Abstract
We have performed a systematic in vivo evaluation of gene expression for the glycoprotein (GP) Ibα subunit of the murine platelet adhesion receptor, GP Ib-IX-V. This study is warranted by in vitro observations of human GP Ibα expression in cells of nonhematopoietic lineage and reports of regulation of the GP Ibα gene by cytokines. However, an in vivo role for a GP Ib-IX-V receptor has not been established beyond that described for normal megakaryocyte/platelet physiology and hemostasis. Our Northern analysis of mouse organs showed high levels of GP Ibα mRNA in bone marrow with a similar expression pattern recapitulated in mice containing a luciferase transgene under the control of the murine GP Ibα promoter. Consistently high levels of luciferase activity were observed in the two hematopoietic organs of mice, bone marrow (1,400 relative light units/μg of protein [RLUs]) and spleen (500 RLUs). Reproducible, but low-levels of luciferase activity were observed in heart, aorta, and lung (30 to 60 RLUs). Among circulating blood cells, the luciferase activity was exclusively localized in platelets. No increase in GP Ibα mRNA or luciferase activity was observed after treatment of mice with lipopolysaccharides (LPS) or tumor necrosis factor-α (TNF-α). We conclude the murine GP Ibα promoter supports a high level of gene expression in megakaryocytes and can express heterologous proteins allowing an in vivo manipulation of platelet-specific proteins in the unique environment of a blood platelet.
PLATELET GLYCOPROTEIN (GP) membrane receptors provide circulating platelets with the ability to recognize distinct adhesive ligands exposed as a result of vascular injury or perturbation to the vascular lining.1 The platelet GP Ib-IX-V receptor complex contributes to this process initiating hemostasis through interactions with the adhesive ligand, von Willebrand factor.2-4 This receptor has a second unrelated role in maintaining circulating platelet morphology, as suggested by the congenital absence of the receptor, the Bernard-Soulier syndrome, and the release of “giant” platelets.1,2 Biochemical characterizations of the GP Ib-IX-V complex have documented the assembly of the complex from four distinct gene products5-7with the ligand binding properties and linkages to the platelet membrane skeleton contained in the α-subunit of GP Ib (GP Ibα).8-13
While an in vivo role for platelet GP Ib-IX-V in hemostasis is well-documented, the expression of the complex in cells of nonhematopoietic lineage has been a controversial subject. A recent report characterizes an endothelial cell form of the GP Ib-IX-V complex14 contrasted by reports concluding endothelial cells lack the GP Ibα subunit and the ligand binding properties of the entire complex.15,16 Whereas the physiologic relevance, if any, of GP Ibα protein or a GP Ib-IX-V receptor complex on the surface of endothelial cells has not been determined, speculation has been fueled by in vitro assays characterizing the ability of cytokines to increase GP Ibα expression by endothelial cells and the possibility of a GP Ib-IX-V–dependent link between thrombotic events and inflammation.17,18 But again, these results are not without controversy, as others have been unable to reproduce the findings.15 Certainly, a systematic evaluation of GP Ibα in vivo expression would aid in ascertaining the biological sequelae that have been suggested from observations on cultured cells.
We have recently cloned and sequenced mouse genomic DNA containing the homologue to the human platelet GP Ibα gene.19 The mouse gene contains a similar exon/intron arrangement to the human gene and has allowed us an opportunity to perform a systematic evaluation of GP Ibα in vivo gene expression in a murine model system. Here we demonstrate high levels of GP Ibα mRNA in mouse bone marrow with a similar expression pattern obtained in transgenic animals containing a fragment of the mouse GP Ibα gene promoter. Our studies identified low-levels of GP Ibα promoter activity in heart, aorta, and lung, but contradictory to in vitro analyses, no increase in gene expression was observed after treatment of mice with cytokines or lipopolysaccharides (LPS). The presence of cis-acting elements in the murine GP Ibα gene promoter common among other megakaryocytic-specific promoters is discussed along with the physiologic relevance, if any, of low-levels of gene expression observed in some nonhematopoietic organs.
MATERIALS AND METHODS
Source of nucleic acids.
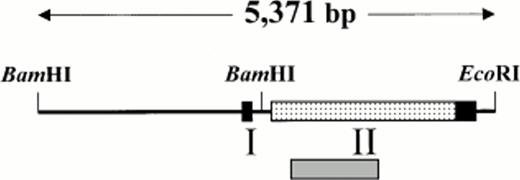
The isolation and characterization of a P1 plasmid containing approximately 80-kb pairs of mouse 129/SvJ genomic DNA has been previously described.19 The sequence of 5,371 bp from this clone has been determined and deposited in GenBank (accession no.U91967). The characterized sequence contains the mouse homologue to the human platelet GP Ibα gene and a schematic organization of the gene is presented in Fig 1.
Northern analysis to detect the murine GP Ibα transcript was performed using a 1-kb radiolabeled fragment corresponding to nucleotides 2880-3871 (numbering according to GenBank U91967) and encoding mature mouse GP Ibα polypeptide sequence from residues 57-386.19 Northern analysis to detect the luciferase transcript in transgenic mice was performed by radiolabeling aBamHI restriction fragment (2.6 kb) containing the complete coding sequence for luciferase present in the plasmid, p19/LUC.20 A mouse cDNA fragment encoding intracellular adhesion molecule (ICAM)-1 was obtained from Genome Systems (St Louis, MO) as a deposited clone in the dbEST database and available through the cDNA consortium (accession no. aa111579). Characterization of the clone showed a 1.2-kb insert with a restriction fragment pattern identical to that for the ICAM-1 sequence and limited sequencing confirmed the presence of a partial cDNA sequence for murine ICAM-1.21 For hybridization studies, the DNA fragments were radiolabeled with [α-32P]deoxyadenosine triphosphate (dATP) using a Prime-It II labeling kit available from Stratagene (La Jolla, CA).
RNA isolation and Northern analysis.
Organs were dissected from mice ranging from 6 to 12 weeks of age. Dissected organs were immediately frozen in liquid nitrogen and manually pulverized in a liquid nitrogen-chilled porcelain mortar using a pestle. Immediately after evaporation of the liquid nitrogen, the organ powder was dissolved in 4 mol/L guanidinium isothiocyanate/0.1 mol/L Tris (pH 7.5)/0.5% n-lauroyl sarcosine. A centrifugation (3,000g) was performed to remove large particulate matter and the supernatant was applied to a cesium chloride cushion to isolate total RNA.22 Gel electrophoresis of RNA was performed through denaturing formaldehyde gels containing 1% agarose.22 Transfer to nitrocellulose was performed by capillary action and the filters were hybridized at 42°C in a solution containing 50% formamide/5× Denhardt's solution/0.75 mol/L NaCl/50 mmol/L NaH2PO4/5 mmol/L EDTA/0.1% sodium dodecyl sulfate (SDS)/100 μg/mL denatured salmon sperm DNA.22 After overnight hybridization filters were washed three times in 0.3 mol/L sodium chloride/0.03 mol/L sodium citrate/0.1% SDS (10 minutes at room temperature [RT]) and one time in 0.03 mol/L sodium chloride/0.003 mol/L sodium citrate/0.1% SDS (30 minutes at 50°C). The washed nitrocellulose filters were analyzed by autoradiography using Kodak X-OMAT film (Eastman-Kodak, Rochester, NY).
Generation of transgenic mice.
A 2.6-kb BamHI fragment from the mouse GP Ibα gene containing exon I, portions of the single intron, and approximately 2.4 kb of sequence 5′ to the transcription initiation site was cloned into the vector p19/LUC immediately upstream of the luciferase coding sequence. p19/LUC is a promoterless reporter plasmid used to assay heterologous promoter activity.20 Before injection into mouse zygotes, the mouse promoter/luciferase cassette was removed from the vector and extensively dialyzed against a buffer composed of 5 mmol/L Tris (pH 7.5)/0.15 mmol/L EDTA. DNA injection and implantation into pseudopregnant B6xSJL females was performed by the Transgenic Core Facility at The Scripps Research Institute. At approximately 5 weeks of age, 30 transgenic offspring were screened for the presence of the luciferase transgene by Southern blot analysis of genomic DNA isolated from the distal 1 cm of tail.23 Ten mice gave positive results for integration of the luciferase transgene. These mice were also characterized by luciferase activity assays (described below) using 10 μL of whole blood obtained from a periorbital sinus bleed. Five of the 10 mice contained luciferase activity in their blood. These five B6xSJL founder mice were expanded into independent transgenic colonies used in the various assays. The two transgenic colonies, designated 32 and 12, exhibiting the highest levels of expression were chosen for the more detailed analyses shown in Figs 2 and 4-7.
Luciferase assays.
Six- to 12-week-old transgenic mice weighing 25 to 30 g were anesthetized by inhalation of metofane (methoxyflurane; Pitman-Moore, Mundelein, IL). All tissues were rapidly removed by standard dissection techniques and placed in liquid nitrogen. Frozen tissue powders were obtained and suspended in lysis buffer (0.1 mol/L potassium phosphate buffer [pH 7.8] containing 1% Triton X-100/1 mmol/L dithiothreitol [DTT]/2 mmol/L EDTA). The samples were lysed by two freeze-thaw cycles and centrifuged for 20 minutes at 4°C (14,000g). The organ/cellular homogenates were stored at −70°C until their use in luciferase activity assays. The luciferase activity present in 20-μL samples was determined by mixing 100 μL of assay buffer (50 mmol/L K2HPO4/50 mmol/L KH2PO4/15 mmol/L MgSO4/5 mmol/L ATP/1 mmol/L DTT) with the substrate D-luciferin and assaying in a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA). Each sample was assayed a minimum of three times. Relative light units (RLUs) were calculated after adjusting for background activity typically 200 to 250 RLUs. Total protein concentration of the organ extract was measured using a bicinchoninic assay reagent (Pierce Chemical Co, Rockford, IL). Luciferase activity was expressed as RLUs per microgram of protein.
Preparation of mouse platelet-rich and platelet-poor plasma.
Mouse blood was drawn from the periorbital sinus into the anticoagulant sodium citrate (1:9 vol/vol). Samples were centrifuged for 5 minutes at 400g and the platelet rich plasma (PRP) was collected. Samples were then centrifuged at 3,000g for 15 minutes to prepare platelet poor plasma (PPP). Platelet numbers were determined manually using Munopette (Becton Dickinson, Rutherford, NJ) and a hemocytometer.
Cytokine treatments.
Recombinant mouse tumor necrosis factor-α (TNF-α; Boehringer Mannheim, Indianapolis, IN) was injected intravenously through periorbital sinus at 20 μg/kg of body weight or intraperitoneally at 4 μg per mouse. LPS (E coli serotype 0111:B4; Sigma, St Louis, MO) was injected intraperitoneally at 25 mg/kg. Control mice were injected with an equivalent volume of sterile saline by the same route of administration.
RESULTS
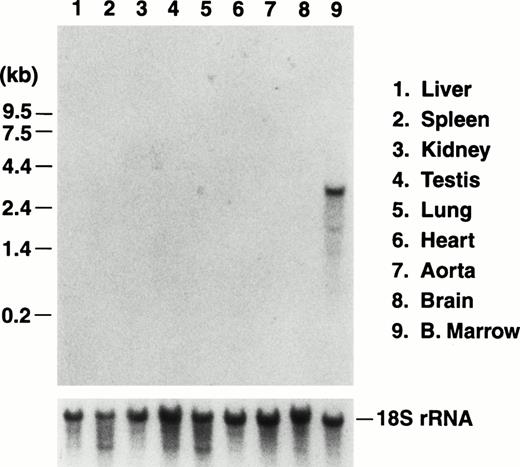
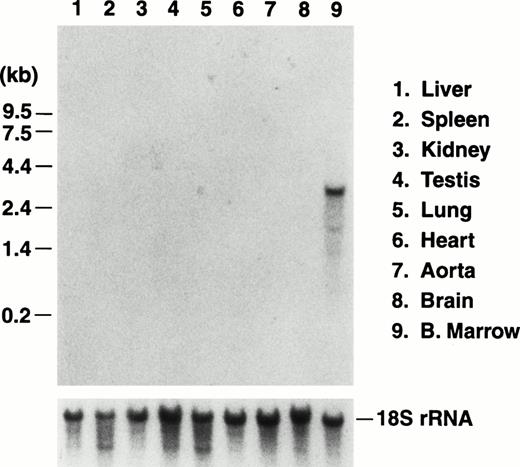
We previously reported the sequence of a 2.8-kbBamHI/EcoRI restriction fragment containing a single open reading frame encoding the mouse GP Ibα precursor polypeptide.19 The sequence of a 5′ contiguous 2.6-kbBamHI fragment has also been determined allowing a complete depiction of the murine GP Ibα gene (Fig1). To examine the in vivo expression of the mouse GP Ibα gene, Northern analysis was performed on RNA isolated from the major murine organs (Fig 2). Using probes from the protein-coding sequence of the murine GP Ibα gene, a single 2.7-kb GP Ibα mRNA was exclusively detected in bone marrow RNA and is consistent with the predicted size of the murine GP Ibα transcript. No hybridization signals were visible in any other organ even after lengthy autoradiographic exposures (2 weeks). The mouse GP Ibα transcript is approximately 300 nucleotides longer than the human mRNA owing to length divergence within the region of the gene encoding the macroglycopeptide domain.19 Thus, within the limits of detection using total RNA isolated from murine organs, GP Ibα gene expression is restricted to bone marrow.
Genomic arrangement of the mouse GP Ibα gene. Restriction enzyme analysis of a P1 plasmid containing a fragment of mouse genomic DNA identified contiguous BamHI andBamHI/EcoRI restriction fragments that were chosen for DNA sequence determination. Based on sequence alignment with the human GP Ibα gene, boundaries for two exons are proposed (boxed regions) with an open reading frame (shaded box) encoding the putative mouse GP Ibα precursor polypeptide.19 A shaded gray region under the schematic representation of the gene identifies the approximate position of a radiolabeled fragment used for Northern analysis (Fig 2). The nucleotide sequence has been deposited in GenBank (accession no.U91967).
Genomic arrangement of the mouse GP Ibα gene. Restriction enzyme analysis of a P1 plasmid containing a fragment of mouse genomic DNA identified contiguous BamHI andBamHI/EcoRI restriction fragments that were chosen for DNA sequence determination. Based on sequence alignment with the human GP Ibα gene, boundaries for two exons are proposed (boxed regions) with an open reading frame (shaded box) encoding the putative mouse GP Ibα precursor polypeptide.19 A shaded gray region under the schematic representation of the gene identifies the approximate position of a radiolabeled fragment used for Northern analysis (Fig 2). The nucleotide sequence has been deposited in GenBank (accession no.U91967).
Northern blot analysis of RNA prepared from the major murine organs. Total RNA was isolated from nine organs of adult mice and electrophoresed through a 1% agarose/formaldehyde denaturing gel. After electrophoresis, the RNA was transferred to a nitrocellulose membrane and hybridized with a radiolabeled probe of the murine GP Ibα coding sequence.19 A representative photograph of the autoradiograph obtained after hybridization and washing documents an RNA species of 2.7 kb in RNA prepared from bone marrow of a mouse femur. The size of the RNA is consistent with the predicted size of the transcript encoding mouse GP Ibα (Fig 1). No other hybridizing signals were observed after a lengthy (2 weeks) exposure to x-ray film. The migrating position of an RNA molecular weight standard is shown to the left. After obtaining the autoradiograph, the nitrocellulose membrane was stripped of radioactivity and rehybridized with a radiolabeled DNA probe from the mouse 18S rRNA gene to confirm similar amounts of RNA were loaded from the different RNA preparations (lower panel).
Northern blot analysis of RNA prepared from the major murine organs. Total RNA was isolated from nine organs of adult mice and electrophoresed through a 1% agarose/formaldehyde denaturing gel. After electrophoresis, the RNA was transferred to a nitrocellulose membrane and hybridized with a radiolabeled probe of the murine GP Ibα coding sequence.19 A representative photograph of the autoradiograph obtained after hybridization and washing documents an RNA species of 2.7 kb in RNA prepared from bone marrow of a mouse femur. The size of the RNA is consistent with the predicted size of the transcript encoding mouse GP Ibα (Fig 1). No other hybridizing signals were observed after a lengthy (2 weeks) exposure to x-ray film. The migrating position of an RNA molecular weight standard is shown to the left. After obtaining the autoradiograph, the nitrocellulose membrane was stripped of radioactivity and rehybridized with a radiolabeled DNA probe from the mouse 18S rRNA gene to confirm similar amounts of RNA were loaded from the different RNA preparations (lower panel).
Sequence alignment between the 5′ regions of human and mouse GP Ibα genes is shown defining potential regulatory elements within the 5′ region of the mouse GP Ibα gene promoter and identifying the intron/exon boundaries of the mouse gene (Fig 3). Specifically, mouse nucleotides 2,387 to 2,466 (numbering corresponds to GenBank accession no. U91967) represent exon I with the thymine at nucleotide position 2,387 corresponding to a similar thymine identified as the transcription initiation site of the human gene.20 Again, based on sequence alignment, exon II was defined as nucleotides 2,659 to 5,313 and contains an open reading frame beginning at nucleotide 2,665 extending through nucleotide 4,869.19 Thus, the characterized mouse gene sequence has an overall genomic arrangement similar to the human GP Ibα gene with an 80 nucleotide 5′ untranslated exon, a single intron of 192 nucleotides, and a single exon encoding the mouse GP Ibα polypeptide.24 25
Promoter alignments of mouse and human GP Ibα gene sequences. Mouse (Mo) and human (Hu) GP Ibα gene sequences are aligned flanking the transcription initiation site of the human GP Ibα gene (nucleotide +1). Negative nucleotide numbering relative to the transcription initiation site is shown to the left of each sequence. The human GP Ibα gene is composed of a 5′ untranslated exon (exon I), a single intron, and a single exon (exon II) containing the initiating methionine codon (ATG). The exons are highlighted by a shaded box with only a limited 5′ portion of exon II displayed. Mouse exon I corresponds to nucleotides 2,387 to 2,466 and exon II begins at nucleotide 2,659 of GenBank accession no.U91967. The human 5′ sequence contains GATA and Etscis-acting elements, which have previously been shown by mutagenesis to be essential for promoter activity in megakaryocytic-like cell lines.20 Mutated bases of the human sequence that eliminated promoter activity are highlighted by black boxes at nucleotides −150 to −142 (Ets) and −92 to −91 (GATA).20 The mouse GP Ibα sequence displays a similar overall arrangement with conserved GATA and Ets elements along with positive regulatory element (MegPos) identified in the rat and human platelet factor 4 promoters.41 Translated sequence for the first 14 residues of the human and mouse GP Ibα signal peptides is shown in exon II with a single-letter notation for each residue except where there exists sequence differences, in which case both species-specific amino acids are shown. A BamHI restriction site is underlined (nucleotides 207-212) and corresponds to the 3′ boundary of the promoter fragment used to generate transgenic mice expressing the reporter protein, luciferase. Human GP Ibα DNA sequence corresponds to GenBank accession no. M22403.
Promoter alignments of mouse and human GP Ibα gene sequences. Mouse (Mo) and human (Hu) GP Ibα gene sequences are aligned flanking the transcription initiation site of the human GP Ibα gene (nucleotide +1). Negative nucleotide numbering relative to the transcription initiation site is shown to the left of each sequence. The human GP Ibα gene is composed of a 5′ untranslated exon (exon I), a single intron, and a single exon (exon II) containing the initiating methionine codon (ATG). The exons are highlighted by a shaded box with only a limited 5′ portion of exon II displayed. Mouse exon I corresponds to nucleotides 2,387 to 2,466 and exon II begins at nucleotide 2,659 of GenBank accession no.U91967. The human 5′ sequence contains GATA and Etscis-acting elements, which have previously been shown by mutagenesis to be essential for promoter activity in megakaryocytic-like cell lines.20 Mutated bases of the human sequence that eliminated promoter activity are highlighted by black boxes at nucleotides −150 to −142 (Ets) and −92 to −91 (GATA).20 The mouse GP Ibα sequence displays a similar overall arrangement with conserved GATA and Ets elements along with positive regulatory element (MegPos) identified in the rat and human platelet factor 4 promoters.41 Translated sequence for the first 14 residues of the human and mouse GP Ibα signal peptides is shown in exon II with a single-letter notation for each residue except where there exists sequence differences, in which case both species-specific amino acids are shown. A BamHI restriction site is underlined (nucleotides 207-212) and corresponds to the 3′ boundary of the promoter fragment used to generate transgenic mice expressing the reporter protein, luciferase. Human GP Ibα DNA sequence corresponds to GenBank accession no. M22403.
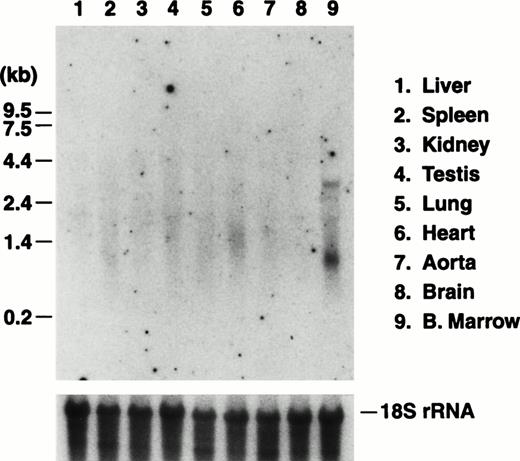
The in vivo activity of a mouse GP Ibα promoter fragment was tested with the generation of transgenic mice expressing the reporter enzyme luciferase under the control of the 5′ 2.6-kb BamHI fragment depicted in Fig 1. Individual founder mice were expanded into independent transgenic colonies and the luciferase mRNA transcript was visualized by Northern analysis of the major murine organs (Fig 4). The expression of the transgene transcript showed an organ-specific pattern similar to that obtained by probing for the endogenous GP Ibα transcript, namely detectable levels of transcript exclusively in bone marrow RNA preparations.
Northern blot analysis of RNA prepared from transgenic murine organs. Transgenic mice were generated expressing the reporter protein, luciferase, under the control of a BamHI promoter fragment of the murine GP Ibα gene (Fig 1). Five mouse colonies were expanded from individual founder mice. Results are presented from one mouse line and are typical of each of the five lines in which expression of the transgene was observed. As described in Fig 2, total RNA was prepared from the major organs of the transgenic mice and analyzed by Northern analysis. The luciferase mRNA transcript of 2.4 kb was detected using a radiolabeled fragment from the coding sequence for luciferase. Similar to results probing for the endogenous GP Ibα transcript, a transcript is only visible in RNA prepared from the marrow of a mouse femur. Subsequent hybridization of the same filter with a portion of the mouse 18S rRNA gene is shown to illustrate similar RNA levels for each lane.
Northern blot analysis of RNA prepared from transgenic murine organs. Transgenic mice were generated expressing the reporter protein, luciferase, under the control of a BamHI promoter fragment of the murine GP Ibα gene (Fig 1). Five mouse colonies were expanded from individual founder mice. Results are presented from one mouse line and are typical of each of the five lines in which expression of the transgene was observed. As described in Fig 2, total RNA was prepared from the major organs of the transgenic mice and analyzed by Northern analysis. The luciferase mRNA transcript of 2.4 kb was detected using a radiolabeled fragment from the coding sequence for luciferase. Similar to results probing for the endogenous GP Ibα transcript, a transcript is only visible in RNA prepared from the marrow of a mouse femur. Subsequent hybridization of the same filter with a portion of the mouse 18S rRNA gene is shown to illustrate similar RNA levels for each lane.
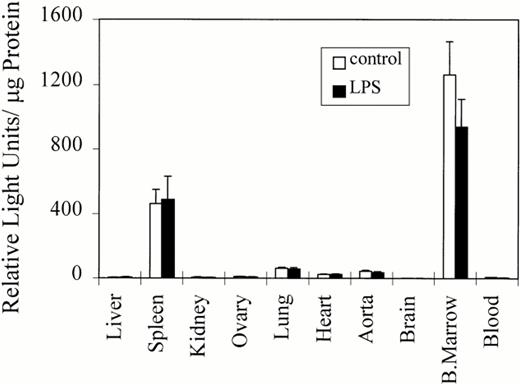
The expression of the luciferase transgene was further evaluated by examining luciferase activity in organ homogenates from two transgenic colonies (Fig 5). In all transgenic mice analyzed, the highest levels of luciferase activity were observed in bone marrow isolated from a femur. Significant luciferase activity was also observed in spleen, consistent with the spleen being a major hematopoietic organ in the mouse. A reproducible, but low-level of transgene expression, was observed in heart, aorta, and lung even after extensive perfusion of the mouse. These expression levels were approximately 2% to 5% of the activity observed in bone marrow. Although the different colonies varied somewhat in the levels of luciferase expression, the tissue distribution of luciferase activity and the ratio of expression among different organs within the same transgenic colony was similar to those results presented in Fig 5. Thus, the integration site of the transgene seems to be an unlikely factor for the generation of luciferase activity in the organ-specific manner illustrated in Fig 5.
Luciferase activity in transgenic mouse colonies. Luciferase activity in organ homogenates from two transgenic colonies (32 [n = 5] and 12 [n = 5]) derived from independent founders is presented. The mean and standard error of luciferase activity (RLUs per μg of protein) are shown. Maximal luciferase activity was observed in bone marrow and spleen, both hematopoietic organs in mice and both containing murine megakaryocytes. Luciferase activity was also detected in blood, but is not apparent in this figure owing to the small contribution of platelet protein to the total protein found in whole blood (see Results). Consistent, albeit low levels, of luciferase activity were observed in lung, heart, and aorta even after extensive perfusion of the mouse. The relevance of low levels of gene activity in these organs is discussed.
Luciferase activity in transgenic mouse colonies. Luciferase activity in organ homogenates from two transgenic colonies (32 [n = 5] and 12 [n = 5]) derived from independent founders is presented. The mean and standard error of luciferase activity (RLUs per μg of protein) are shown. Maximal luciferase activity was observed in bone marrow and spleen, both hematopoietic organs in mice and both containing murine megakaryocytes. Luciferase activity was also detected in blood, but is not apparent in this figure owing to the small contribution of platelet protein to the total protein found in whole blood (see Results). Consistent, albeit low levels, of luciferase activity were observed in lung, heart, and aorta even after extensive perfusion of the mouse. The relevance of low levels of gene activity in these organs is discussed.
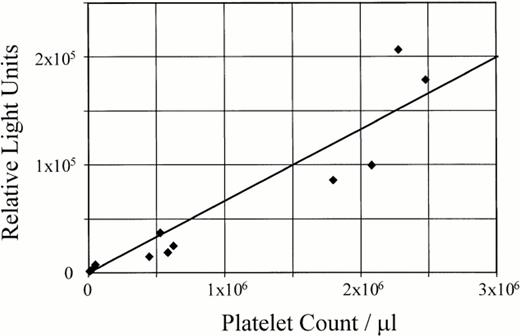
Typically, positive transgenic mice were identified by a direct assay of luciferase activity in whole blood with values ranging from 15,000 to 30,000 light units from 10 μL of whole blood, as compared with 200 to 250 light units using 10 μL of whole blood from nontransgenic mice. This strategy was routinely used to identify positive mice and simplify the creation of the transgenic colonies. Yet, as shown in Fig5, the luciferase activity present in whole blood is not apparent if the activity is normalized to light units per μg of total protein. Experiments assaying PRP and PPP from different transgenic lines confirmed the luciferase activity in whole blood coincides with the presence of platelets exhibiting a linear correlation between luciferase activity and the number of platelets (Fig 6). Thus, the apparent absence of luciferase activity in whole blood as presented in Fig 5 reflects the minuscule contribution of platelet proteins to the complete protein composition of whole blood rather than an absence of luciferase activity. Indeed, mouse platelets represent 0.53% of the unit volume of whole blood.26
Luciferase activity in transgenic platelets. A linear correlation exists between the number of platelets and luciferase activity in the transgenic mice. PRP and PPP from transgenic mice (colony 32) was used to resuspend blood cells and subsequently determine luciferase activity. These reconstitution experiments confirmed the luciferase activity in whole blood coincides completely with the presence of platelets with a linear correlation between luciferase activity and the number of platelets (r2 = 0.88).
Luciferase activity in transgenic platelets. A linear correlation exists between the number of platelets and luciferase activity in the transgenic mice. PRP and PPP from transgenic mice (colony 32) was used to resuspend blood cells and subsequently determine luciferase activity. These reconstitution experiments confirmed the luciferase activity in whole blood coincides completely with the presence of platelets with a linear correlation between luciferase activity and the number of platelets (r2 = 0.88).
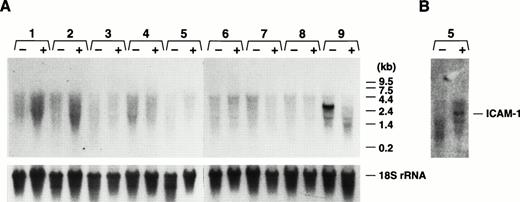
Having characterized the normal in vivo expression pattern of the GP Ibα gene and the ability of transgenic colonies expressing a reporter protein to recapitulate this expression pattern, we performed experiments to determine whether the endogenous GP Ibα gene or the GP Ibα/luciferase transgene were responsive to cytokines. As a model of gram-negative sepsis, LPS was administered to mice by intraperitoneal injection (25 mg/kg). The wide range of LPS-induced effects and toxicity in mice is well-documented and the doses administered to individual mice were sufficient to achieve maximal levels for a variety of cytokines within 3 to 6 hours.27 28 Examining luciferase activity in transgenic mice organs, no increase in GP Ibα expression was detected (Fig 7). Assaying for the presence of the endogenous GP Ibα transcript in mouse organs, no increase in GP Ibα expression was detected (Fig 8). In fact, the situation was just the opposite in bone marrow with an absence of GP Ibα transcript 24 hours post-LPS treatment (Fig 8A). The induction of an LPS-induced inflammatory state was confirmed with an increase in ICAM-1 mRNA (Fig8B). Additionally, mice were administered recombinant murine TNF-α either intraperitoneally (20 μg/kg maximal dose) or intravascular (4 μg/kg maximal dose). At time points up to 24 hours postinjection, organs were assayed for luciferase activity and levels of GP Ibα mRNA were determined by Northern analysis. At all doses and time points examined, administration of TNF-α had no demonstrable effects on GP Ibα gene or luciferase transgene expression (data not shown).
Transgene activity in an animal model of gram-negative sepsis. LPS was administered to mice (colony 32) containing a luciferase transgene under the control of the mouse GP Ibα promoter. The wide range of LPS-induced effects and toxicity in mice is well documented and doses administered to individual mice were sufficient to achieve maximal levels for a variety of cytokines. LPS was administered by intraperitoneal injection (25 mg/kg) and luciferase activity in the major murine organs was determined. Results are shown for assays performed 24 hours postinjection, although similar results were obtained at 4-hour intervals leading up to the 24-hour assay shown. The mean and standard error of the mean are shown for each organ (n = 4). Control mice were injected intraperitoneally with an equal volume of saline buffer. No differences in the levels of luciferase activity were observed at any time point.
Transgene activity in an animal model of gram-negative sepsis. LPS was administered to mice (colony 32) containing a luciferase transgene under the control of the mouse GP Ibα promoter. The wide range of LPS-induced effects and toxicity in mice is well documented and doses administered to individual mice were sufficient to achieve maximal levels for a variety of cytokines. LPS was administered by intraperitoneal injection (25 mg/kg) and luciferase activity in the major murine organs was determined. Results are shown for assays performed 24 hours postinjection, although similar results were obtained at 4-hour intervals leading up to the 24-hour assay shown. The mean and standard error of the mean are shown for each organ (n = 4). Control mice were injected intraperitoneally with an equal volume of saline buffer. No differences in the levels of luciferase activity were observed at any time point.
Northern analysis of GP Ibα expression in mice administered LPS. As a model of gram-negative sepsis, LPS was administered to mice (colony 32) by intraperitoneal injection (25 mg/kg). As described in Fig 2, total RNA was prepared from treated (+) and control (−) mice. (A) Lanes 1 to 9 correspond to the same organs listed in the legend for Fig 2. Again, a visible GP Ibα transcript is only present in bone marrow (lane 9) with no increase in GP Ibα expression detected in any organ, but an absence of the GP Ibα bone marrow transcript 24 hours post-LPS treatment. (B) The nitrocellulose filter of (A) was rehybridized with an ICAM-1–radiolabeled cDNA fragment and confirmed an inflammatory state in the treated mice with an increase in the lung ICAM-1 mRNA (lane 5).
Northern analysis of GP Ibα expression in mice administered LPS. As a model of gram-negative sepsis, LPS was administered to mice (colony 32) by intraperitoneal injection (25 mg/kg). As described in Fig 2, total RNA was prepared from treated (+) and control (−) mice. (A) Lanes 1 to 9 correspond to the same organs listed in the legend for Fig 2. Again, a visible GP Ibα transcript is only present in bone marrow (lane 9) with no increase in GP Ibα expression detected in any organ, but an absence of the GP Ibα bone marrow transcript 24 hours post-LPS treatment. (B) The nitrocellulose filter of (A) was rehybridized with an ICAM-1–radiolabeled cDNA fragment and confirmed an inflammatory state in the treated mice with an increase in the lung ICAM-1 mRNA (lane 5).
DISCUSSION
Among platelet receptors, the GP Ib-IX-V complex is important because it initiates thrombus formation through a tethering of the circulating platelet to von Willebrand factor, an adhesive ligand of the subendothelial matrix.4 The importance of this receptor-ligand interaction for normal platelet biology is best exemplified by congenital bleeding disorders resulting from the lack of either the receptor or the ligand, the Bernard-Soulier syndrome, and von Willebrand disease, respectively. In addition, the absence of a GP Ib-IX-V complex coincides with the release of giant platelets, leading to speculations that the synthesis and assembly of the complex is directly linked to normal platelet morphology. Thus, the importance of the complex in normal megakaryocyte and platelet physiology is clear and well-documented. Nevertheless, expression of GP Ibα and other subunits of the complex by nonmegakaryocytic cells has been suggested by a number of in vitro observations documenting expression of the individual subunits of the complex in cultured human endothelial and smooth muscle cells.17,18,29-31 Despite these observations on cultured cells, little is known concerning the regulation of GP Ibα expression in vivo, the only corollary is an immunologic identification of GP Ibα in tonsillar endothelium.14 17
The current study examined GP Ibα gene expression to determine in a systematic manner the relative abundance of the GP Ibα transcript among the major organs of the mouse. We conclude the GP Ibα mRNA is a dominant transcript in bone marrow, as identified by Northern analysis of total mouse RNA. No other major organ of the mouse expresses GP Ibα to the same extent. We also used a transgenic model of GP Ibα expression using the highly-sensitive reporter gene product, luciferase, and observed the highest levels of luciferase activity in bone marrow and spleen (Fig 5), both hematopoietic organs in the mouse. Consistent and reproducible luciferase activity was observed in lung, heart, and aorta, yet the gene expression in these satellite organs was, on average, 20 to 50 times lower than the activity present in bone marrow preparations. Northern blots examining GP Ibα transcript levels, luciferase transcript levels, and assays of luciferase protein activity showed that differences in technique sensitivity can bias conclusions on the expression of a particular gene product. In fact, we did not observe GP Ibα or luciferase mRNA in spleen, yet luciferase activity assays showed the luciferase transgene was capable of expressing protein to a level nearly one third that of bone marrow.
Our results identifying a low level of in vivo GP Ibα gene activity within some nonhematopoietic organs, namely, heart, aorta, and lung, forces a more compelling question as to whether low levels of gene activity provide previously unrecognized functional properties for GP Ibα or a GP Ib-IX-V complex? No phenotypic abnormalities beyond hemostatic deficiency and giant platelets have been described for patients with the Bernard-Soulier syndrome. Others have suggested from in vitro experiments there exists a GP Ib-IX-V link between the release of cytokines and thrombotic potential.17,18 However, our in vivo results do not support such a link. At LPS doses sufficient to achieve maximal levels for a variety of cytokines,27,28 we observed no increase in GP Ibα gene expression (Figs 7 and 8). Direct administration of murine TNF-α did not increase GP Ibα gene expression. In fact, in the LPS-induced model of gram-negative sepsis, the GP Ibα mRNA was absent 24 hours posttreatment (Fig 8A). This result may reflect the more global changes, such as platelet depletion, which is reported to occur after LPS-induced toxicity in mice.32 Of course, one caveat in our experiment is the use of a mouse model, which may or may not, extrapolate to expression of the human GP Ibα gene.
The expression of GP Ibα mRNA and protein in cells other than megakaryocytes is not without controversy. Some investigators have been unable to document expression of GP Ibα by cultured endothelial cells15 or identify von Willebrand factor binding to endothelial cells dependent on a GP Ib-IX-V receptor complex.16 Reconciling these discrepant results from different laboratories can be difficult. Complicating factors for the interpretation of GP Ibα expression by endothelial cells could include the apparent changes in gene expression occurring during the transition of an endothelial cell from an in vivo to an in vitro environment,33 endothelial cell heterogeneity,34 or even interlaboratory variations using similar techniques, but achieving different levels of sensitivity. An alternative explanation might be low levels of transcription for a tissue-specific gene in nonspecific cells.35,36 Two of the best documented examples are the presence of clotting cofactor VIII mRNA in lymphocytes37 and the detection of the Duchenne muscular dystrophy transcripts in nonmuscle cells.38 The transcription of a cell-specific gene in a variety of other cell types can be accounted for by two different mechanisms.39 The first is a basal level of transcription that has been referred to as “illegitimate” or “leaky” transcription.35 The second is expression by a subset of cells within a given tissue and can be particularly confusing if the vascular cells of a tissue or organ are contributing “leaky” activity. Thus, in light of no identified physiologic relevance for GP Ibα gene activity beyond that described for megakaryocytes and platelets, it is conceivable the low level of gene activity observed in some cells and organs may represent “leaky” transcription. Perhaps studies characterizing a targeted disruption of the GP Ibα gene in mice may provide more definitive answers.
While characterizing the endogenous in vivo expression pattern of GP Ibα, this study has identified a murine promoter fragment capable of directing the synthesis of heterologous gene products in megakaryocytes. Thus, the promoter must contain all necessary elements for in vivo expression of the GP Ibα gene. The human GP Ibα promoter has been characterized and contains megakaryocytic-specificcis-acting elements, specifically GATA and Ets motifs, common among megakaryocytic-specific genes.20,40-43 Both GATA and Ets motifs are conserved within the murine GP Ibα promoter along with a positive megakaryocytic regulatory element identified in the rat and human platelet factor 4 promoters41 (Fig 3). Previous in vitro analysis of the human GP Ibα promoter identified 253 nucleotides 5′ to the transcription initiation site as essential for maximum activity in human erythroleukemia cells.20 The alignment between the mouse and human GP Ibα sequences displays some highly conserved sequences within this region of both promoters along with conserved sequences in both exon I and the single intron of each gene (Fig 3). Indeed, the identification of conserved elements between the human and mouse promoter sequences may have merit. We have previously shown that the human GP Ibα promoter functions in vivo to express the human GP Ibα polypeptide on the surface of murine transgenic platelets.44 Thus, conserved mechanisms must exist for megakaryocytic gene expression between the two species. Alignment of genomic sequences extending 5′ to that shown in Fig3 did not display long stretches of sequence similarity between the human and murine promoters, but without further in vivo tests of promoter activity, it is impossible to suggest that the relatively short promoter regions displayed in Fig 3 are sufficient for in vivo activity.
Overall, these studies support the long-term objective of manipulating membrane receptors in the unique cellular characteristics of a platelet. The expression of megakaryocytic genes and the identification of promoter fragments supporting expression in a manner that mirrors the endogenous gene product provide crucial information to achieve this objective, namely, the in vivo expression of variant receptors. Such studies should provide new information relevant to the in vivo physiology and pathophysiology of platelet receptor function.
ACKNOWLEDGMENT
The authors acknowledge the Sam and Rose Stein Charitable Trust Fund in the Department of Molecular and Experimental Medicine at The Scripps Research Institute for the generation of a DNA core facility. The authors are grateful for the technical support of James Roberts and Hon Tran during the completion of this project.
Supported by Grant No. HL-50545 from the Heart, Lung, and Blood Institute of the National Institutes of Health, Bethesda, MD.
Address reprint requests to Jerry Ware, PhD, Mail Drop SBR8, The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail: jware@scripps.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.



![Fig. 5. Luciferase activity in transgenic mouse colonies. Luciferase activity in organ homogenates from two transgenic colonies (32 [n = 5] and 12 [n = 5]) derived from independent founders is presented. The mean and standard error of luciferase activity (RLUs per μg of protein) are shown. Maximal luciferase activity was observed in bone marrow and spleen, both hematopoietic organs in mice and both containing murine megakaryocytes. Luciferase activity was also detected in blood, but is not apparent in this figure owing to the small contribution of platelet protein to the total protein found in whole blood (see Results). Consistent, albeit low levels, of luciferase activity were observed in lung, heart, and aorta even after extensive perfusion of the mouse. The relevance of low levels of gene activity in these organs is discussed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/2/10.1182_blood.v92.2.488/5/m_blod41403005x.jpeg?Expires=1769422812&Signature=oXMZAOKCWTghYq~e7Pj0xf~~5JUNwQ~pl-edrqiXDSvZsAtLj~3hBwjh89stwbjHzj~-eJ0hrTwEUx8wBlGkOtgjeqjQoHGP9SOLwUEA-9WG8fSjV8mc8KhU3~dha4t7aktg8iGYfmbExfzkO5qVk6mhAg3Z9IHkTbN~xLTFpvBsFTqG~fYQB8aGwqwXor-qg5CPtHUlKwpMuhJQq5IiZd-rixLc66Ayg5GufhjbKYfWfP2ifypew1qIWE959gEDkzdFTdvgCMLKWk7ZsYA8fsa5~jti3f9LzPg~VmqPEli-kz-EKyhSNpD9NIaAEAW416dshmA66PjFejksqLe~lA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Fig. 5. Luciferase activity in transgenic mouse colonies. Luciferase activity in organ homogenates from two transgenic colonies (32 [n = 5] and 12 [n = 5]) derived from independent founders is presented. The mean and standard error of luciferase activity (RLUs per μg of protein) are shown. Maximal luciferase activity was observed in bone marrow and spleen, both hematopoietic organs in mice and both containing murine megakaryocytes. Luciferase activity was also detected in blood, but is not apparent in this figure owing to the small contribution of platelet protein to the total protein found in whole blood (see Results). Consistent, albeit low levels, of luciferase activity were observed in lung, heart, and aorta even after extensive perfusion of the mouse. The relevance of low levels of gene activity in these organs is discussed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/2/10.1182_blood.v92.2.488/5/m_blod41403005x.jpeg?Expires=1769742625&Signature=wI9lzGz7G2wcRprZFNtAB62CqQ9WIEdwb1CUJhUCCqOcgS6sfjm7NgFf2I4ZhImnmKXJBpP~sptoYtYl86DEeqWI2hz03kdf7HJHuLwqX-c-bJKWlD-nvpV-y2CjKq0JFWflNnnjkU2tIzQQ3A9aehzxkQUGX5B8SEpcrlATaIXIXRscrCQXx8oHdw0dGv2CQNyzw7mP9Qm1H5y7r1xON-q3VYFEh6BcJv8aoHAp5unqayssy3Ik~FhTCH2G2SdVsQKnDlpZAhEknGwQHRMIFlbvgvSyHpSDgXM13bkYc-ZeS-rs5gh6r1Ce2gXhRGQ3DiE9PR~FDjDq9mZvhNtoOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)