Abstract
In erythrocytes, 80-kD protein 4.1R regulates critical membrane properties of deformability and mechanical strength. However, previously obtained data suggest that multiple isoforms of protein 4.1, generated by alternative pre-mRNA splicing, are expressed during erythroid differentiation. Erythroid precursors use two splice acceptor sites at the 5′ end of exon 2, thereby generating two populations of 4.1 RNA: one that includes an upstream AUG-1 in exon 2′ and encodes high molecular weight isoforms, and another that skips AUG-1 in exon 2′ and encodes 4.1 by initiation at a downstream AUG-2 in exon 4. To begin an analysis of the complex picture of protein 4.1R expression and function during erythropoiesis, we determined the number and primary structure of 4.1R isoforms expressed in erythroblasts. We used reverse-transcription polymerase chain reaction to amplify and clone full-length coding domains from the population of 4.1R cDNA containing AUG-1 and the population excluding AUG-1. We observed an impressive repertoire of 4.1R isoforms that included 7 major and 11 minor splice variants, thus providing the first definitive characterization of 4.1R primary structures in a single-cell lineage. 4.1R isoforms, transfected into COS-7 cells, distributed to the nucleus, cytoplasm, plasma membrane, and apparent centrosome. We confirmed previous studies showing that inclusion of exon 16 was essential for efficient nuclear localization. Unexpectedly, immunochemical analysis of COS-7 cells transfected with an isoform lacking both AUG-1 and AUG-2 documented that a previously unidentified downstream translation initiation codon located in exon 8 can regulate expression of 4.1R. We speculate that the repertoire of primary structure of 4.1R dictates its distinct binding partners and functions during erythropoiesis.
THE 80-kD protein 4.1R is a crucial structural component of the plasma membrane of circulating erythrocytes. Plasma membranes of these terminally differentiated red cells consist of a lipid bilayer, integral proteins, and an underlying skeletal protein latticework, which plays a critical role in supporting the bilayer and in regulating membrane properties of deformability and mechanical strength.1,2 Well-characterized 80-kD protein 4.1R is a critical constituent of the membrane skeleton because it participates in both horizontal associations within the skeletal protein network and in vertical associations linking the skeletal network to integral proteins embedded in the lipid bilayer.3 The N-terminal 30-kD region of 4.1R functions as a membrane binding domain via specific interactions with integral proteins glycophorin C,4-9 band 3,10,11 and CD44.12 A 10-kD domain further downstream contains the binding sites for spectrin and actin and is essential for formation of the ternary complex of 4.1R, spectrin, and actin, which regulates membrane mechanical strength.13-16
However, 80-kD 4.1R represents only one member of a family of 4.1R isoforms generated by complex alternative pre-mRNA splicing and expressed in both nucleated erythroid precursors and nonerythroid tissues.17-21 Although mature erythrocytes express only a restricted repertoire of 4.1R isoforms,18 we have obtained evidence that 4.1R expression in erythroblasts is more complicated and includes a much broader variety of 4.1R isoforms. Indeed, analysis of 4.1R mRNA structure during erythropoiesis showed that erythroid precursors use two splice acceptor sites at the 5′ end of exon 2, thereby generating two populations of 4.1R RNA: one that includes an upstream AUG (AUG-1) and encodes high molecular weight isoforms of 4.1R, and another that skips AUG-1 and encodes 4.1R by initiation at a downstream AUG (AUG-2) in exon 4. Initial characterization of protein expression by Western blot analysis using antipeptide antibodies revealed four prominent bands between ∼69-135 kD.20 By immunofluorescence microscopy, protein 4.1 epitopes localized predominantly to the plasma membrane, cytoplasm, and apparent centrosome in erythroblasts.20 Further studies in mammalian nonerythroid cells substantiate that 4.1 localizes to intracellular organelles. In centrosomes, protein 4.1 epitopes distribute along centriolar cylinders and on pericentriolar fibers.22 In addition, 4.1 is a component of the nuclear matrix and appears to be disbursed throughout nuclear domains involved in RNA and DNA metabolism.23-25 Moreover, the 80-kD isoform of protein 4.1 expressed in transfected COS cells localizes within the nucleus.26 It therefore seems likely that the various protein 4.1 isoforms expressed in erythroblasts may contribute significantly not only to plasma membrane structure but also to nuclear and centrosomal architecture and function.
The spectrin-based membrane skeleton is in a dynamic state during erythropoiesis. Kinetic studies of membrane skeletal protein synthesis and assembly in a number of mammalian systems have led to two important generalizations.27-31 Firstly, synthesis and assembly of the various protein constituents are asynchronous; secondly, regulation of assembly depends not only on the quantity of a given protein synthesized but also on the availability of binding partners with which it must interact for assembly. However, characterization of membrane composition during erythropoiesis has become increasingly complicated because it has recently become clear that a number of red cell skeletal proteins are either products of complex alternative pre-mRNA splicing events or are encoded by genes that are members of gene families.21 32-38 Because the majority of studies detailing membrane protein synthesis and assembly were performed before our knowledge of the larger number of potential players, there remain important unanswered questions. One of these questions concerns the expression and function of protein 4.1R since previous kinetic studies focused on synthesis and membrane assembly of only the 80-kD polypeptide. Furthermore, protein expression in intracellular compartments to which 4.1 localizes, such as nucleus and centrosome, has not been extensively characterized.
To initiate analysis of the complex picture of protein 4.1R expression and function during erythropoiesis, we determined the number and primary structure of 4.1R isoforms expressed in well-hemoglobinized erythroblasts. We observed a total of 18 isoforms, 9 encoded from AUG-1, 7 encoded from AUG-2, and, surprisingly, 2 lacking both of the previously identified translation initiation sites. Of the 18 isoforms identified, 7 isoforms comprised 87% of all of the clones characterized, with 4 isoforms including AUG-1 and 3 isoforms excluding AUG-1 and initiating translation at AUG-2. These 7 major isoforms varied from one another in their patterns of inclusion and exclusion of exons encoding the functionally critical spectrin/actin binding domain and membrane binding domain. In transfected COS-7 cells, isoforms distributed to the nucleus, cytoplasm, plasma membrane, and apparent centrosome. Interestingly, individual isoforms segregated to more than one subcellular compartment. Inclusion of exon 16 appeared to be critical for nuclear localization, as had been previously reported.26 Finally, the two isoforms lacking both AUG-1 and AUG-2 were expressed as proteins with translation initiated at a previously unsuspected initiation codon located in exon 8. These two ∼ 68-kD polypeptides, which included sequences encoded by exon 16, showed strong nuclear localization. We theorize that the existence of such a diverse repertoire of 4.1R primary structure may reflect preferential interaction of 4.1R isoforms with distinct binding partners and varied functions of 4.1R during erythropoiesis.
MATERIALS AND METHODS
Erythroid progenitor isolation.
Normal human bone marrow cells were obtained after informed consent from patients at the time of hip replacement surgery. CD34+cells were isolated by positive selection using an immunomagnetic bead procedure (Dynabeads M-450; Dynal, Oslo, Norway) as previously described.39 CD34+ erythroid progenitor cells were grown in standard methylcellulose colony assays with optimal concentrations of recombinant stem cell factor (SCF; 25 ng/mL), recombinant interleukin-3 (IL-3; 50 U/mL), and recombinant erythropoietin (Epo; 2 U/mL) to stimulate the proliferation of colony-forming unit–erythroid (CFU-E) and burst-forming unit–erythroid (BFU-E). BFU-E–derived colonies identified as erythroid were plucked at day 11. Cells were washed twice and incubated overnight in the presence of Epo in a modified Eagle’s medium (MEM) + 20% fetal calf serum (FCS) to eliminate the rare contaminating macrophages. The nonadherent cell suspension contained over 95% basophilic and early polychromatophilic erythroblasts as determined by examination of May-Grunwald-Giemsa–stained cytospin slides.
RNA preparation and DNA cloning.
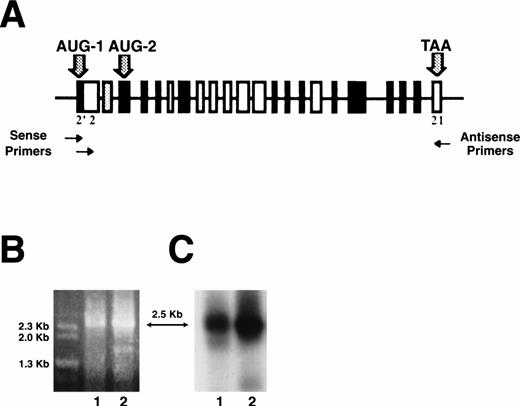
Total RNA from BFU-E–derived erythroblasts was prepared as previously described.40 Full-length 4.1R cDNAs were cloned by reverse transcriptase-polymerase chain reaction (RT-PCR) of total RNA as previously described.24 Primers used for PCR are shown in Table 1 and Fig 1A. 4.1R cDNAs including AUG-1 were amplified with a pair of primers complementary to exon 2′ and to exon 21 (Fig 1A). Amplification of 4.1R cDNAs excluding AUG-1 was achieved by using a primer complementary to the end of the region upstream of exon 2′ and to the beginning of exon 2, thus selecting cDNAs excluding exon 2′, in conjunction with the primer complementary to exon 21 (Fig 1A). 4.1R cDNAs were then inserted into pSP72 vector (Promega Corp, Madison, WI). Before cloning, the pSP72 vector was modified by insertion of a KT3-epitope tag derived from a sequence of the C-terminal domain of the Simian Virus 40 (SV40) large T antigen.41 Alternatively, the KT3-epitope tag was replaced by either an HA-epitope tag from the influenza viral hemagglutinin or a c-myc–epitope tag. cDNA clones were screened by Southern dot blotting using 32P-labeled probes specific for each alternatively spliced exon of 4.1R, and screening was confirmed by DNA sequencing (Sequenase Version 2.0 DNA Sequencing Kit, USB, Cleveland, OH). Finally, epitope-tagged 4.1R cDNAs were inserted into the expression vector pSV2neoCMV (kindly provided by Dr P. Yaswen, Lawrence Berkeley National Laboratory, Berkeley, CA) using anEcoRI restriction site and used for cell transfection. In some experiments, various ATGs throughout the 4.1R coding sequence were mutated to CTGs using the Quikchange kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. Mutated clones were then processed as described above.
Analysis of RT-PCR products of full-length 4.1R erythroblast mRNA. Total RNA was purified from well-hemoglobinized erythroblasts, transcribed into cDNA, and amplified by RT-PCR using primer sets to amplify full-length coding domains from two populations of 4.1R cDNA: those that contained AUG-1 and those that deleted AUG-1 (A). Erythroblast cDNA amplified with exon 2′ (sense) and exon 21 (antisense) primers gave a product of ∼2.5 kb when analyzed on a 0.7% agarose gel (B, lane 1). Eythroblast cDNA amplified with exon 2 (sense) and exon 21 (antisense) primers also gave a product of ∼2.5 kb (B, lane 2). These amplification products were consistent with the predicted sizes of protein 4.1R cDNAs either containing or deleting AUG-1. By Southern blot analysis of amplified cDNA products, both RT-PCR products hybridized with 80-kD full-length 4.1R DNA, identifying them as 4.1R (C).
Analysis of RT-PCR products of full-length 4.1R erythroblast mRNA. Total RNA was purified from well-hemoglobinized erythroblasts, transcribed into cDNA, and amplified by RT-PCR using primer sets to amplify full-length coding domains from two populations of 4.1R cDNA: those that contained AUG-1 and those that deleted AUG-1 (A). Erythroblast cDNA amplified with exon 2′ (sense) and exon 21 (antisense) primers gave a product of ∼2.5 kb when analyzed on a 0.7% agarose gel (B, lane 1). Eythroblast cDNA amplified with exon 2 (sense) and exon 21 (antisense) primers also gave a product of ∼2.5 kb (B, lane 2). These amplification products were consistent with the predicted sizes of protein 4.1R cDNAs either containing or deleting AUG-1. By Southern blot analysis of amplified cDNA products, both RT-PCR products hybridized with 80-kD full-length 4.1R DNA, identifying them as 4.1R (C).
Cell culture and transfections.
NIH/3T3 cells were obtained from ATCC (Rockville, MD),and COS-7 cells were kindly provided by Dr C. Collins (UCSF, San Francisco, CA). Cells were transiently transfected by lipofection as previously described with minor modifications.24 Briefly, cells on coverslips were grown for 24 hours, washed with phosphate-buffered saline (PBS) and with Opti-MEM I medium (Life Technologies Inc, Gaithersburg, MD) and incubated for 8 to 14 hours in 1 mL of Opti-MEM I medium containing 6 μL of LipofectAMINE (Life Technologies Inc) and 2.5 μg of cDNAs encoding various epitope-tagged 4.1R isoforms. After transfection, cells were washed in PBS and incubated in growth medium for 34 to 40 hours.
Immunofluorescence microscopy.
Samples were processed for immunofluorescence microscopy as previously described with minor modifications.24 All steps were performed at room temperature. Forty-eight hours after transfection, cells were fixed with 3% paraformaldehyde for 20 minutes and permeabilized with 0.5% Triton X-100 for 10 minutes. After blocking in 10% (vol/vol) goat serum for 1 hour, samples were probed with either an anti-KT3–epitope tag monoclonal antibody (kindly provided by Dr B. Schwer, Cornell University Medical College, New York, NY) or with an affinity purified anti-HA–epitope tag polyclonal antibody (Zymed Laboratories Inc, South San Francisco, CA) or with a monoclonal anti–c-myc–epitope tag antibody (Boehringer Mannheim, Indianapolis, IN) diluted 1/10, 1/300, and 1/200, respectively. In some experiments, samples were double stained with a monoclonal anti–HA-epitope tag antibody (Babco, Richmond, CA) diluted 1/1,000 and a polyclonal 4.1R antibody raised against the 21- amino acid (aa) sequence encoded by exon 16,24 used at 10 μg/mL. After incubation with primary antibodies, samples were incubated for 1 hour, either with anti-mouse IgG conjugated to Texas red or anti-rabbit IgG conjugated to fluorescein isothiocyanate (FITC; Molecular Probes Inc, Eugene, OR), diluted 1/2,000 and 1/5,000, respectively. Coverslips were mounted using Vectorshield containing 4′,6-diamidino-2-phenylindole (DAPI) as a nuclear DNA staining probe (Vector Laboratories, Burlingame, CA). Microscopic analysis of the samples and image processing were performed as previously described.24
Immunoprecipitation.
COS-7 cells, seeded at 106 in 100-mm cell culture dishes, were grown for 24 hours, transfected as described above using 24 μL of LipofectAMINE and 10 μg of DNA per dish and 48 hours after transfection, cells were washed twice in PBS and lyzed in 750 μL of ice cold RIPA buffer (10 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 1% IGEPAL CA-630 [Sigma Chemical Co, St Louis, MO], 0.1% sodium dodecyl sulfate (SDS), 2 mmol/L Pefabloc (Boehringer Mannheim), 2 mmol/L benzamidine, 10 μg/mL aprotinin, 5 μg/mL leupeptin, and 2 μg/mL pepstatin A) for 30 minutes on ice. All steps were performed at 4°C. Lysate from two dishes was centrifuged and supernatant was precleared for 1 hour with 100 μL of protein A agarose beads (Life Technologies). Precleared supernatant was incubated overnight with 100 μL of protein A agarose beads, which had been preincubated overnight with 10 μg of polyclonal anti–HA-epitope tag antibody. After centrifugation, beads were extensively washed with RIPA buffer and denatured by boiling for 10 minutes.
Western blotting.
SDS-polyacrylamide gel electrophoresis (PAGE) of samples was performed on 6% or 7% acrylamide gels. The proteins were transferred onto nitrocellulose membrane using a semidry electroblotter (Integrated Separation Systems Inc, Natick, MA). All steps were performed at room temperature. After blocking for 1 hour in Tris buffered saline (TBS), 0.1% Tween-20, 4% non-fat dry milk, 1% bovine serum albumin (BSA), 0.02% sodium azide, blots were washed in TBS, 0.1% Tween-20 then probed for 1 hour with polyclonal anti–HA-epitope tag antibody diluted at 1μg/mL in TBS, 0.1% Tween-20, 3% BSA, 0.02% sodium azide. After several washes, blots were incubated with anti-rabbit IgG coupled to horseradish peroxidase (Amersham, Arlington Heights, IL) diluted at 1/1,000, as described for primary antibody, except sodium azide was excluded. After several washes, blots were developed using the Renaissance chemiluminescence detection kit (NEN Life Science Products, Boston, MA).
RESULTS
Erythroblast RNA contains seven major protein 4.1R isoforms.
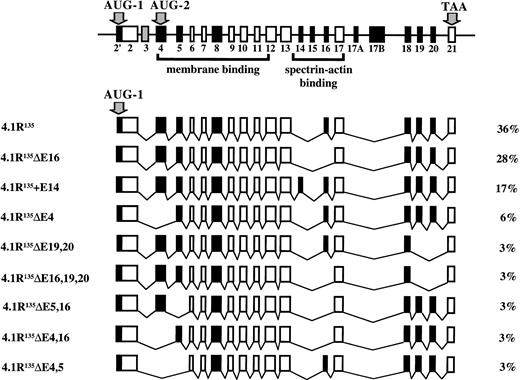
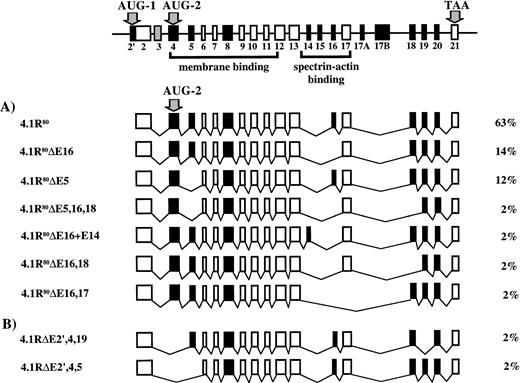
To determine the number and primary structure of 4.1R isoforms expressed in well-hemoglobinized erythroblasts, CD 34+cells were isolated from normal human bone marrow cells, grown in methylcellulose with SCF, IL-3, and Epo, and BFU-E–derived colonies plucked at day 11. Total RNA was purified, transcribed into cDNA, and amplified by RT-PCR using primer sets to amplify full-length coding domains from two populations of 4.1R cDNA: those that contained AUG-1 and those that deleted AUG-1 (Table 1 and Fig 1A). Erythroblast cDNA amplification products obtained with both sets of primers were ∼2.5 kilobases (kb), consistent with the predicted sizes of protein 4.1R cDNAs either containing or deleting AUG-1 (2,569 bp and 2,552 bp, respectively; Fig 1B). Moreover, both RT-PCR bands hybridized with 80-kD full-length 4.1R DNA by Southern blot analysis, identifying them as 4.1R (Fig 1C). Because each PCR band presumably represented a heterogeneous population of alternatively spliced mRNAs of similar size, a cDNA library was constructed from each band to facilitate characterization of individual subclones. To determine the number and primary structure of erythroblast 4.1R isoforms, these subclones were categorized into distinct classes using both Rsa I restriction enzyme digestions and hybridization with exon-specific oligonucleotide probes. Representative clones from each pattern were then completely sequenced. A total of 18 distinct isoforms were found. Of 87 independent 4.1 cDNAs subcloned, 36 clones (41%) included AUG-1, whereas 51 clones (59%) lacked AUG-1. Nine isoforms were encoded from AUG-1; however, 4 isoforms comprised 87% of this set (Fig 2). The most common form was the exon pattern encoding the 135-kD isoform (4.1R135) previously described in erythroblasts and nucleated nonerythroid cells.18-20,23-25 The second major pattern differed from the first in that it excluded exon 16 (4.1R135ΔE16), while the third major pattern was the 135-kD form plus exon 14 (4.1R135+E14). The final major isoform of this set was the 135-kD isoform missing exon 4 (4.1R135ΔE4). Thus, the significant variations among the four major isoforms encoded from AUG-1 involved inclusion or exclusion of exon 14, exon 16, encoding the functionally critical spectrin-actin binding domain, and exon 4, encoding a region of the NH2 extension and a region of the membrane binding domain. Seven isoforms were encoded from AUG-2 with 3 patterns dominating and comprising 89% of this group (Fig 3A). The most common form was the exon pattern encoding the 80-kD isoform (4.1R80), well-characterized in mature red cells.18-20 The second major pattern differed from the first in that it excluded exon 16 (4.1R80ΔE16), whereas the third major pattern was the 80-kD form minus exon 5 (4.1R80ΔE5). Therefore, in the predominant isoforms encoded from AUG-2 the significant distinctions among patterns involved exon 16, encoding the spectrin-actin binding domain, and exon 5, encoding a region of the membrane binding domain. Minor isoform patterns observed in erythroblasts varied from one another not only in the membrane binding and spectrin/actin binding domains but also in exons 18, 19, and 20, encoding the C-terminal domain of 4.1R that interacts with nuclear mitotic apparatus protein (NuMA) and elongation factor 1-α.42 43Interestingly, 2 isoforms (4.1RΔE2′,4,19 and 4.1RΔE2′,4,5) lacked the two exons containing, respectively, AUG-1 and AUG-2 (Fig 3B). This observation was of great surprise since AUG-1 and AUG-2 are, to date, the only documented active translation initiation sites.
Diagram of protein 4.1R isoforms produced by initiation of translation at AUG-1. At the top is pictured 4.1R mRNA depicting the alternatively spliced exons, constitutive exons, and noncoding exons. The two translation initiation sites, AUG-1 and AUG-2, are in exon 2′ and exon 4, respectively. The nine distinct protein 4.1R isoforms produced by initiation of translation at AUG-1 are depicted below, along with the percentage that each splice variant represented of the total clones analyzed.
Diagram of protein 4.1R isoforms produced by initiation of translation at AUG-1. At the top is pictured 4.1R mRNA depicting the alternatively spliced exons, constitutive exons, and noncoding exons. The two translation initiation sites, AUG-1 and AUG-2, are in exon 2′ and exon 4, respectively. The nine distinct protein 4.1R isoforms produced by initiation of translation at AUG-1 are depicted below, along with the percentage that each splice variant represented of the total clones analyzed.
Diagram of protein 4.1R isoforms lacking AUG-1 and protein 4.1R isoforms lacking AUG-1 and AUG-2. At the top is pictured 4.1R mRNA depicting the alternatively spliced exons, constitutive exons, and noncoding exons. The two translation initiation sites, AUG-1 and AUG-2, are in exon 2′ and exon 4, respectively. (A) The seven distinct protein 4.1R isoforms produced by initiation of translation at AUG-2 are shown, along with the percentage that each splice variant represented of the total clones analyzed. (B) The two 4.1R isoforms that lacked both exon 2′ and exon 4, the exons containing, respectively, AUG-1 and AUG-2.
Diagram of protein 4.1R isoforms lacking AUG-1 and protein 4.1R isoforms lacking AUG-1 and AUG-2. At the top is pictured 4.1R mRNA depicting the alternatively spliced exons, constitutive exons, and noncoding exons. The two translation initiation sites, AUG-1 and AUG-2, are in exon 2′ and exon 4, respectively. (A) The seven distinct protein 4.1R isoforms produced by initiation of translation at AUG-2 are shown, along with the percentage that each splice variant represented of the total clones analyzed. (B) The two 4.1R isoforms that lacked both exon 2′ and exon 4, the exons containing, respectively, AUG-1 and AUG-2.
Major 4.1R isoforms are expressed in transfected COS-7 cells.
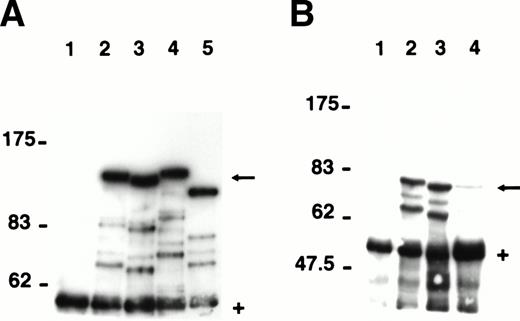
To characterize the proteins encoded by the seven major 4.1R mRNA isoforms expressed in erythroblasts, we transfected COS-7 cells with HA-epitope tagged constructs corresponding to each isoform. Immunoprecipitates of transfected cell lysates were analyzed by Western blots probed with a polyclonal antibody specific for the HA-epitope tag (Fig 4). 4.1R135, the predominant isoform translated from AUG-1, migrated at ∼136 kD (Fig 4A; lane 2), a molecular weight very close to the 135 kD previously reported for this isoform.18-20 The deletion of exon 16 resulted in a slight decrease in protein size with 4.1R135ΔE16 migrating as a protein of ∼131 kD (Fig 4A; lane 3). A third isoform, similar to 4.1R135 but including exon 14 (4.1R135+E14), showed a molecular weight of ∼138 kD (Fig 4A; lane 4), whereas a fourth isoform missing exon 4 (4.1R135ΔE4) was expressed as a protein of ∼116 kD (Fig4A; lane 5). In parallel experiments, the three major isoforms translated from AUG-2 were analyzed. The predominant isoform of this group, 4.1R80, showed an expected size of ∼79 kD (Fig 4B; lane 2). Deletion of exon 16 (4.1R80ΔE16) or of exon 5 (4.1R80ΔE5) resulted in a small decrease in protein size, both polypeptides migrating at ∼77 kD (Fig 4B; lanes 3 and 4). The expression of each of these proteins was specific because immunoprecipitates of lysates of cells transfected with the empty expression vector showed no expressed protein (Fig 4A and B; lane 1).
Expression of the seven major protein 4.1R isoforms in transfected COS-7 cells. The major 4.1R isoforms identified by RT-PCR of erythroblast total RNA were expressed as HA-epitope–tagged proteins in transfected COS-7 cells. Proteins were immunoprecipitated from precleared cell lysates using a polyclonal anti–HA-epitope tag antibody and analyzed by SDS-PAGE on a 6% (A) or 7% (B) acrylamide gel. (A) Cells transfected with empty expression vector (lane 1); HA-tagged 4.1R135 (lane 2); HA-tagged 4.1R135▵E16 (lane 3); HA-tagged 4.1R135+E14 (lane 4); and HA-tagged 4.1R135▵E4 (lane 5). (B) Cells transfected with empty expression vector (lane 1); HA-tagged 4.1R80 (lane 2); HA-tagged 4.1R80▵E16 (lane 3); HA-tagged 4.1R80▵E5 (lane 4). HA-tagged 4.1R isoforms (←). IgG heavy chains migrating ∼55 kD (+). Fainter bands migrating between the HA-epitope–tagged proteins and IgG heavy chains are likely to represent proteolytic fragments of the epitope tagged proteins (see Results). Note that 4.1R80▵E5 is expressed at a much lower level than the other 4.1R isoforms.
Expression of the seven major protein 4.1R isoforms in transfected COS-7 cells. The major 4.1R isoforms identified by RT-PCR of erythroblast total RNA were expressed as HA-epitope–tagged proteins in transfected COS-7 cells. Proteins were immunoprecipitated from precleared cell lysates using a polyclonal anti–HA-epitope tag antibody and analyzed by SDS-PAGE on a 6% (A) or 7% (B) acrylamide gel. (A) Cells transfected with empty expression vector (lane 1); HA-tagged 4.1R135 (lane 2); HA-tagged 4.1R135▵E16 (lane 3); HA-tagged 4.1R135+E14 (lane 4); and HA-tagged 4.1R135▵E4 (lane 5). (B) Cells transfected with empty expression vector (lane 1); HA-tagged 4.1R80 (lane 2); HA-tagged 4.1R80▵E16 (lane 3); HA-tagged 4.1R80▵E5 (lane 4). HA-tagged 4.1R isoforms (←). IgG heavy chains migrating ∼55 kD (+). Fainter bands migrating between the HA-epitope–tagged proteins and IgG heavy chains are likely to represent proteolytic fragments of the epitope tagged proteins (see Results). Note that 4.1R80▵E5 is expressed at a much lower level than the other 4.1R isoforms.
Of note, several bands migrating between the 4.1R isoforms and the IgG heavy chains were present on the Western blot analysis (Fig 4A and B). These bands were likely comprised of proteolytic fragments of the expressed 4.1R isoforms because they were not present in cells transfected with empty vector. However, another possibility was that these additional bands might represent smaller 4.1R isoforms translated from alternative AUGs located downstream of the first translation initiation codon in the transfected cDNA. Such a phenomenon has been reported for the generation of both glycophorin C and glycophorin D from glycophorin C mRNA.44 To investigate this hypothesis, we mutated the two in-frame AUGs downstream of AUG-1 in 4.1R135, one located in exon 4 and the other in exon 8. In addition, in 4.1R80 we mutated the AUG in exon 8, which represents the first initiation codon downstream of AUG-2, the normal site of translation initiation in 4.1R80. Western blot analysis of immunoprecipitates of lysates of cells transfected with the mutant cDNAs showed that the AUG → CUG (Met → Leu) mutations did not alter the pattern of polypeptide bands (data not shown). These observations strongly argue against the possibility that the transfected cells expressed several 4.1R isoforms derived from the unique 4.1R cDNA used for transfection and confirmed that the additional bands observed on the Western blots were indeed proteolytic fragments of the expressed proteins.
4.1R isoforms distribute broadly within the cell.
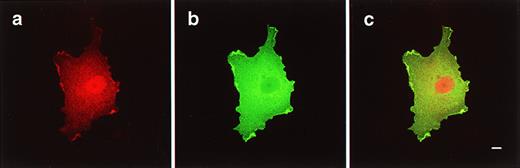
To test the hypothesis that individual 4.1R isoforms segregate to distinct subcellular compartments, we studied the distribution pattern of the seven major isoforms in transfected COS-7 cells by immunofluorescence microscopy. We observed that 4.1R135localized to the cytoplasm and to a similar extent to the nucleus with exclusion of the nucleoli (Fig 5a through c). In contrast, 4.1R135ΔE16 was predominantly expressed in the cytoplasm with no staining of the nucleus (Fig 5d through f). This result suggested that exon 16 encoded a key determinant for nuclear localization. Cells transfected with both isoforms also showed an accumulation of protein in a distribution evoking membrane ruffles that appeared as bright regions located at the periphery of the cell (particularly obvious in the cells displayed in Fig 5b and f).
Expression of HA-epitope–tagged 4.1R135 and 4.1R135▵E16 isoforms in transfected COS-7 cells. COS-7 cells were transfected with HA-epitope–tagged 4.1R135 or 4.1R135▵E16 isoforms, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a polyclonal anti–HA-epitope tag antibody as primary antibody and anti–rabbit IgG coupled to FITC as secondary antibody. Three fields of cells transfected with each 4.1R isoform are displayed. 4.1R135localized to the cytoplasm and to a lesser extent to the nucleus with exclusion of the nucleoli (a through c). Because the intensity of nuclear and cytoplasmic staining is very similar in cells transfected with 4.1R135, we highlighted the nucleus of the cell shown in (c) by showing DAPI staining superimposed with the FITC staining (see inset in lower left corner of [c]). In contrast, 4.1R135▵E16 was exclusively expressed in the cytoplasm with no staining of the nucleus (d through f). Scale bar: 10 μm.
Expression of HA-epitope–tagged 4.1R135 and 4.1R135▵E16 isoforms in transfected COS-7 cells. COS-7 cells were transfected with HA-epitope–tagged 4.1R135 or 4.1R135▵E16 isoforms, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a polyclonal anti–HA-epitope tag antibody as primary antibody and anti–rabbit IgG coupled to FITC as secondary antibody. Three fields of cells transfected with each 4.1R isoform are displayed. 4.1R135localized to the cytoplasm and to a lesser extent to the nucleus with exclusion of the nucleoli (a through c). Because the intensity of nuclear and cytoplasmic staining is very similar in cells transfected with 4.1R135, we highlighted the nucleus of the cell shown in (c) by showing DAPI staining superimposed with the FITC staining (see inset in lower left corner of [c]). In contrast, 4.1R135▵E16 was exclusively expressed in the cytoplasm with no staining of the nucleus (d through f). Scale bar: 10 μm.
Similar observations were made in COS-7 cells cotransfected with 4.1R80 and 4.1R80ΔE16, two distinct 80-kD 4.1R isoforms either containing or lacking exon 16, respectively. To allow independent visualization of these isoforms, when coexpressed in the same cell, 4.1R80 was tagged with the c-myc–epitope tag while 4.1R80ΔE16 was tagged with the HA-epitope tag. As shown in Fig 6, c-myc–epitope tagged 4.1R80 was strongly expressed in the nucleus as well as in the cytoplasm and membrane ruffles (Fig 6a). In marked contrast, HA-epitope tagged 4.1R80ΔE16 localized poorly to the nucleus but was expressed in the cytoplasm as well as in membrane ruffles (Fig 6b). Superimposition of these images confirmed the distinct nuclear distribution of these two isoforms (Fig 6c). Thus, deletion of exon 16 resulted in a dramatic decrease in nuclear expression of both 4.1R80 and 4.1R135.
Coexpression of c-myc–epitope–tagged 4.1R80and HA-epitope–tagged 4.1R80▵E16 isoforms in transfected COS-7 cells. COS-7 cells were cotransfected with c-myc–epitope–tagged 4.1R80 and HA-epitope–tagged 4.1R80▵E16 isoforms, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a monoclonal anti-c-myc–epitope tag antibody and a polyclonal anti–HA-epitope tag antibody as primary antibodies and anti-mouse IgG coupled to Texas red and rabbit IgG coupled to FITC as secondary antibodies. A cell probed with antibody to the c-myc–epitope tag showed that 4.1R80was strongly expressed in the nucleus as well as in the cytoplasm and membrane ruffles (a). In marked contrast, the same cell probed with antibody to the HA-epitope tag (b) showed that 4.1R80▵E16 localized poorly to the nucleus but was strongly expressed in the cytoplasm as well as in membrane ruffles. Superimposed images of (a) and (b) are shown in (c). Scale bar: 10 μm.
Coexpression of c-myc–epitope–tagged 4.1R80and HA-epitope–tagged 4.1R80▵E16 isoforms in transfected COS-7 cells. COS-7 cells were cotransfected with c-myc–epitope–tagged 4.1R80 and HA-epitope–tagged 4.1R80▵E16 isoforms, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a monoclonal anti-c-myc–epitope tag antibody and a polyclonal anti–HA-epitope tag antibody as primary antibodies and anti-mouse IgG coupled to Texas red and rabbit IgG coupled to FITC as secondary antibodies. A cell probed with antibody to the c-myc–epitope tag showed that 4.1R80was strongly expressed in the nucleus as well as in the cytoplasm and membrane ruffles (a). In marked contrast, the same cell probed with antibody to the HA-epitope tag (b) showed that 4.1R80▵E16 localized poorly to the nucleus but was strongly expressed in the cytoplasm as well as in membrane ruffles. Superimposed images of (a) and (b) are shown in (c). Scale bar: 10 μm.
In addition, we consistently observed stronger nuclear staining with 4.1R80 than with 4.1R135 (compare Fig 5a through c with Fig 6a). Furthermore, the percentage of cells showing nuclear staining was higher in cells transfected with 4.1R80 than in cells transfected with 4.1R135(90% and 60%, respectively). These observations suggest that the presence of the N-terminal extension, encoded when translation is initiated at AUG-1, impacts the extent of 4.1R expression in the nucleus.
Considered in aggregate, these data suggest that individual 4.1R isoforms can be expressed in various cell compartments.
A single 4.1R isoform can localize to more than one subcellular compartment.
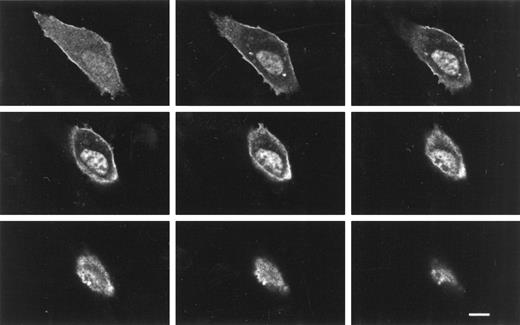
Because conventional microscopy cannot decisively discriminate cytoplasmic from nuclear localization or plasma membrane from cytoplasmic expression, we analyzed transfected cells by confocal immunofluorescence microscopy. Analysis of sequential sections of a 3T3 cell transfected with a KT3-epitope tagged 4.1R80 construct confirmed that the 4.1R80 isoform was strongly expressed in the nucleus (Fig 7), as had been observed in transfected COS-7 cells. In addition, these sections showed clearly that the protein was also expressed in the plasma membrane, which appeared as a bright rim outlining the contour of the cell. Finally, several of these sections revealed two bright immunofluorescent spots located in the cytoplasm on either side of the nucleus. The size of these spots and their position in relation to the nucleus and to one another in an interphase cell suggest that they represent centrosomal expression of 4.1R80. This observation is consistent with our prior data that 4.1 antipeptide antibodies localize to the pericentriolar matrix of the centrosome.20 22 Hence, confocal microscopy confirmed our surprising findings that a single 4.1R isoform can localize to more than one subcellular compartment.
Confocal z-sections of a NIH/3T3 cell transfected with KT3-epitope–tagged 4.1R80. NIH/3T3 cells were transfected with KT3-epitope–tagged 4.1-R80, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a monoclonal anti–KT3-epitope tag antibody as a primary antibody and anti-mouse IgG coupled to Texas red as a secondary antibody. Cells were analyzed by confocal microscopy. The 4.1R80 isoform was strongly expressed in the nucleus, the plasma membrane, and apparent centrosomes. Every other z-section of a series of 18 0.5-μm z-sections are displayed. Scale bar: 10 μm.
Confocal z-sections of a NIH/3T3 cell transfected with KT3-epitope–tagged 4.1R80. NIH/3T3 cells were transfected with KT3-epitope–tagged 4.1-R80, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a monoclonal anti–KT3-epitope tag antibody as a primary antibody and anti-mouse IgG coupled to Texas red as a secondary antibody. Cells were analyzed by confocal microscopy. The 4.1R80 isoform was strongly expressed in the nucleus, the plasma membrane, and apparent centrosomes. Every other z-section of a series of 18 0.5-μm z-sections are displayed. Scale bar: 10 μm.
4.1R isoforms lacking both AUG-1 and AUG-2 can still be expressed.
Two of the eighteen 4.1R mRNA isoforms identified by RT-PCR of erythroblast total RNA lacked the AUGs encoded by exon 2′ and exon 4. Because these AUGs represent the only two translation initiation codons thought to be active in 4.1R expression, the presence of isoforms lacking both AUG-1 and AUG-2 was totally unexpected. To determine whether an unsuspected downstream initiation codon might be functional, COS-7 cells were transfected with one of these two isoforms, HA-epitope tagged 4.1RΔE2′,4,5 (Fig 8A [see page 4410]), and analyzed immunohistochemically. In immunoprecipitates of lysates of transfected COS-7 cells the HA-epitope–tagged protein migrated as an ∼68-kD protein (Fig 8B). Transfected cells were then analyzed by double-label immunofluorescence microscopy using a monoclonal HA-epitope tag antibody and a polyclonal antibody raised against the 21 amino acids of exon 16 of 4.1R (Fig 8C). Probing with the HA-epitope tag antibody (Fig 8C, panel a) showed very strong nuclear expression of this ∼68-kD isoform with exclusion of the nucleoli, and weak cytoplasmic staining. An identical pattern of staining was obtained with the 4.1R antipeptide antibody (Fig 8C; panel b), confirming that the expressed protein was, indeed, a 4.1R isoform.
Expression of HA-epitope–tagged 4.1R▵E2′,4,5 isoform in transfected COS-7 cells. COS-7 cells were transfected with HA-epitope–tagged 4.1R80▵E2′,4,5 isoform (exon map shown in [A]). (B) Protein was immunoprecipitated from precleared cell lysates using a polyclonal anti–HA-epitope tag antibody and analyzed by SDS-PAGE on a 7% acrylamide gel. The HA-epitope–tagged protein migrated at ∼68 kD. Lane 1, cells transfected with empty expression vector; lane 2, cells transfected with HA-tagged 4.1-R80▵E2′,4,5. HA-tagged 4.1R isoforms (←). IgG heavy chains migrating ∼55 kD (+). (C) In other experiments, transfected cells were analyzed by immunofluorescence microscopy. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and double-stained using a monoclonal anti–HA-epitope tag antibody and a polyclonal anti-4.1R antibody raised against the 21 amino acids of exon 16 as primary antibodies and anti-mouse IgG coupled to Texas red and anti-rabbit IgG coupled to FITC as secondary antibodies. Cells, reacting with the HA-epitope tag antibody, showed very strong nuclear expression of 4.1R▵E2′,4,5 with exclusion of the nucleoli and weak cytoplasmic staining (a). An identical pattern of staining was obtained with the anti-4.1R peptide antibody (b). Superimposed images of (a) and (b) are shown in (c). Scale bar: 10μm.
Expression of HA-epitope–tagged 4.1R▵E2′,4,5 isoform in transfected COS-7 cells. COS-7 cells were transfected with HA-epitope–tagged 4.1R80▵E2′,4,5 isoform (exon map shown in [A]). (B) Protein was immunoprecipitated from precleared cell lysates using a polyclonal anti–HA-epitope tag antibody and analyzed by SDS-PAGE on a 7% acrylamide gel. The HA-epitope–tagged protein migrated at ∼68 kD. Lane 1, cells transfected with empty expression vector; lane 2, cells transfected with HA-tagged 4.1-R80▵E2′,4,5. HA-tagged 4.1R isoforms (←). IgG heavy chains migrating ∼55 kD (+). (C) In other experiments, transfected cells were analyzed by immunofluorescence microscopy. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and double-stained using a monoclonal anti–HA-epitope tag antibody and a polyclonal anti-4.1R antibody raised against the 21 amino acids of exon 16 as primary antibodies and anti-mouse IgG coupled to Texas red and anti-rabbit IgG coupled to FITC as secondary antibodies. Cells, reacting with the HA-epitope tag antibody, showed very strong nuclear expression of 4.1R▵E2′,4,5 with exclusion of the nucleoli and weak cytoplasmic staining (a). An identical pattern of staining was obtained with the anti-4.1R peptide antibody (b). Superimposed images of (a) and (b) are shown in (c). Scale bar: 10μm.
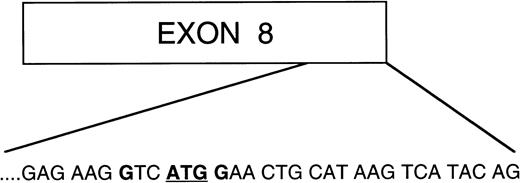
Analysis of the 4.1R sequence revealed a candidate translation initiation site located at the end of exon 8 (Fig 9). A number of observations argued in favor of its being the relevant codon used to express the ∼68-kD protein from 4.1RΔE2′,4,5. This particular AUG was the first AUG downstream of exon 4 that was in frame with the 4.1R amino acid sequence. In addition, it was surrounded by an optimal consensus Kozak sequence (GTCATGG) required for proper translation initiation.45 Utilization of this codon for translation initiation would result in a polypeptide of ∼65 kD, thus corresponding closely to the observed molecular weight of expressed 4.1RΔE2′,4,5 protein. Significantly, when this AUG was mutated to CUG (Met → Leu), the ∼68 kD polypeptide was no longer detectable by Western blot analysis of immunoprecipitates of lysates of cells transfected with the mutant cDNA (data not shown). We, therefore, speculate that in the absence of the two previously characterized translation initiation codons, a third AUG located in exon 8 can effectively initiate translation of smaller 4.1R isoforms.
Schematic diagram of exon 8 of protein 4.1R. Amino acid sequence at the 3′ end of exon 8 consisting of an AUGcodon surrounded by an optimal consensus Kozak sequence with a purine (G) in position −3 and a G in position +4. This is the first AUG codon downstream of AUG-2, which is in frame with the 4.1R coding sequence, therefore making it a good candidate codon for initiation of translation of the two isoforms lacking both AUG-1 and AUG-2.
Schematic diagram of exon 8 of protein 4.1R. Amino acid sequence at the 3′ end of exon 8 consisting of an AUGcodon surrounded by an optimal consensus Kozak sequence with a purine (G) in position −3 and a G in position +4. This is the first AUG codon downstream of AUG-2, which is in frame with the 4.1R coding sequence, therefore making it a good candidate codon for initiation of translation of the two isoforms lacking both AUG-1 and AUG-2.
DISCUSSION
A major finding of the current study is that the well-hemoglobinized erythroblast expresses an impressive repertoire of 4.1R isoforms, which includes 7 major and 11 minor splice variants. Although prior reports have suggested the presence of more than a single 4.1R isoform in various cells and tissues,18-21;24;46-52 the current studies provide the first definitive characterization of the array of 4.1R primary structures in a single-cell lineage. Interestingly, the predominant splicing event between the major isoforms initiated at both AUG-1 and AUG-2 involved inclusion or exclusion of exon 16. This particular exon, along with exon 17, encodes key residues for efficient interaction of 4.1R with spectrin and actin.13 In the mature erythrocyte, formation of the spectrin/actin/4.1 ternary complex is critical for normal membrane mechanical strength.14-16 53 Our current data emphasize that expression of exon 16 could also play an important functional role in erythroblasts.
Also striking was the finding that transfected protein 4.1R isoforms show a very broad distribution within the cell with localization to the plasma membrane, membrane ruffles, cytoplasm, centrosomes, and nucleus. These results confirm earlier immunofluorescent observations showing endogenous 4.1 in similar subcellular compartments.22-24,26,47,50 However, because antibodies to 4.1R can recognize more than one isoform, it has previously been impossible to track either the distribution of a single isoform or to determine the presence of more than one 4.1R isoform within the same subcellular compartment. Of great surprise to us was the observation that a single isoform can distribute to multiple sites. Indeed, the confocal microscopic analysis of 3T3 cells transfected with the KT3-epitope–tagged 4.1R80 construct showed conclusively that 4.1R80 was expressed in the nucleus, plasma membrane, and apparent centrosome. Earlier this year, Luque and colleagues reported that 4.1R80 expressed in transfected COS cells distributed in most cells within the nucleus and along a network of cytoplasmic filaments and that a smaller population of cells showed overexpression of the protein within the cytoplasm.26Although we cannot completely rule out the possibility that overexpression might have altered the pattern of localization that we observed, it is important to note that localization patterns in the plasma membrane, the nucleus, and the centrosome similar to those described here for epitope-tagged 4.1R in transfected cells have definitely been observed for endogenous protein 4.1. We speculate that diverse posttranslational modifications of the 4.1R80isoform may contribute to the differential subcellular sorting.
Several exons appeared to influence 4.1R localization in the nucleus. Clearly, exclusion of exon 16 resulted in a dramatic decrease in nuclear expression of the protein. This was true for the 80-kD isoform translated from AUG-2, as previously reported by Luque et al,26 but also for higher molecular weight isoforms translated from AUG-1. We conclude that a motif present in exon 16 performs a critical role in nuclear targeting, as we and others have previously suggested.26,54 Such a motif (KKKRER) has been shown to be necessary but not sufficient for nuclear targeting of 4.1R.26 In addition, we observed that nuclear expression of the AUG-1 isoform 4.1R135, containing exon 16, was lower than that of its AUG-2 counterpart, 4.1R80. These results strongly suggest that the 209 amino acid N-terminal extension has a negative impact on nuclear localization of the polypeptide. It is unlikely that this effect results solely from the increase in protein size because even larger proteins have been shown to be efficiently transported to the nucleus.55 56 A more probable explanation is that expression of the N-terminal extension produces an overall protein conformation that either masks recognition of nuclear localization sequence(s) or inhibits binding of 4.1R to a protein involved in nuclear import of polypeptides.
A number of different 4.1R isoforms localized to the plasma membrane of transfected cells. Interestingly, this was observed even for isoforms lacking a fully functional spectrin/actin binding domain (ie, lacking exon 16). These data emphasize that in nucleated cells, sequence motifs in addition to those encoded by exon 16 can regulate assembly of 4.1R onto the plasma membrane. We were also interested to observe an accumulation of protein 4.1R in structures evoking membrane ruffles, regions of the plasma membrane overlying dynamically reorganizing bundles of actin-containing microfilaments. These specialized subdomains of the plasma membrane have been described, for example, at leading lamellae of migrating cells,57,58 sites of injury,59 or following cell activation by various growth factors.60-65 It is pertinent that Bessis has previously described membrane motility in developing erythroblasts.66Membrane ruffles are enriched in the actin-associated proteins, alpha-actinin, and fimbrin.62 Of particular note, the 4.1R80 binding partner, spectrin, has also been identified in these structures,60,62 and it has been proposed that spectrin links growth factor receptors to actin microfilaments of ruffling membranes.60 In nucleated cells, protein 4.1R may anchor the spectrin network to membrane receptors in a manner analogous to the way it anchors the spectrin-actin network to glycophorin C, band 3 (and possibly CD44) in the erythrocyte plasma membrane. Ruffling in nucleated cells appears to be regulated by both protein phosphorylation61,62,64-67 and calmodulin binding.67 Because 4.1R is a substrate for various protein kinases68 and its interactions with the plasma membrane are tightly regulated by calmodulin,11,12 69 it is tempting to speculate that protein 4.1R may participate in the dynamic interaction of actin microfilaments with various components of the plasma membrane in membrane ruffles.
The 30-kD plasma membrane binding domain, encoded by exons 4 through 12, has been shown to mediate the interaction of 4.1R with transmembrane proteins glycophorin C, band 3, and CD44.4-7,10-12 Exon 5, one of the alternatively spliced exons within this domain, contains the LEEDY sequence implicated in 4.1R-band 3 interaction.70 In the current study, we showed that the 4.1R80ΔE5 isoform was expressed poorly when transfected into COS-7 cells. This was also true for another isoform lacking exon 5, 4.1R80ΔE5,16,18 (data not shown). Moreover, both proteins accumulated in cytoplasmic inclusion bodies in a large proportion of transfected cells (unpublished data). We hypothesize that deletion of exon 5 may result in an overall misfolding of the protein, thus altering its processing and targeting to the plasma membrane. Alternatively, the absence of binding sequences, such as the LEEDY motif, may impair either sorting of the protein to the plasma membrane or assembly onto the membrane.
The two previously defined translation initiation codons responsible for 4.1R expression are located within exon 2′ and exon 4. Identification of two isoforms (4.1RΔE2′,4,5 and 4.1RΔE2′,4,19) lacking both of these translation initiation codons was, therefore, unexpected. However, our immunofluorescence observations in COS-7 cells transfected with 4.1RΔE2′,4,5 documented that an additional translation initiation codon can potentially regulate expression of 4.1R. Immunoprecipitates of lysates of transfected COS-7 cells confirmed the expression of a 68-kD 4.1R polypeptide. These data clearly show that a formerly unsuspected downstream initiation codon can be functional. Analysis of the 4.1R sequence reveals the presence of a candidate AUG located at the end of exon 8. This is the first AUG codon downstream of AUG-2, which is in frame with the 4.1R coding sequence and which is surrounded by an optimal consensus Kozak sequence with a purine in position −3 and a G in position +4 (GTCATGG).45 Of note, the use of this codon for translation initiation is in accordance with the observed molecular weight of 4.1RΔE2′,4,5 expressed in transfected COS-7 cells, and mutation of this codon results in the disappearance of the protein. We have previously described a protein of ∼69 kD on Western blots of human erythroblasts, further suggesting the possibility that isoforms lacking both AUG-1 and AUG-2 may be physiologically relevant.
The terminally differentiating erythroblast and the circulating mature erythrocyte differ dramatically from one another both structurally and functionally. The principal functions of the erythroblast are to synthesize hemoglobin and, by the conclusion of terminal differentiation, to have assembled a mechanically strong yet deformable plasma membrane. To perform these functions an erythroblast possesses the subcellular organelles and biochemical machinery essential for protein synthesis and mitosis. To import the iron essential for hemoglobin synthesis, the erythroblast plasma membrane undergoes repeated receptor-mediated endocytosis.71 Plasma membrane vesiculation, in the form of exocytosis, also plays a crucial role in remodeling of the reticulocyte membrane.72-74 On the other hand, the primary function of the terminally differentiated erythrocyte is to efficiently deliver oxygen. In dramatic contrast to the erythroblast plasma membrane, the plasma membrane of the mature erythrocyte undergoes neither endocytosis nor exocytosis. Indeed, vesicle formation is extremely detrimental to the erythrocyte, resulting in loss of surface area and loss of normal membrane deformability.75 These marked differences in the cellular functions of the erythroblast and erythrocyte are reflected in substantial differences in their subcellular biochemistry. We report here that one of these biochemical differences is clearly protein 4.1R isoform expression. Although a single 4.1R isoform, 4.1R80, is the predominant form of 4.1R in erythrocytes, erythroblasts express additional 4.1R isoforms with functional activities potentially relevant to microtubule nucleation, nuclear function, and plasma membrane vesiculation. We speculate that the repertoire of primary structure of 4.1R described in this report dictates distinct binding partners and functions of 4.1R during erythropoiesis.
ACKNOWLEDGMENT
We thank Dr S.W. Krauss (LBNL, Berkeley, CA) for helpful discussions. We also thank Dr B. Schwer (Cornell University Medical College, New York, NY) for providing us with the anti–KT3-epitope tag antibody and Dr C. Collins (UCSF, San Francisco, CA) for providing us with COS-7 cells. We are very grateful to Drs D. Callahan and K. Benson (LBNL, Berkeley, CA) for their invaluable help in cell imaging and to Derek Clark for his help in figure processing.
Supported by National Institutes of Health Grants DK26263 and DK32094 and by the Director, Office of Health and Environment Research Division, U.S. Department of Energy, under Contract DE-AC03-76SF00098.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to J.A. Chasis, MD, Lawrence Berkeley National Laboratory, Mail Stop 74-157, Cyclotron Road, Berkeley, CA 94720; e-mail: jachasis@lbl.gov.

![Fig. 5. Expression of HA-epitope–tagged 4.1R135 and 4.1R135▵E16 isoforms in transfected COS-7 cells. COS-7 cells were transfected with HA-epitope–tagged 4.1R135 or 4.1R135▵E16 isoforms, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a polyclonal anti–HA-epitope tag antibody as primary antibody and anti–rabbit IgG coupled to FITC as secondary antibody. Three fields of cells transfected with each 4.1R isoform are displayed. 4.1R135localized to the cytoplasm and to a lesser extent to the nucleus with exclusion of the nucleoli (a through c). Because the intensity of nuclear and cytoplasmic staining is very similar in cells transfected with 4.1R135, we highlighted the nucleus of the cell shown in (c) by showing DAPI staining superimposed with the FITC staining (see inset in lower left corner of [c]). In contrast, 4.1R135▵E16 was exclusively expressed in the cytoplasm with no staining of the nucleus (d through f). Scale bar: 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/11/10.1182_blood.v92.11.4404/5/m_blod423gas05z.jpeg?Expires=1767927645&Signature=lvJ-Hy-AyRhctYgc2kJEPzKNC1lyXpSe2o2iwOb6SUyOkAX5T-AJezfEfcBvKLdaY3CopoENL7puVyYOsYlJ~RMo5U3CXQUsUMuzKzCdto0kChWlBKb4AfB2E9Q4yyvBnoAdmO8nRiKPkG7aSSpsFinkY0FsJK8R6JLbWLth6nUawiANnR1SUkbofYXxPC5AhcWlIC7lEkptpO3I5A2XW~-HrALyz8yykMeiyI3wV05MkmtyIUnxBSbteBSzur5rHsuBIpcIff47pXC74~fPLNfoth4Pm3yzUi43mim~jPjQ6QDUVVkfUyMT~FT-lQgJT8RY6ASId0FqkOoVz2bBgQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. Expression of HA-epitope–tagged 4.1R▵E2′,4,5 isoform in transfected COS-7 cells. COS-7 cells were transfected with HA-epitope–tagged 4.1R80▵E2′,4,5 isoform (exon map shown in [A]). (B) Protein was immunoprecipitated from precleared cell lysates using a polyclonal anti–HA-epitope tag antibody and analyzed by SDS-PAGE on a 7% acrylamide gel. The HA-epitope–tagged protein migrated at ∼68 kD. Lane 1, cells transfected with empty expression vector; lane 2, cells transfected with HA-tagged 4.1-R80▵E2′,4,5. HA-tagged 4.1R isoforms (←). IgG heavy chains migrating ∼55 kD (+). (C) In other experiments, transfected cells were analyzed by immunofluorescence microscopy. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and double-stained using a monoclonal anti–HA-epitope tag antibody and a polyclonal anti-4.1R antibody raised against the 21 amino acids of exon 16 as primary antibodies and anti-mouse IgG coupled to Texas red and anti-rabbit IgG coupled to FITC as secondary antibodies. Cells, reacting with the HA-epitope tag antibody, showed very strong nuclear expression of 4.1R▵E2′,4,5 with exclusion of the nucleoli and weak cytoplasmic staining (a). An identical pattern of staining was obtained with the anti-4.1R peptide antibody (b). Superimposed images of (a) and (b) are shown in (c). Scale bar: 10μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/11/10.1182_blood.v92.11.4404/5/m_blod423gas08z.jpeg?Expires=1767927645&Signature=lbAbGzQX44aujs3PZEHGYnYjp5akGe8QxAJN7yWGl0rDdS0IEoKF~fOO0c7UcsemTcsfqkQQQIZVytgpdfoV9WeP-~okqwcWG2hD8UNfgdEjVsMetXRrhX-THHDgDweYTJraQ5KrMGgAGWmywdetXk5UAwtv4djuVJLmootdP4lup0BcAXUpNZPiAm9QuS4FHv1axzsMCc9267yPluhjBtzg1kWAe8q6byfipLvb3nIZuuIGyTE7iOLPwkQvR6ouKT-wOeRlxiG8iYxSwYYxGU2emlg8xDAyT8HM9MXAnSdVVOf6b8eDLcL8AT3pp9-skAgsv8SlUpJS77SWtSFFOA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)