DNA was isolated using Qiagen-tip 100 (Qiagen, Victoria, Australia) columns as recommended by the manufacturer. Stable, G418-resistant Ψ2 virus-producing cell lines were produced by calcium phosphate transfection and used to infect FDC-P1 cells by cocultivation, as described previously.34G418-resistant FDC-P1 cells were then analyzed for antigen expression by indirect immunofluorescence and flow cytometry.
Detection of CD164 mRNA by Northern Blot Analysis
Total RNA was extracted from cultured HBMSCs and the primitive myeloid cell line, KG1a, using the RNAzol B Method (Biotecx Laboratories, Inc, Houston, TX), as recommended by the manufacturer. Total RNA (20 μg) was separated by electrophoresis on a 1% formaldehyde-agarose gel and transferred overnight by capillary action in 10× SSC onto Hybond N+ (Amersham, Poole, UK) filters. After transfer, the filters were washed in 2× SSC and air-dried, and the RNA was covalently cross-linked to the filters by exposure to 0.4 J/cm2 of UV radiation in a Stratagene UV Stratalinker 1800. Filters were prehybridized for 16 hours at 42°C in a prehybridization solution composed of 50% deionized formamide, 5× SSC (0.34 mol/L NaCl, 75 mmol/L sodium citrate, pH 7.0), 5× Denhardt’s solution, 0.1% SDS, 10 mmol/L HEPES, 1 mmol/L EDTA, 0.05% (2 mmol/L) sodium pyrophosphate, and 200 μg/mL of sheared salmon sperm DNA. Hybridization was performed at 42°C for 16 hours with 50 ng of BamHI/Xho I CD164 cDNA restriction fragment previously labeled with 50 μCi of (α-32P)-ATP (Bresatec, Adelaide, Australia) using the GIGAprime DNA Labelling Kit (Bresatec) kit, as recommended by the manufacturer. The filters were subsequently washed twice in 2× SSC, 0.1% SDS for 10 minutes at room temperature, followed by a single wash in 0.5× SSC, 0.1% SDS at 65°C for a further 10 minutes. After washing, the filter was air-dried and exposed to Kodak X-Omat autoradiography film (Eastman Kodak, Rochester, NY) at −80°C for 72 hours with a Cronex intensifying screen (du Pont, Wilington, DE).
Statistical Analysis
Data points derived from multiple experiments are reported, except where stated, as the mean ± 1 standard error of the mean (SEM). Analysis of the variance to determine significant differences between treatments was performed using ANOVA Factorial analyses. Statistical significance (P ≤ .05, .01) between data sets at each time point was determined using the Fisher PLSD test.
RESULTS
The MoAbs 103B2/9E10 and 105.A5 Identify an Antigen Expressed by Primitive and Committed Human Hematopoietic Progenitors
The murine MoAbs, 103B2/9E10 and 105.A5, were generated by immunization of BALB/c mice with the megakaryocytic cell line MOLM-1. Although exhibiting extensive reactivity with the immunizing cell line, both MoAbs were found to bind only weakly to peripheral blood mononuclear cells (data not shown). In addition, 103B2/9E10 and 105.A5 were found to react with cultured HBMSCs (Fig 1) and a minor subpopulation of BMMNCs (Fig 2) and were therefore selected for further examination.
MoAbs 103B2/9E10 and 105.A5 identify an antigen expressed by cultured HBMSCs. The expression of the antigen identified by MoAbs 103B2/9E10 and 105.A5 was assessed by indirect in situ immunofluorescence (as described in Materials and Methods). Cytoplasmic and membrane staining was detected for both 103B2/9E10 (B) and 105.A5 (D). IgG3 (A) and IgM (C) nonbinding, control antibodies demonstrate no detectable levels of immunofluorescence.
MoAbs 103B2/9E10 and 105.A5 identify an antigen expressed by cultured HBMSCs. The expression of the antigen identified by MoAbs 103B2/9E10 and 105.A5 was assessed by indirect in situ immunofluorescence (as described in Materials and Methods). Cytoplasmic and membrane staining was detected for both 103B2/9E10 (B) and 105.A5 (D). IgG3 (A) and IgM (C) nonbinding, control antibodies demonstrate no detectable levels of immunofluorescence.
(A and B) Dual-parameter immunofluorescence analysis demonstrating the expression of CD34 and 103B2/9E10 (A) or 105.A5 (B) epitopes by BMMNCs. SBA-depleted BMMNCs were stained with the directly conjugated MoAb HPCA-2-PE (-CD34) and 103B2/9E10 (detected with anti-IgG3-FITC). Data are displayed as dual-parameter histograms of 5 × 104 light-scatter-gated events collected as list-mode data. Stat-markers were established with the use of nonbinding, isotype-matched control antibodies as described in Materials and Methods. (C and D) Assay of clonogenic progenitors in populations sorted on the basis of CD34 and MoAb 103B2/9E10 (C) or 105.A5 (D). FACS of the CD34+, CD34+103B2/9E10+, and CD34+103B2/9E10lo/− or CD34+105.A5+, and CD34+105.A5lo/− subpopulations demonstrates that clonogenic progenitors ([▪] CFU-GM, [□] BFU-E, and [▧] CFU-Mix) are present almost exclusively in the CD34+103B2/9E10+ (C) and CD34+105.A5+ (D) subpopulation. Results are expressed as the number of CFU-GM at day 14 per 1 × 103cells plated. Data represent the mean ± SE (n = 3).
(A and B) Dual-parameter immunofluorescence analysis demonstrating the expression of CD34 and 103B2/9E10 (A) or 105.A5 (B) epitopes by BMMNCs. SBA-depleted BMMNCs were stained with the directly conjugated MoAb HPCA-2-PE (-CD34) and 103B2/9E10 (detected with anti-IgG3-FITC). Data are displayed as dual-parameter histograms of 5 × 104 light-scatter-gated events collected as list-mode data. Stat-markers were established with the use of nonbinding, isotype-matched control antibodies as described in Materials and Methods. (C and D) Assay of clonogenic progenitors in populations sorted on the basis of CD34 and MoAb 103B2/9E10 (C) or 105.A5 (D). FACS of the CD34+, CD34+103B2/9E10+, and CD34+103B2/9E10lo/− or CD34+105.A5+, and CD34+105.A5lo/− subpopulations demonstrates that clonogenic progenitors ([▪] CFU-GM, [□] BFU-E, and [▧] CFU-Mix) are present almost exclusively in the CD34+103B2/9E10+ (C) and CD34+105.A5+ (D) subpopulation. Results are expressed as the number of CFU-GM at day 14 per 1 × 103cells plated. Data represent the mean ± SE (n = 3).
Dual-parameter flow cytometric analysis showed that a significant proportion of the 103B2/9E10+ and 105.A5 BMMNCs characterized by low perpendicular light scatter (PLS) and low to moderate forward light scatter (FLS) also coexpressed the CD34 antigen. A mean of 65.4% ± 9.25% (range, 49.25% to 74.90%; n = 6; Fig2A) and 53.5% ± 4.8% (range, 21.5% to 62%; n = 6; Fig 2B) of the CD34+ cells were found to bind 103B2/9E10 and 105A5, respectively. Moreover, MoAb 105.A5 consistently reacted with approximately 11% to 21% more of the BMMNC fraction that lacked the expression of the CD34 antigen (CD34−105.A5+). Subsequent morphological analysis showed that the CD34−105.A5+fraction harbored cells of all the erythroid stages, including normoblasts (S.M.W., in press).
The BMMNC-derived CD34+ cell subset that stained more highly with the 103B2/9E10 (CD34+103B2/9E10+fraction) was isolated by FACS and assayed for its content of clonogenic cells in in vitro methylcellulose cultures. This fraction was compared both with the unfractionated CD34+ population (CD34+) and with the CD34+ subset that expressed low to negligible levels of the 103B2/9E10 epitope (CD34+103B2/9E10lo/− fraction). Collectively, these assays demonstrated that virtually all the detectable myeloid (CFU-GM), erythroid (BFU-E), and multipotential colony-forming cells (CFU-Mix) were recovered in the population of cells that expressed the epitope recognized by the MoAb 103B2/9E10 (Fisher PLSD; P ≥ .05; Fig 2C).
To ascertain whether MoAb 105.A5, like 103B2/9E10, bound to lineage-restricted HPCs, in vitro clonogenic assays were performed on BMMNC-derived, FACS-isolated CD34+105.A5+ and CD34+105.A5lo/− fractions. In accord with the results obtained with 103B2/9E10, virtually all detectable myeloid (CFU-GM), erythroid (BFU-E), and multipotential colony-forming cells (CFU-Mix) were recovered in the population of cells that expressed the antigen recognized by the MoAb 105.A5 (Fisher PLSD; P ≥ .05; Fig 2D).
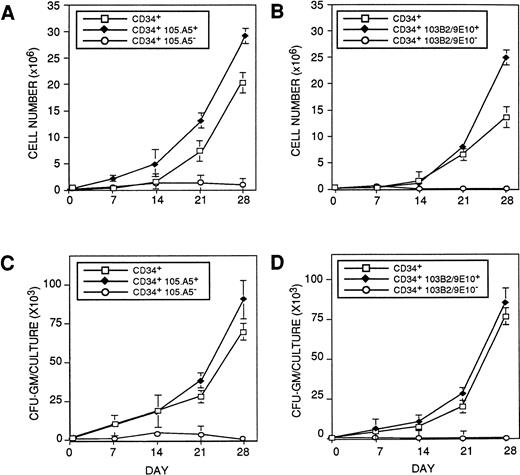
Previous studies have demonstrated that primitive multipotential blast colony-forming cells and cells that initiate long-term hematopoiesis in vitro are restricted to a minor proportion of CD34+ cells that coexpress Thy-1 (CD90)48,49 and c-kit (CD117)50 but lack lineage-restricted antigens48,51 and show low to undetectable expression of CD33,52 CD38,53 and HLA-DR.54 Therefore, to determine if primitive HPCs in human BM would express the 103B2/9E10 and 105.A5 epitopes, functional studies were performed. CD34+103B2/9E10+ and CD34+103B2/9E10lo/− or CD34+105.A5+ and CD34+105.A5lo/− fractions were isolated from normal adult BMMNCs and assayed for their ability for de novo generation of CFU-GM and nucleated cell production in the cytokine-driven stromal cell-free suspension culture (pre-CFU) assay. Figure3A to 3D show, respectively, the production of total hematopoietic cells and CFU-GM over time in culture from the various populations sorted with the 103B2/9E10 (Fig 3A and B) and 105.A5 (Fig 3C and D). The CD34+103B2/9E10lo/− cells failed to generate CFU-GM in excess of those present in the input population, whereas an 85-fold expansion in the number of clonogenic cells was observed with the CD34+103B2/9E10+ sorted population, within the 28-day period of the pre-CFU culture. Moreover, the CD34+ 103B2/9E10+ population displayed a twofold greater capacity to produce nucleated cells than the comparable CD34+ population (Fisher PLSD; P ≥ .05; Fig 3A and B). These data therefore demonstrate that, in addition to the directly clonogenic HPCs, the majority, if not all of the pre-CFU in adult human BM also express the 103B2/9E10 antigen (Fisher PLSD; P ≥ .05). Similar data were obtained for the CD34+105A5+and CD34+105A5− fractions, ie, primitive hematopoietic cells with the capacity to initiate and maintain hematopoiesis in this culture system were also restricted to the CD34+105.A5+ subpopulation, whereas the CD34+105.A5lo/− subpopulation failed to generate CFU-GM within the 28-day period of the pre-CFU assay (Fisher PLSD; P ≥ .05; Fig 3C and D).
CD34+ cells initiating hematopoiesis in the cytokine-supplemented (pre-CFU) assay express the 103B2/9E10 and 105.A5 epitopes. BMMNC were sorted into (□) CD34+, (⧫) CD34+ 103B2/9E10+, (○) CD34+103B2/9E10lo/− (A and C) or (□) CD34+, (⧫) CD34+105.A5+, (○) CD34+ 105.A5lo/− subpopulations (B and D) (as described in Materials and Methods) and assayed for their ability to initiate and maintain hematopoiesis in a stroma-independent, cytokine-supplemented culture. Cultures were established in triplicate using 1 × 103 sorted cells per well in medium supplemented with 10 ng/mL each of purified recombinant human IL-1β, IL-3, IL-6, G-CSF, GM-CSF, and SCF. Additional factors were added at the same concentrations on days 7, 14, and 21. On days 7, 14, and 21, the cells were harvested, washed, and assayed for nucleated cell number and CFU-GM as previously described. The results are expressed as the mean number ± SE of nucleated cell number (A and B) and CFU-GM (C and D) recovered at days 7, 14, 21, and 28 for each group. A representative experiment (1 of 3) is shown.
CD34+ cells initiating hematopoiesis in the cytokine-supplemented (pre-CFU) assay express the 103B2/9E10 and 105.A5 epitopes. BMMNC were sorted into (□) CD34+, (⧫) CD34+ 103B2/9E10+, (○) CD34+103B2/9E10lo/− (A and C) or (□) CD34+, (⧫) CD34+105.A5+, (○) CD34+ 105.A5lo/− subpopulations (B and D) (as described in Materials and Methods) and assayed for their ability to initiate and maintain hematopoiesis in a stroma-independent, cytokine-supplemented culture. Cultures were established in triplicate using 1 × 103 sorted cells per well in medium supplemented with 10 ng/mL each of purified recombinant human IL-1β, IL-3, IL-6, G-CSF, GM-CSF, and SCF. Additional factors were added at the same concentrations on days 7, 14, and 21. On days 7, 14, and 21, the cells were harvested, washed, and assayed for nucleated cell number and CFU-GM as previously described. The results are expressed as the mean number ± SE of nucleated cell number (A and B) and CFU-GM (C and D) recovered at days 7, 14, 21, and 28 for each group. A representative experiment (1 of 3) is shown.
Given the similarity in distribution of the epitopes identified by 103B2/9E10 and 105A5, these data suggested the possibility that similar glycoprotein antigens were identified by the two MoAbs on CD34+ cells. This was subsequently confirmed by three-color flow cytometric analysis with MoAbs HPCA2 (α-CD34), 103B2/9E10, and 105A5, which resulted in colinear staining of approximately 60% of the CD34+ population (data not shown).
Isolation of a Novel cDNA Clone With the 103B2/9E10 and 105.A5 MoAbs
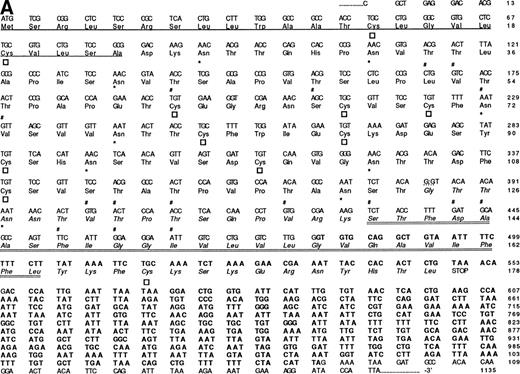
Given their reactivity with cultured HBMSCs (Fig 1), MoAbs 103B2/9E10 and 105.A5 were used to screen an HBMSC cDNA expression library in the retroviral vector pRUF.neo, as previously described.34 Subsequent to the specific-isolation of 103B2/9E10 and 105.A5 antigen-expressing FDC-P1 clones, genomic DNA was isolated and the corresponding cDNA was rescued by PCR. After partial-sequence analysis, the resultant nucleotide sequences were compared with entries submitted to the Genbank/EMBL databases via standard FASTA alignment analysis and found to share some homology to a previously reported cDNA isolated from KATO-III human gastric carcinoma cells.55 However, complete sequence analysis of the region encompassing the coding sequence (and the immediate flanking regions) showed a number of differences to the sequence previously identified by Masuzawa et al55(Fig 4A). Although sharing complete identity from nucleotide 1 to 383, the 103B2/9E10 and 105.A5 cDNA clones exhibited a deletion of 58 nucleotides at this site. This loss is presumably due to the use of alternate 5′ and 3′ splice junction sites present within this region. In accordance to the GT-AG rule described by Mount,56 analysis of the MGC-24 (multiglycosylated core protein of 24 kD) sequence showed two splice junction consensus motifs. Seven of the nine nucleotides of the MGC-24 sequence (CAG:GTAAAC) conform with the well-recognized 5′ splice junction consensus site (CAG:GTAAGT). Moreover, the short consensus sequence for the 3′ splice junction (CAG:G) is also present, suggesting that the 103B2/9E10 and 105.A5-derived cDNAs represent an alternative-spliced variant of MGC-24 which we have termed MGC-24v. In accord with this, recent studies of the genomic structure of the MGC-24 gene (Watt et al, unpublished data) showed that the MGC-24 gene does harbor sequence that can encode for this alternately spliced exon.
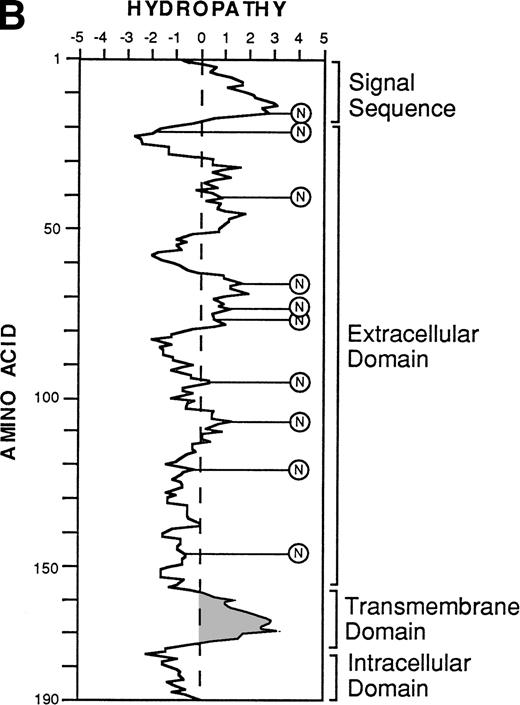
The nucleotide, deduced amino acid sequence, and hydropathy plot of CD164. (A) The putative signal sequence is underlined. Potential sites of N-linked and O-linked glycan attachment are indicated by (*) asterisks and (#) hatch-markings, respectively. Cysteine residues are indicated by boxes. Nucleotide (and amino acid) sequences that differ from the published sequences of MGC-2455 are in bold. The putative transmembrane domain (amino acid 161 to 180) is italicized. Please note that the nucleotide and amino acid sequence numbering is based on that of Masuzawa et al.55 (B) The hydropathy plot of CD164 polypeptide backbone, according to the method of Kyte and Doolittle.61Various regions are bracketed and identified at the right of the figure. Circled Ns represent potential N-linked glycosylation sites.
The nucleotide, deduced amino acid sequence, and hydropathy plot of CD164. (A) The putative signal sequence is underlined. Potential sites of N-linked and O-linked glycan attachment are indicated by (*) asterisks and (#) hatch-markings, respectively. Cysteine residues are indicated by boxes. Nucleotide (and amino acid) sequences that differ from the published sequences of MGC-2455 are in bold. The putative transmembrane domain (amino acid 161 to 180) is italicized. Please note that the nucleotide and amino acid sequence numbering is based on that of Masuzawa et al.55 (B) The hydropathy plot of CD164 polypeptide backbone, according to the method of Kyte and Doolittle.61Various regions are bracketed and identified at the right of the figure. Circled Ns represent potential N-linked glycosylation sites.
Translation of the new open reading frame (ORF) gave rise to a 178 amino acid polypeptide divergent from MGC-24 at the COOH-terminus. Examination of sequences 3′ to the 103B2/9E10 and 105A5 ORFs also showed the presence of 595 bp of sequence unique to the MGC-24v cDNA (Fig 4A, bold script). Because this region lies outside the ORF, the functional significance of this additional sequence remains to be defined. Furthermore, analysis of the MGC-24v cDNA showed a translation start site in the context of a Kozak57 consensus sequence (ACACGATGT), with a putative initiator methionine score of 60. Computer analysis (MacDNASIS, Version 2.0; Hitachi Software Engineering Co, Tokyo, Japan) of the resultant polypeptide showed a putative molecular weight of approximately 19.1 kD. This polypeptide sequence was not significantly related to that of any other core protein so far reported (Swiss-PROT search, National Centre for Biological Information, National Institutes of Health, Bethesda, MD). The initiation methionine is followed by a putative 22 amino acid signal sequence containing a hydrophobic core (amino acids Ala23-Asp24; Fig 4A).58 Following this putative signal peptide, 9 potential sites of N-linked glycosylation (consensus sequence: AsnXaaThr/Ser, with the exception of AsnProThr/Ser or AsnXaaThr/Ser-Pro58) are observed on Asn residues at positions 26, 32, 41, 72, 77, 94, 104, 121, and 127 (Fig4A). Moreover, the mature 178 amino acid protein is extremely rich in serine and threonine, with 37 (or ∼20%) of the encoded amino acids made up of these residues. At least 16 of these residues can serve as attachment sites for O-linked glycans (Fig 4A)28,59 and in combination with N-linked glycans make up more than 70% of the molecular mass of the mature protein. In addition, the MGC-24v sequence lacks the acceptor site for the addition of a glycosaminoglycan (GAG) chain60 at position Serine142 and Glycine143 (SG) present within the MGC-24 polypeptide sequence reported by Masuzawa et al.55 Hydropathy analysis (Kyte and Doolittle,61 GENETYX-MAC/1 3.0.1) of this MGC-24 isoform showed that the majority of the encoded protein was highly hydrophilic, consistent with the high hydroxyl amino acid content of the protein (Fig 4B). However, in contrast to the findings of Masuzawa et al,55 examination of the C-terminus showed a region (amino acids 140 to 164) of high hydrophobicity that may represent a putative transmembrane-anchoring motif. This putative transmembrane domain is followed by a very short COOH-terminal hydrophilic domain (amino acid residues 165 to 178), consistent with the requirements of a type I transmembrane protein. A schematic diagram demonstrating the salient features of the hematopoietic form of MGC-24 is presented in Fig 5.
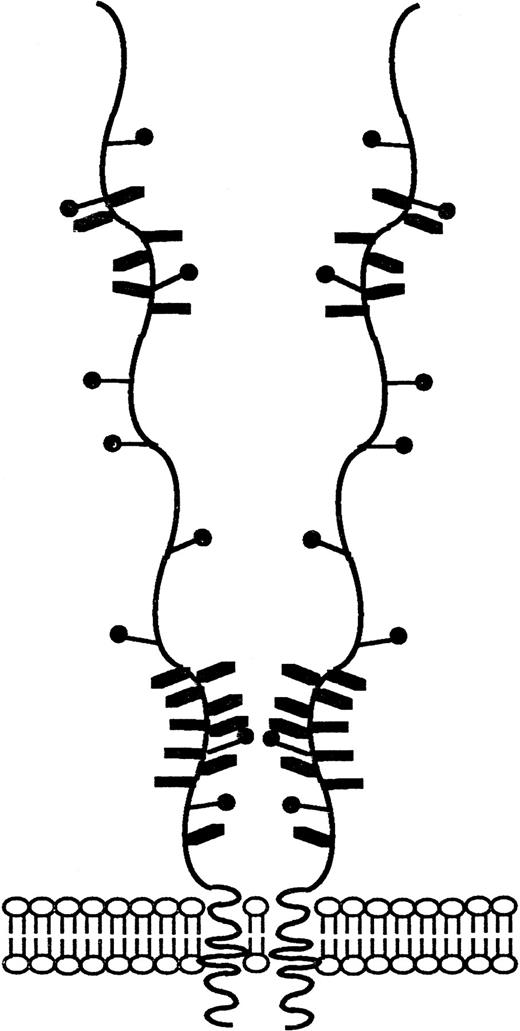
Schematic representation of the CD164 glycoprotein. Schematic diagram of CD164 based on biochemical and nucleotide data described in previous figures. The salient features of this molecule include (1) the numerous (16 predicted) potential sites of O-linked glycan atachment, (2) the nine possible N-linked glycosylation sites, and (3) the putative site of glycosaminoglycan attachment.
Schematic representation of the CD164 glycoprotein. Schematic diagram of CD164 based on biochemical and nucleotide data described in previous figures. The salient features of this molecule include (1) the numerous (16 predicted) potential sites of O-linked glycan atachment, (2) the nine possible N-linked glycosylation sites, and (3) the putative site of glycosaminoglycan attachment.
To confirm that MoAb 103B2/9E10 and 105.A5 identified the product of the MGC-24v cDNA, a 2.7-kb BamHI-Xho I restriction fragment (harboring both the entire coding sequence and the 5′ and 3′ noncoding regions) was subcloned into the pRUF.neovector and subsequently introduced into FDC-P1 cells by retroviral transduction.34 The resultant G418-resistant cell population and the parental cell line were then tested (by indirect immunofluorescence and flow cytometry) for their ability to bind MoAb 103B2/9E10 and 105.A5. As demonstrated in Fig 6, both MoAbs reacted specifically with this transfectant. In accord with this data, these MoAbs were instrumental in the recent clustering of MGC-24v as CD164 at the VI International Workshop and Conference of Human Leukocyte Differentiation Antigens (HLDA) in Kobe, Japan.33 MGC-24v will hereon be referred to as CD164.
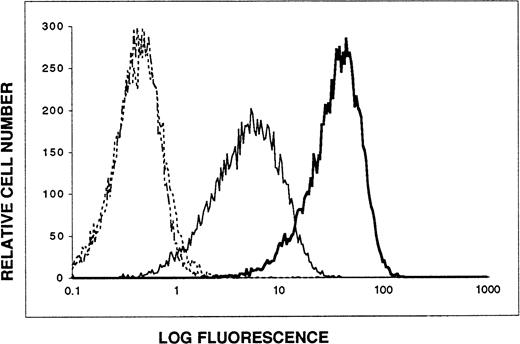
103B2/9E10 and 105.A5 MoAbs recognize the same glycoprotein antigen on transfectants expressing the CD164 cDNA. A 2.7-kb BamHI-Xho I restriction fragment of the CD164 cDNA (harboring both the entire coding sequence and the 5′ and 3′ noncoding regions) was subcloned into the pRUFneovector and subsequently introduced into FDC-P1 cells by retroviral transduction (refer to Materials and Methods). The resultant G418-resistant cell population was stained by indirect immunofluorescence and analyzed by flow cytometry. Data are displayed as single-parameter fluorescence (FITC) histograms of 1 × 104 light-scatter gated events, collected as list mode data. (···) IgG3 control MoAb; (-·-) IgM control MoAb; (—) MoAb 103B2/9E10; () MoAb 105.A5
103B2/9E10 and 105.A5 MoAbs recognize the same glycoprotein antigen on transfectants expressing the CD164 cDNA. A 2.7-kb BamHI-Xho I restriction fragment of the CD164 cDNA (harboring both the entire coding sequence and the 5′ and 3′ noncoding regions) was subcloned into the pRUFneovector and subsequently introduced into FDC-P1 cells by retroviral transduction (refer to Materials and Methods). The resultant G418-resistant cell population was stained by indirect immunofluorescence and analyzed by flow cytometry. Data are displayed as single-parameter fluorescence (FITC) histograms of 1 × 104 light-scatter gated events, collected as list mode data. (···) IgG3 control MoAb; (-·-) IgM control MoAb; (—) MoAb 103B2/9E10; () MoAb 105.A5
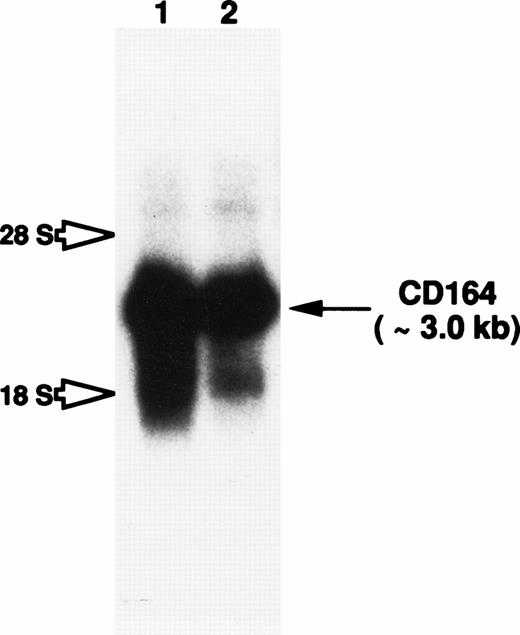
Total RNA isolated from cultured HBMSCs and the candidate myeloid progenitor cell line KG1a (characterized by its high levels of CD34+ surface antigen expression62) was examined by Northern blot analysis for the presence of CD164 transcripts. A prominent CD164 transcript of approximately 3.0 kb was observed in RNA blots of both cell preparations (Fig 7). Although weak hybridization with two diffuse species of 4.8 kb and 1.9 kb was also observed in both lanes, this most likely represents cross-hybridization of the MGC-24v probe with the 28s and 18s ribosomal RNAs, respectively. Therefore, the consistent detection of a single strong hybridizing mRNA species, argues against a significant level of alternative RNA splicing, at least in hematopoietic tissues.
Northern blot analysis to examine CD164 expression in both hematopoietic progenitor and BM stromal cells. Total RNA derived from HBMSCs (lane 1) and the primitive myeloid cell line, KG1a (lane 2), were subjected to electrophoresis on a 1.0% formaldehyde-agarose gel and transferred to a nylon membrane by capillary action. CD164 mRNA expression was examined by overnight hybridization at 42°C with a32P-radiolabeled full-length CD164 probe. The membrane was washed as described in Materials and Methods. Membranes were exposed to X-Omat AR film for 48 hours with intensifying screens. Hybridization to the 3.0-kb CD164 transcript is observed in both cell lines (indicated by arrow), whereas possible cross-hybridization with the 18s and 28s ribosomal RNA is indicated by open arrows.
Northern blot analysis to examine CD164 expression in both hematopoietic progenitor and BM stromal cells. Total RNA derived from HBMSCs (lane 1) and the primitive myeloid cell line, KG1a (lane 2), were subjected to electrophoresis on a 1.0% formaldehyde-agarose gel and transferred to a nylon membrane by capillary action. CD164 mRNA expression was examined by overnight hybridization at 42°C with a32P-radiolabeled full-length CD164 probe. The membrane was washed as described in Materials and Methods. Membranes were exposed to X-Omat AR film for 48 hours with intensifying screens. Hybridization to the 3.0-kb CD164 transcript is observed in both cell lines (indicated by arrow), whereas possible cross-hybridization with the 18s and 28s ribosomal RNA is indicated by open arrows.
Characterization of the CD164 Protein Identified by MoAbs 103B2/9E10 and 105.A5
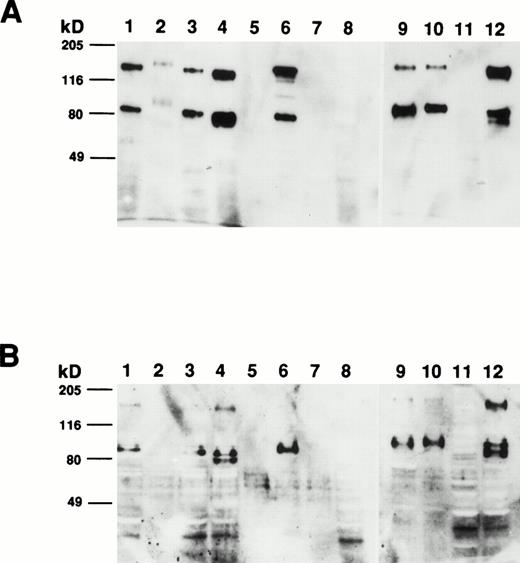
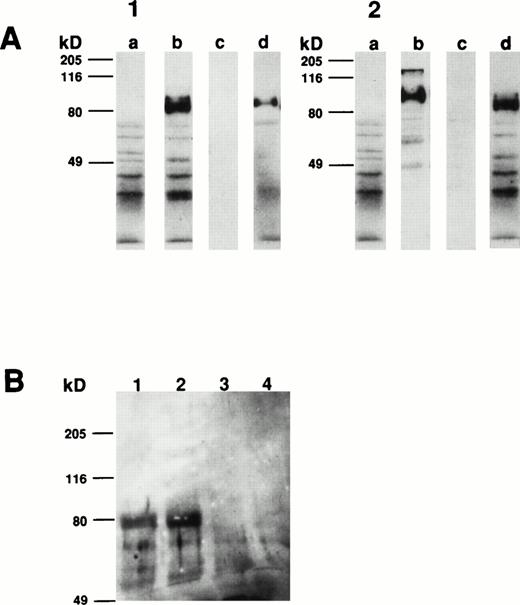
To determine the size of the CD164 protein identified by MoAb 103B2/9E10, membrane proteins from a variety of hematopoietic and nonhematopoietic cell lines and cell preparations were subjected to Western blot analysis. As demonstrated in Fig 8A, MoAb 103B2/9E10 identified two differentially migrating species, with apparent molecular weights of 80 and 160 kD, respectively, under nonreducing conditions. However, upon reduction, the intensity of the 160-kD immunoreactive protein was considerably reduced, suggesting that CD164 exists as a homodimer of two 80-kD monomers (Fig 8B). The protein recognized by the 103B2/9E10 MoAb was expressed in a majority of the cells tested, with the exception of peripheral blood erythrocytes, keratinocytes, Jurkat T cells, and PB-derived B cells. Interestingly, the molecular mass of the 103B2/9E10 cell surface molecule (CSM) differed marginally among the different cells tested.
Determination of the molecular mass of the CSM identified by MoAb 103B2/9E10. Membrane preparations from a variety of hematopoietic and nonhematopoietic cell lines/preparations were separated by 10% SDS-polyacrylamide gel electrophoresis under nonreducing and reducing conditions and transferred to nitrocellulose. The filters were successively incubated with 103B2/9E10 MoAb supernatant and antimouse-HRPO, and the immunoreactive proteins were detected by ECL as described in Materials and Methods. Under nonreducing conditions (A), MoAb 103B2/9E10 identifies two differentially migrating species with an estimated molecular mass of 80 and 160 kD. However, upon reduction (B), the lower migrating 80-kD band is the major immunoreactive protein. (A and B) Lane 1, KG1a (progenitor cell line); lane 2, HL-60 (promyelocytic cell line); lane 3, Hel-DR (erythroleukemic); lane 4, K562 (erythroleukemic); lane 5, red blood cells; lane 6, Mo7e (megakaryocyte); lane 7, Jurkat (T-cell line); lane 8, CD19+ B cells; lane 9, HBMSCs; lane 10, human umbilical vein endothelial cells (HUVECs); lane 11, T1 (keratinocytes); lane 12, MG63 (osteosarcoma cell line).
Determination of the molecular mass of the CSM identified by MoAb 103B2/9E10. Membrane preparations from a variety of hematopoietic and nonhematopoietic cell lines/preparations were separated by 10% SDS-polyacrylamide gel electrophoresis under nonreducing and reducing conditions and transferred to nitrocellulose. The filters were successively incubated with 103B2/9E10 MoAb supernatant and antimouse-HRPO, and the immunoreactive proteins were detected by ECL as described in Materials and Methods. Under nonreducing conditions (A), MoAb 103B2/9E10 identifies two differentially migrating species with an estimated molecular mass of 80 and 160 kD. However, upon reduction (B), the lower migrating 80-kD band is the major immunoreactive protein. (A and B) Lane 1, KG1a (progenitor cell line); lane 2, HL-60 (promyelocytic cell line); lane 3, Hel-DR (erythroleukemic); lane 4, K562 (erythroleukemic); lane 5, red blood cells; lane 6, Mo7e (megakaryocyte); lane 7, Jurkat (T-cell line); lane 8, CD19+ B cells; lane 9, HBMSCs; lane 10, human umbilical vein endothelial cells (HUVECs); lane 11, T1 (keratinocytes); lane 12, MG63 (osteosarcoma cell line).
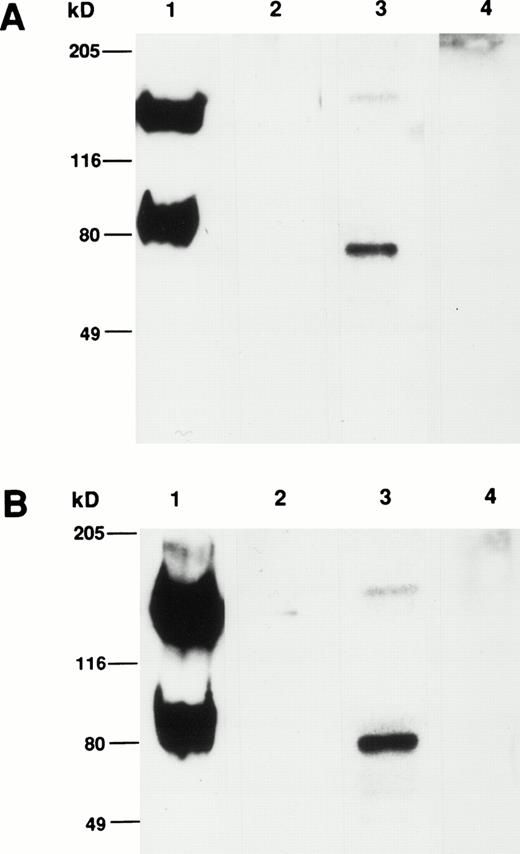
Comparison of the apparent molecular weight of the CSM identified by MoAbs 105.A5 and 103B2/9E10 by Western blotting showed that, under both reducing and nonreducing conditions, both MoAbs identified identical protein species from cultured HBMSCs (Fig9A and B). Furthermore, Western blotting analysis of membrane extracts isolated from FDC-P1 cells selected with the 105.A5 and 103B2/9E10 MoAbs (105.A5 and 103B2/9E10 FDC-P1 clones) showed that similar recombinant proteins of 80 kD were detected with both MoAbs (Fig 10A). Moreover, immune-precipitation of biotinylated-FDC-P1 lysates showed that, although the IgM MoAb 105.A5 was unable to selectively precipitate the CD164 protein, MoAb 103B2/9E10 identified a protein of the appropriate molecular mass from lysates derived from both 105.A5 and 103B2/9E10 FDC-P1 cells (Fig 10B).
(A and B) Western blot analysis of cultured HBMSCs with MoAb 103B2/9E10 and 105.A5. Membrane preparations from cultured HBMSCs were separated by 7.5% SDS-polyacrylamide gel electrophoresis under nonreducing and reducing conditions and transferred to nitrocellulose. The filters were successively incubated with either the 103B2/9E10 (A) or 105.A5 MoAb (B) supernatant (or isotype-matched, nonbinding controls) and antimouse-HRPO. The immunoreactive proteins were detected by ECL as described in Materials and Methods. Under nonreducing conditions, MoAbs 103B2/9E10 and 105.A5 identify two differentially migrating species with an apparent molecular weight of 80 and 160 kD (A and B, lane 1). However, upon reduction, the lower migrating 80-kD band is the major immunoreactive protein (A and B, lane 3). (A) Lane 1, nonreduced: 103B2/9E10 MoAb; lane 2, nonreduced: IgG3negative control; lane 3, reduced: 103B2/9E10 MoAb; lane 4, IgG3 negative control. (B) Lane 1, nonreduced: 105.A5 MoAb; lane 2, nonreduced: IgM negative control; lane 3, reduced: 105.A5 MoAb; lane 4, IgM negative control.
(A and B) Western blot analysis of cultured HBMSCs with MoAb 103B2/9E10 and 105.A5. Membrane preparations from cultured HBMSCs were separated by 7.5% SDS-polyacrylamide gel electrophoresis under nonreducing and reducing conditions and transferred to nitrocellulose. The filters were successively incubated with either the 103B2/9E10 (A) or 105.A5 MoAb (B) supernatant (or isotype-matched, nonbinding controls) and antimouse-HRPO. The immunoreactive proteins were detected by ECL as described in Materials and Methods. Under nonreducing conditions, MoAbs 103B2/9E10 and 105.A5 identify two differentially migrating species with an apparent molecular weight of 80 and 160 kD (A and B, lane 1). However, upon reduction, the lower migrating 80-kD band is the major immunoreactive protein (A and B, lane 3). (A) Lane 1, nonreduced: 103B2/9E10 MoAb; lane 2, nonreduced: IgG3negative control; lane 3, reduced: 103B2/9E10 MoAb; lane 4, IgG3 negative control. (B) Lane 1, nonreduced: 105.A5 MoAb; lane 2, nonreduced: IgM negative control; lane 3, reduced: 105.A5 MoAb; lane 4, IgM negative control.
Western blot and immunoprecipitation analysis of transfectant-derived recombinant CD164 protein. Membrane extracts isolated from FDC-P1 cells selected with the 105.A5 and 103B2/9E10 MoAbs (105.A5 and 103B2/9E10 FDC-P1 clones) were separated by 10% SDS-polyacrylamide gel electrophoresis under reducing conditions and transferred to nitrocellulose. The filters were successively incubated with either the 103B2/9E10 (panel 1) or 105.A5 MoAb (panel 2) supernatant (or isotype-matched, nonbinding controls), antimouse-HRPO and the immunoreactive proteins were detected by ECL as described in Materials and Methods. Under reducing conditions, MoAbs 103B2/9E10 and 105.A5 identify similar recombinant proteins of 80 kD. (A1) and (A2) contain reduced 103B2/9E10 and 105.A5 FDC-P1 protein blotted, respectively. Lanes (a) IgG3 negative control, (b) MoAb 103B2/9E10, (c) IgM negative control, and (d) MoAb 105.A5. (B) Biotinylated membrane preparations of FDC-P1 cells expressing CD164 (ie, 103B2/9E10 and 105.A5-selected FDC-P1 cells) were coincubated with the indicated MoAb for 16 hours at 4°C. Immune complexes were precipitated using goat antimouse sepharose, resuspended in reducing SDS-PAGE buffer, resolved on a 7.5% SDS-polyacrylamide gel, and visualized with biotin-streptavidin-HRPO complex and ECL. MoAb 103B2/9E10 immunoprecipitate a protein of the appropriate molecular mass from lysates derived from both 105.A5 and 103B2/9E10 FDC-P1 cells. Lane 1, 103B2/9E10 FDC-P1 lysate immunoprecipitated with MoAb 103B2/9E10; lane 2, 105.A5 FDC-P1 lysate immunoprecipitated with MoAb 103B2/9E10; lane 3, 103B2/9E10 FDC-P1 lysate immunoprecipitated with IgG3 negative control; lane 4, 105.A5 FDC-P1 lysate immunoprecipitated with IgM negative control.
Western blot and immunoprecipitation analysis of transfectant-derived recombinant CD164 protein. Membrane extracts isolated from FDC-P1 cells selected with the 105.A5 and 103B2/9E10 MoAbs (105.A5 and 103B2/9E10 FDC-P1 clones) were separated by 10% SDS-polyacrylamide gel electrophoresis under reducing conditions and transferred to nitrocellulose. The filters were successively incubated with either the 103B2/9E10 (panel 1) or 105.A5 MoAb (panel 2) supernatant (or isotype-matched, nonbinding controls), antimouse-HRPO and the immunoreactive proteins were detected by ECL as described in Materials and Methods. Under reducing conditions, MoAbs 103B2/9E10 and 105.A5 identify similar recombinant proteins of 80 kD. (A1) and (A2) contain reduced 103B2/9E10 and 105.A5 FDC-P1 protein blotted, respectively. Lanes (a) IgG3 negative control, (b) MoAb 103B2/9E10, (c) IgM negative control, and (d) MoAb 105.A5. (B) Biotinylated membrane preparations of FDC-P1 cells expressing CD164 (ie, 103B2/9E10 and 105.A5-selected FDC-P1 cells) were coincubated with the indicated MoAb for 16 hours at 4°C. Immune complexes were precipitated using goat antimouse sepharose, resuspended in reducing SDS-PAGE buffer, resolved on a 7.5% SDS-polyacrylamide gel, and visualized with biotin-streptavidin-HRPO complex and ECL. MoAb 103B2/9E10 immunoprecipitate a protein of the appropriate molecular mass from lysates derived from both 105.A5 and 103B2/9E10 FDC-P1 cells. Lane 1, 103B2/9E10 FDC-P1 lysate immunoprecipitated with MoAb 103B2/9E10; lane 2, 105.A5 FDC-P1 lysate immunoprecipitated with MoAb 103B2/9E10; lane 3, 103B2/9E10 FDC-P1 lysate immunoprecipitated with IgG3 negative control; lane 4, 105.A5 FDC-P1 lysate immunoprecipitated with IgM negative control.
CD164 Mediates the Adhesion of CD34+ Cells to BM Stroma
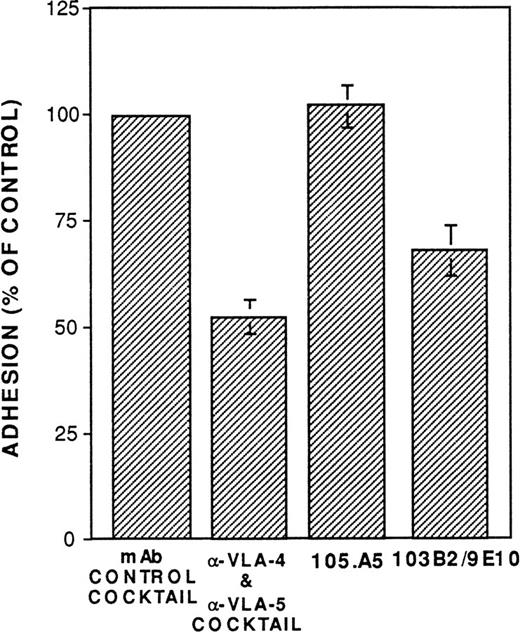
To date, no function has been ascribed to either CD164 or the MGC-24 mucin-like glycoproteins. As described above, a number of studies suggest that mucin-like molecules represent adhesive glycoproteins. To examine the possible adhesive function of CD164 on CD34+cells, assays were performed to examine whether MoAbs 103B2/9E10 or 105A5 were able to perturb the adhesion of CD34+ cells to cultured marrow stromal cells in vitro. As shown in Fig 11, MoAb 103B2/9E10 (but not 105.A5) significantly blocked (by approximately 30%; Fisher PLSD;P ≥ .01) the adhesion of chromium-labeled CD34+cells to allogeneic BM stromal cells in vitro, suggesting an adhesion function for CD164. Further functional studies are in progress.
The CD164 mucin-like glycoprotein functions as an adhesion molecule expressed by CD34+ and BM stromal cells. Allogeneic BM stromal cells (2 × 104) were transferred to each well of a 96-well plate 24 hours before performing the assay. BM CD34+ cells isolated with 561-Dynabeads were labeled with 51Cr and subsequently incubated on ice at 1 × 105/mL in RPMI (supplemented with 2% FCS) containing 20 μg/mL of the MoAb 105.A5, 103B2/9E10, IgM, and IgG3 nonbinding control MoAbs, or the anti-VLA4 (P4C2) and VLA-5 MoAbs (PHM2), as indicated. Cells (1 × 104 in a volume of 100 μL) were subsequently transferred (without washing) to each well of the 96-well plate containing stromal cells. Incubation was performed for 30 minutes at 37°C, after which unbound cells were removed by washing. Adhesion was quantitated by liquid scintillation counting of solubilized lysates. Data are presented as the percentage of control adhesion obtained in the presence of a cocktail of IgG3 and IgM nonbinding control MoAbs and represents the mean ± SE of three experiments.
The CD164 mucin-like glycoprotein functions as an adhesion molecule expressed by CD34+ and BM stromal cells. Allogeneic BM stromal cells (2 × 104) were transferred to each well of a 96-well plate 24 hours before performing the assay. BM CD34+ cells isolated with 561-Dynabeads were labeled with 51Cr and subsequently incubated on ice at 1 × 105/mL in RPMI (supplemented with 2% FCS) containing 20 μg/mL of the MoAb 105.A5, 103B2/9E10, IgM, and IgG3 nonbinding control MoAbs, or the anti-VLA4 (P4C2) and VLA-5 MoAbs (PHM2), as indicated. Cells (1 × 104 in a volume of 100 μL) were subsequently transferred (without washing) to each well of the 96-well plate containing stromal cells. Incubation was performed for 30 minutes at 37°C, after which unbound cells were removed by washing. Adhesion was quantitated by liquid scintillation counting of solubilized lysates. Data are presented as the percentage of control adhesion obtained in the presence of a cocktail of IgG3 and IgM nonbinding control MoAbs and represents the mean ± SE of three experiments.
Antibody Ligation of CD164 on Primitive CD34+CD38− HPC Suppresses Cytokine-Induced Recruitment of Cellular Division
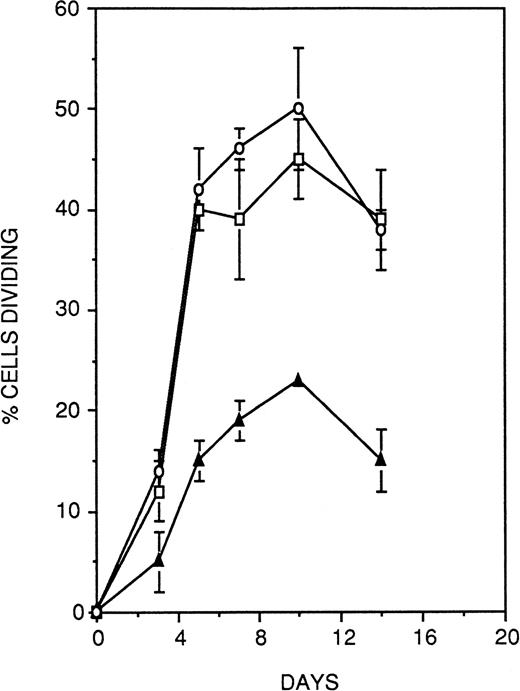
To investigate the possible functional contribution of CD164 to the regulation of hematopoiesis, we chose to examine the effect of antibody-mediated ligation of CD164 on the growth of CD34+CD38− hematopoietic progenitors under stromal cell-free, cytokine-supported assay conditions. For these experiments, single CD34+CD38− cells were deposited on wells of a Terasaki plate coated with equal concentrations of either the anti-CD164 antibody 103.B2 or isotype-matched binding and nonbinding control antibodies. Proliferation of cells was stimulated by the combination of IL-3, IL-6, G-CSF, and SCF (36GS), and cultures were monitored at regular intervals over 14 days to assess cell growth. As shown in Fig 12, culture of CD34+CD38− cells on either of the two control antibodies led to the recruitment of approximately 40% of the population into division over the 14-day time period of the assay. In contrast, the CD164 antibody resulted in a specific and significant (Fisher PLSD; P ≥ .01) reduction by approximately 50% at each of the timepoints measured in the proportion of CD34+CD38− cells recruited into division under these assay conditions. At the termination of the assays, nonresponding wells (ie, wells originally seeded with a single cell but exhibiting no division) were examined to determine the viability of the cells they contained. A mean of 52% ± 11% (n = 3) of cells grown on CD164 antibody were nonviable at this day-14 time point, as evidenced by complete cellular breakdown, a markedly higher proportion than that observed on either the nonbinding control antibody (8.7% ± 4.5%) or the binding isotype-matched antibody (12.3% ± 3.3%). Thus, antibody ligation of CD164 on CD34+CD38− cells both suppresses cytokine-induced recruitment of the cells into division and also induces death of a high proportion of the population.
Antibody ligation of CD164 on primitive CD34+CD38− HPCs suppresses cytokine-induced recruitment of cellular division. Single CD34+CD38− cells were deposited on wells of a Terasaki plate coated with equal concentrations of either the anti-CD164 antibody 103.B2 (▴) or isotype-matched binding (□) and nonbinding control antibodies (○). Cellular proliferation was stimulated with IL-3, IL-6, G-CSF, and SCF (36GS). Cultures were monitored at regular intervals over 14 days to assess cell growth. Culture of CD34+CD38− cells on either of the two control antibodies led to the recruitment of approximately 40% of the population into division, whereas the CD164 antibody resulted in a specific and significant (P < .05) reduction by approximately 50% at each of the time points measured in the proportion of CD34+CD38− cells recruited into division under these assay conditions. Thus antibody ligation of CD164 on CD34+CD38− cells suppresses cytokine-induced recruitment of the cells into division.
Antibody ligation of CD164 on primitive CD34+CD38− HPCs suppresses cytokine-induced recruitment of cellular division. Single CD34+CD38− cells were deposited on wells of a Terasaki plate coated with equal concentrations of either the anti-CD164 antibody 103.B2 (▴) or isotype-matched binding (□) and nonbinding control antibodies (○). Cellular proliferation was stimulated with IL-3, IL-6, G-CSF, and SCF (36GS). Cultures were monitored at regular intervals over 14 days to assess cell growth. Culture of CD34+CD38− cells on either of the two control antibodies led to the recruitment of approximately 40% of the population into division, whereas the CD164 antibody resulted in a specific and significant (P < .05) reduction by approximately 50% at each of the time points measured in the proportion of CD34+CD38− cells recruited into division under these assay conditions. Thus antibody ligation of CD164 on CD34+CD38− cells suppresses cytokine-induced recruitment of the cells into division.
DISCUSSION
Cellular interactions between primitive hematopoietic progenitors and the stromal microenvironment of the BM are of major importance in the regulation of hematopoiesis.63-66 Accordingly, cell surface antigens that are selectively expressed by either or both components may play an important role in mediating these interactions. The recent cloning of four counter-receptors for the selectin family (GlyCAM-1, CD34, MAdCAM-1, and PSGL-1) has led to the identification of a family of adhesion molecules variously termed mucin-like proteins or sialomucins.67 Mucin-like molecules are characteristically serine- and threonine-rich proteins that are heavily decorated with O-linked glycans. This dense array of O-linked carbohydrate results in an extended, nonglobular thread-like structure that provides an optimal platform for the presentation of multiple terminal sugar moieties.
The present study describes two MoAbs, namely 103B2/9E10 and 105.A5, that exhibit reactivity to both a subpopulation of the CD34+ HPC population and BM stromal fibroblasts. Molecular cloning and sequence analyses showed that both MoAbs identify a novel transmembrane isoform of the mucin-like glycoprotein termed MGC-24.55 MGC-24 (for multi-glycosylated core protein of 24 kD) was originally cloned from KATO-III human gastric carcinoma cells and found to encode a novel polypeptide backbone highly decorated with both O-and N-linked carbohydrate side chains. Complete sequence analysis of the ORFs of both the 103B2/9E10- and 105.A5-derived cDNA clones demonstrated that, although sharing complete identity with MGC-24 from nucleotide 1 to 383, the next 58 nucleotides of MGC-24 cDNA sequence were deleted. This was followed by 595 bp of sequence unique to the CD164 cDNA. This sequence has been confirmed by genomic cloning studies (Watt et al, unpublished data). In contrast to the epithelial-derived MGC-24 protein, computer modelling and hydropathy analysis of the 178 amino acid translated region showed the presence of both a putative transmembrane-anchoring motif (residues 140-164) and a short cytoplasmic domain (residues 165-178).
Using a polyclonal antiserum to the native deglycosylated core protein, Masuzawa et al55 demonstrated that the mature, epithelial-derived MGC-24 glycoprotein behaved as a high molecular mass, principally soluble, polymorphic mucin-like molecule that contained numerous peanut agglutinin (PNA)-binding sites. In contrast, using MoAbs 103B2/9E10 and 105.A5, Western blot and immunoprecipitation analyses of membrane extracts from cells representing various hematopoietic cell lineages showed that CD164 exists in its native state as disulphide-linked homodimers of two 80- to 85-kD subunits. CD164 lacks the consensus glycosaminoglycan (GAG)-attachment site found in MGC-24, and it is possible to speculate that GAG-association is responsible for the high molecular weight of the epithelial-derived MGC-24 glycoprotein. These results, together with our more recent genomic cloning studies (Watt et al, unpublished data), suggest that CD164 and MGC-24 represent splice variants of a single gene and are unlikely to represent members of a multigene family that, like CD66 molecules, share a significant degree of homology in their extracellular domains.68 69
Because CD164 was molecularly cloned from a BM stromal cell library, it is unclear whether an alternate isoform distinct from CD164 (but reactive with 103B2/9E10 and 105.A5) may be expressed by hematopoietic progenitors and their precursors. Current data do not permit definitive conclusions to be drawn; however, several observations suggest that the isoform expressed by hematopoietic cells may be equivalent to that expressed by BM stromal cells. A 3-kb transcript, for example, was observed in both BM stromal cells and the candidate multipotential progenitor cell line KG1a by Northern blot analyses. In addition, both 103B2/9E10 and 105.A5 immunoreactive proteins, derived from various cell preparations (including BM stromal cells and various myeloid cell lines), were of approximately equivalent size. Studies examining the mRNA species present in hematopoietic cells will be required to address this question.
Using a variety of in vitro hematopoietic cell assays, it was also found that clonogenic myeloid (CFU-GM) and erythroid (BFU-E) progenitors were restricted to the subpopulations of CD34+cells that expressed CD164. Moreover, cells that coexpress the progenitor cell antigen CD34 and CD164 glycoprotein were found to be the most primitive progenitors, as demonstrated by their capacity to initiate and sustain hematopoiesis in the in vitro cytokine stimulated pre-CFU assay.
Although MoAbs 105.A5 and 103B2/9E10 exhibit disparate reactivity profiles in normal tissues (Butler and Watt, unpublished data), it is clear from the studies presented herein that both MoAbs identify the product of the CD164 cDNA. The mechanism responsible for this differential reactivity is being determined. However, due to the high degree of glycosylation, it is possible that the MoAbs 105.A5 and 103B2/9E10 identify glycosylation-dependent epitopes and/or heterogeneous glycosylation of CD164 molecules. Future studies will examine their capacity to bind CD164 after treatment with a variety of carbohydrate-modifying enzymes to specifically remove O-linked and N-linked glycans.
A number of studies have attributed an antiadhesive function to mucin-like molecules by virtue of their negative charge and extended configuration. This was first demonstrated for the neural cell adhesion molecule (NCAM; CD56)70-72 and more recently for CD43,15 episialin,73-75, and epiglycanin.76,77 Despite this, the preponderance of recent data suggests that mucin-like molecules also function in a proadhesive manner.5,7-10 Adhesion assays to examine the possible adhesive role of CD164 showed that this molecule appears to facilitate the adhesion of CD34+ hematopoietic precursors to BM stroma. Given its mucin-like nature, one would speculate that the CD164 protein backbone may thus serve as a platform or scaffold for carbohydrate presentation. Such a scaffolding function for a mucin-like glycoprotein is plausible in view of what is currently known about mucin structure. The high carbohydrate content in mucins and mucin-like domains of glycoproteins dominates the physiochemical properties, such that the substituted peptide is highly extended into a rigid rod with all the secondary and tertiary structure precluded.27,28,78It is presumed that the role of the mucin domains is to extend the functional domain of these cell surface molecules well beyond the glycocalyx of the cell and that these provide easy access to the opposing counter-receptor or extracellular macromolecules.28
A number of recent reports have demonstrated that, like integrins,79 mucin-like molecules are capable of integrating external signals into the cytoplasm via their cytoplasmic domains. This was well exemplified by a recent study by Majunith et al,15 who demonstrated using CD43 knockout mice that this cell-surface sialoglycoprotein was able to negatively regulate T-cell activation. Moreover, work by Fackler et al80 demonstrated that enforced expression of the full-length form (but not the truncated form) of CD34 inhibited terminal differentiation of murine myeloid M1 cells, strongly suggesting that a region(s) in the cytoplasmic domain is responsible for the observed maturation arrest phenotype.
These studies prompted the investigation of whether ligation of CD164 on primitive progenitor cells would have an inhibitory effect on the recruitment into cycle of these quiescent cells. Because the physiological counter-receptor(s) for CD164 remains to be determined, MoAb 103B2/9E10 was used as a surrogate ligand. Using this experimental approach, ligation of CD164 was found to markedly suppress both the recruitment of a proportion of the quiescent CD34+CD38− population into division and to induce cell death in a significant proportion of the remainder. Although the mechansims responsible for these effects of anti-CD164 antibody remain unknown, some insights may be gained from studies recently reported by Bazil et al22,23 that examined the effects of antibody mediated cross-linking of an additional mucin-like molecule, CD43, on the growth of human CD34+ HPCs. These studies showed that multipotential progenitors (CFU-GEMM) and erythroid progenitors (BFU-E) were substantially more sensitive to CD43-induced apoptosis than myeloid progenitors (CFU-GM).22 Although expressing CD43 at high level, candidate hematopoietic stem cells (HSCs) were apparently refractory to the apoptosis-inducing effects of the anti-CD43 antibody.23
Thus, mucin-like molecules on HPC appear to be negative regulators of hematopoiesis. The question of whether the growth inhibitory properties of these molecules occurs in vivo requires further study. One possible role of these mucin-mediated interactions in vivo might be as a powerful negative regulatory mechanism to dampen excessive expansion of HPCs. An alternative possibility is that mucin-like molecules may deliver a positive proliferative stimulus to HPCs if combined with an appropriate, as yet unknown, stimulus. This hypothesis is suggested by studies with CD43 antibody that, in contrast to its ability to induce apoptosis of HPCs, provides a costimulatory proliferative stimulus to T cells.81 Thus, in the absence of this additional stimulus, CD43-mediated signalling in HPCs could result to the elimination of these improperly activated cells through the induction of apoptosis. Other scenarios are clearly possible, but for the present studies will be required to elucidate both the physiological significance of these observations and the signal transduction pathways involved.
Although beyond the scope of this submission, valuable information regarding the function and significance of CD164 could be obtained with studies designed to determine the chromosomal location of CD164, which may identify closely linked molecules of known function. In addition, the determination of the genomic organization and exon structure of CD164 may be predictive of the existence of additional splice variants. In fact, our preliminary studies in this respect suggest that the CD164 gene is composed of five exons that span approximately 20 kb of genomic DNA (S.M.W., unpublished data). Finally, the identification of the murine homolog will enable the generation of CD164 knockout mice that should ultimately indicate function for this novel mucin-like molecule.
ACKNOWLEDGMENT
The authors thank Dr L.B. To for the provision of normal human BM samples and for his ongoing support. We also thank Prof Sir D. Weatherall and Prof L. Kanz for their encouragement and assistance.
Supported by grants from the Anti-Cancer Foundation of the Universities of South Australia, the National Health and Medical Research Council of Australia (P.J.S. and A.C.W.Z.), Medical Research Council and Leukaemia Research Fund of Great Britain (S.M.W.), E.U. Concerted Action Programme Grants on the Human Haematopoietic Stem Cell (S.M.W. and H.J.B.), an E.U. Biotech Grant (S.M.W.), a Kay Kendall Leukaemia Fellowship (M.A.B.), and the Deutsche Forschungsgemeinschaft (SFB 510; project A1; H.J.B.). Genbank Bankit No. 195477.
Address reprint requests to Andrew C.W. Zannettino, PhD, Matthew Roberts Laboratory, Leukaemia Research Unit, Hanson Centre for Cancer Research, I.M.V.S., PO Box 14, Rundle Mall, Adelaide 5000, S.A., Australia; e-mail: andrew.zannettino@imvs.sa.gov.au.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

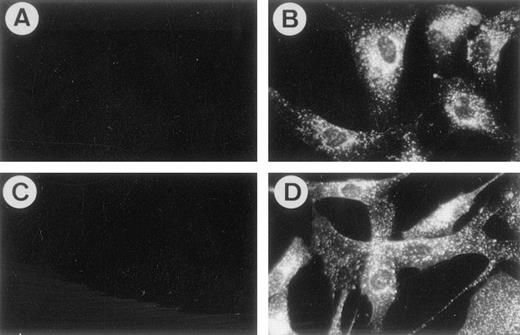
![Fig. 2. (A and B) Dual-parameter immunofluorescence analysis demonstrating the expression of CD34 and 103B2/9E10 (A) or 105.A5 (B) epitopes by BMMNCs. SBA-depleted BMMNCs were stained with the directly conjugated MoAb HPCA-2-PE (-CD34) and 103B2/9E10 (detected with anti-IgG3-FITC). Data are displayed as dual-parameter histograms of 5 × 104 light-scatter-gated events collected as list-mode data. Stat-markers were established with the use of nonbinding, isotype-matched control antibodies as described in Materials and Methods. (C and D) Assay of clonogenic progenitors in populations sorted on the basis of CD34 and MoAb 103B2/9E10 (C) or 105.A5 (D). FACS of the CD34+, CD34+103B2/9E10+, and CD34+103B2/9E10lo/− or CD34+105.A5+, and CD34+105.A5lo/− subpopulations demonstrates that clonogenic progenitors ([▪] CFU-GM, [□] BFU-E, and [▧] CFU-Mix) are present almost exclusively in the CD34+103B2/9E10+ (C) and CD34+105.A5+ (D) subpopulation. Results are expressed as the number of CFU-GM at day 14 per 1 × 103cells plated. Data represent the mean ± SE (n = 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/8/10.1182_blood.v92.8.2613/5/m_blod42015002x.jpeg?Expires=1769099158&Signature=ixyYwHtyF-ff0ZRfploWTxzF1X2p5KZgU3K8vxnXpSZxRbiAS-bBfrZSK5YFQ6DejkzpMBmJiu1ZidGAY-RGwxUDloyDCtIgZF-x-2rNEA6mu3OY4sfnkFMflhh0FmLcFebSc0fsB1ySPAkNAJVoCh1e~sn4AuxO8PF3HXUky-EjXxFuUVORJe9tBsd~hjME1ZNT2v~Ceg7mcMu~srcnQvshckaR6LidVnM5LlaGNZW-yEtFSK35XL3XTn5J0D~mJ51B~Trn3m80wkQt6qyEqKRSE41PP~4bmsB3dfWW1udTgnzvvoRTPoOl6MOBcV2TKL3u4yf1mBxjJEKYSnBH2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)