Abstract
The human β-globin gene complex spans a region of 70 kb and contains numerous sequence variants. These variant sites form a 5′ cluster (5′ β-haplotype) and a 3′ cluster (3′ β-haplotype) with strong linkage disequilibrium among the sites within each cluster, but not between the two clusters. The 9-kb region between the 5′ and 3′ clusters has been estimated to have rates of recombination that are 3 to 30 times normal, and the region has therefore been proposed as a ‘hotspot’ of recombination. We describe three families with evidence of meiotic recombination within this ‘hotspot’ of the β-globin gene cluster and in which the cross-over breakpoints have been defined at the sequence level. In one family, the recombination has occurred in the maternal chromosome within a region of 361 bp between positions −911 and −550 5′ to the β-globin gene. In the other two families, the recombination has occurred in the paternal chromosome within a region of approximately 1,100 bp between positions −542 and +568 relative to the β-globin gene cap site. Both regions occur within the 2-kb region of replication initiation (IR) in the β-globin gene domain with no overlap. The IR region contains a consensus sequence for a protein (Pur), which binds preferentially to single-stranded DNA, a role implicated in recombination events.
RECOMBINATION BETWEEN homologous DNA sequences plays an important role in generating genetic diversity in all organisms. Although meiotic recombination occurs throughout the human genome, it does not occur randomly but appears to be concentrated in specific regions.1 Such areas of relatively increased recombination frequency are present in the major histocompatibility complex (MHC),2,3 where they are responsible for Ig class switching, near or within the Duchenne muscular dystrophy, insulin, collagen, and the β-globin gene loci.4-8 The human β-globin gene region on chromosome 11p is one of the most intensively studied of all human loci; mutations here cause β-thalassemia and sickle cell anemia, which are among the most common genetic diseases in the world.9 The earliest prenatal diagnosis of these disorders using DNA analysis relied on linkage disequilibrium of β-globin gene mutations with adjacent restriction fragment length polymorphisms (RFLPs).10,11 This procedure always carries some degree of risk of recombination between the mutant allele and the RFLP. Indeed a meiotic recombination in the paternal β chromosome in one family has caused an error in the prenatal diagnosis of β-thalassemia using this approach.12
Over 20 RFLPs have now been identified in the β-globin locus.13 These polymorphisms fall into two groups: a 34-kb 5′ cluster that includes the HindII-ε,HindIII-Gγ,HindIII-Aγ, HindII-Ψβ,HindII-3′Ψβ, and Taq I-5′δ polymorphic sites, and a 19-kb 3′ cluster that includes the Hgi AI-β,AvaII-β and BamHI-β sites14 (see Fig 1). Population genetic analysis showed that while sites within the two clusters show strong linkage disequilibrium to each other, no linkage disequilibrium exists between these two clusters and the polymorphic sites within the 9-kb region separating the two clusters are randomly associated.15 Indeed, a recombination rate of 3 to 30 times greater than expected was found in the region, which is the site of 75% of the recombination events in the entire β-globin cluster.8 Thus, this region has been implicated as a ‘hotspot’ for genetic recombination. Studies have identified four separate families with evidence of recombination events within this ‘hotspot’.12,16-18 The localization of the crossover events in these families has been limited by the number of informative RFLPs available, and the narrowest localization was approximately 10 kb.18
Several sequence elements have been proposed as candidate recombination signals, some of which have been identified in the 9-kb ‘hotspot.’ The ‘hotspot’ encompasses the 2-kb region of replication origin in the β-globin gene domain within which is a 16-bp consensus sequence for the Pur element.19,20 In addition, upstream of the initiation replication region, but still within the ‘hotspot,’ lies a 21-bp consensus sequence (with two base mismatches) that is unique to potential initiation regions.21 It is proposed that both the Pur and the 21-bp element facilitates DNA replication and recombination by initiating the unwinding of duplex DNA or maintaining an open duplex. Here we report on three families with evidence of recombination events within the ‘hotspot’ in the β-globin cluster. Two (Greek Cypriot and Asian Indian) are previously unreported, while the third family of Amish origin was previously reported by Gerherd et al.16 In an attempt to address the role of the various putative recombination signals in the ‘hotspot,’ we have undertaken direct sequence analysis to delineate the crossover region. Two distinct regions of crossover within the 9-kb ‘hotspot’ were defined, which suggests that recombination sites in the β-globin complex show a clustering pattern also observed in the human HLA class II region.3 The breakpoint in each family occurs within the 2 kb of replication initiation in the β-globin gene domain.
FAMILIES AND METHODS
Recombinant families.
Three families (Greek Cypriot, Asian Indian, and Amish) were previously identified by typing of the RFLPs (β-haplotype analysis) in the β-globin gene cluster. All families analyzed included both parents and at least three offspring. The Amish family is part of a Centre d’Etude du Polymorphisme Humain (CEPH) family. Informed consent was obtained in all cases before the collection of blood samples.
β-Haplotype analysis.
DNA was extracted from peripheral blood leukocytes or lymphoblastoid cell lines using standard procedures and analyzed by restriction enzyme digestion. β Haplotypes were derived from the following RFLPs in the β-globin gene cluster—HindII-ε,HindIII-Gγ,HindIII-Aγ, HindII-Ψβ,HindI-3′ Ψβ, AvaII-β, BamHI-β,Taq I-5′ δ, Pst I-3′ δ, HinfI-5′ β, Rsa I-5′ β, and HgiAI-β. The RFLPs were analyzed using a combination of standard Southern blot hybridization15 and restriction enzyme analysis of DNA that was specifically amplified by the polymerase chain reaction (PCR) (see Fig 1). PCR amplification of the various polymorphic sites was performed in a total volume of 100 μL containing 10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl, 1.5 to 2.5 mmol/L MgCl2, 0.2 mmol/L each of dTTP, dGTP, dCTP, and dATP, 10 pmol of each primer, and 2.5 U of Taq polymerase (Cetus, Perkin Elmer, Warrington, UK). The sequences of the primers used and details of the conditions of amplification are available on request.
DNA sequence analysis.
A 4.5-kb DNA fragment encompassing the 3′ δ-β globin gene region, exons 1 and 2, and intron 1 of the β-globin gene was enzymatically amplified using two sets of primers, 5′R3K and 3′R5K and 5′β422 and 3′β9. Primers 5′ R 3K (5′-AAT CTG TAC ATC AAG ACC CAG TGA TAT G-3′) and 3′ R5K (5′-GAC ATC TAA CTG TTT CTG CCT GGA CT-3′) corresponding to GenBank (HUMHBB U01317) coordinates 58299-58326 and 61315-61290, respectively, direct the amplification of a 3,016-bp fragment in the δ-β region (Fig 1). PCR amplification was performed in a 100-μL reaction volume containing 0.2 mmol/L of each dNTP, 50 mmol/L KCl, 10 mmol/L Tris-HCl pH 8.3, 2.0 mmol/L MgCl2, 2.5 U Taq polymerase (Cetus), and 20 pmol of each primer 5′ R3K and 3′ R5K. After an initial denaturation of 4 minutes at 95°C, 30 cycles of denaturation at 95°C for 1 minute, annealing at 64°C for 2 minutes, and extension at 72°C for 3 minutes were performed, the last extension reaction was prolonged for 8 minutes. An aliquot of the PCR product was examined on a 1.0% agarose gel. The 3.0-kb amplification product was isolated and purified using a QIAEX II Gel extraction kit (Qiagen Ltd, Surrey, UK) after electrophoresis in a 1.0% agarose gel. The purified PCR product was resuspended in 40 μL 10 mmol/L Tris-HCl pH 8.3. A small aliquot (3/40 μL) of the DNA was examined in a 1% agarose gel to check for purity and concentration. Fifty to 500 ng (generally 6 of the 40 μL) of the product was directly sequenced using the thermal cycle sequencing technique with 33P-labeled terminators andThermosequenase (Amersham, Buckinghamshire, UK). A total sequence of 3,016 nucleotides was determined using a series of forward and reverse primers.
Map of the β-globin gene cluster with the 9-kb ‘hotspot’ delineated by the TaqI-5′δ (5′) and theHgiAI-β (3′) sites. RFLP sites are indicated by the arrows. The 4.5-kb fragment encompassing the 5′ flanking β region is amplified using two sets of primers: 5′R3K versus 3′R5K and 5′β422 and 3′β9. The other polymorphic sites are indicated by vertical lines. The pur, chi, and the 21-bp element at position −355021 are represented by thick horizontal lines. The crossover regions and the region of initiation replication (IR) are indicated below. A total of 11 and 14 primers were used to sequence the forward and reverse strand of the 4.5-kb region, respectively. The sequences and locations of these primers are available on request.
Map of the β-globin gene cluster with the 9-kb ‘hotspot’ delineated by the TaqI-5′δ (5′) and theHgiAI-β (3′) sites. RFLP sites are indicated by the arrows. The 4.5-kb fragment encompassing the 5′ flanking β region is amplified using two sets of primers: 5′R3K versus 3′R5K and 5′β422 and 3′β9. The other polymorphic sites are indicated by vertical lines. The pur, chi, and the 21-bp element at position −355021 are represented by thick horizontal lines. The crossover regions and the region of initiation replication (IR) are indicated below. A total of 11 and 14 primers were used to sequence the forward and reverse strand of the 4.5-kb region, respectively. The sequences and locations of these primers are available on request.
Primers 5′ β422 (5′-TCC AGG CAG AAA CAG TTA GAT GTC-3′) and 3′ β9 (5′-CAT TCG TCT GTT TCC CAT TCT A-3′) corresponding to GenBank coordinates 61292-61315 and 62745-62724, respectively, direct the amplification of a 1,453-bp fragment encompassing the 5′ end of the β-globin gene from position −845 through to position 609 relative to the cap site including exon 1, intron 1, and exon 2 of the β-globin gene (Fig 1). PCR amplification was performed in a 100-μL reaction volume containing 0.2 mmol/L of each dNTP, 10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl, 2.5 mmol/L MgCl2, 2.5 U ofTaq polymerase (Cetus), and 20 pmol of each primers 5′β422 and 3′β9. After an initial denaturation of 4 minutes at 94°C, 30 cycles of denaturation at 94°C for 1 minute, annealing at 59°C for 2 minutes, and extension at 72°C for 3 minutes were performed, the last extension reaction was prolonged for 7 minutes. After checking for amplification, the PCR product was isolated using a QIAEXII Gel extraction kit (Qiagen Ltd, Surrey, UK) as before. The purified DNA fragment was directly sequenced by the thermal cycle sequencing reaction using 33P-labeled terminators and a series of forward and reverse primers.
DNA fingerprinting.
To exclude false parentage, DNA from each member of the families was digested with HinfI, Southern blotted, and hybridized with a panel of seven probes known to be specific for hypervariable minisatellites (MS1, MS8, MS29, MS31, MS43, MS51, Pλg3).22 These hypervariable loci are extremely variable with heterozygosities ranging from 90% for MS8 to 99% for MS31 and are dispersed over four autosomes. The locus-specific minisatellites act as very sensitive hybridization probes for these loci and can be pooled to detect the hypervariable (HVR) loci simultaneously giving rise to multilocus Southern blot patterns that are highly individual-specific (DNA fingerprints). Based on the heterozygosities and the average number of DNA fragments (8.4) resolved using a mixture of five probes, the chance that all of the fragments in one individual are present in a second randomly selected individual has been estimated at <6 × 10−7.22 Therefore, these patterns provide a high level of individual specificity.
RESULTS
Greek Cypriot family.
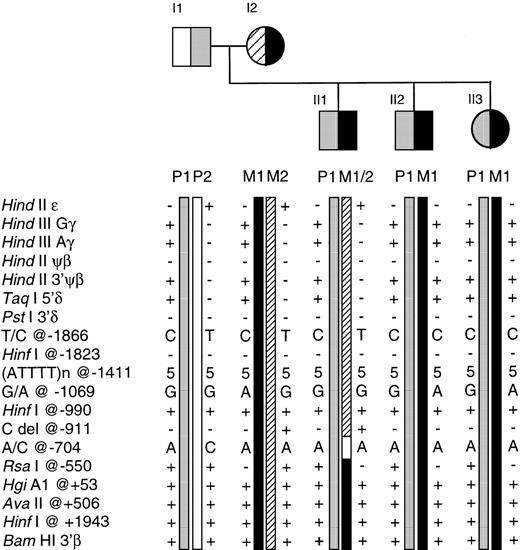
In this family, both parents were heterozygous for different β-thalassemia mutations (β+ 33 C → G in I1 and βo 39 C → T in I2), and all three offspring were compound heterozygotes23 (Fig2).
The Greek Cypriot family illustrating segregation of the β haplotypes and informative recombination events. Both parents are heterozygotes (β+ 33 C → G in I1 and βo39 C → T in I2) and all three offspring, compound heterozygotes for β-thalassemia. β Haplotypes P1 and M1 are associated with the paternal and maternal β-thalassemia alleles, respectively. While individuals II2 and II3 have inherited the P1 and M1 haplotypes intact, individual II1 has inherited P1 and a maternal recombinant chromosome, as identified by the haplotype M1/M2. The site of recombination is located within the 361-bp interval between positions −911 and −550 relative to the β-globin gene cap site.
The Greek Cypriot family illustrating segregation of the β haplotypes and informative recombination events. Both parents are heterozygotes (β+ 33 C → G in I1 and βo39 C → T in I2) and all three offspring, compound heterozygotes for β-thalassemia. β Haplotypes P1 and M1 are associated with the paternal and maternal β-thalassemia alleles, respectively. While individuals II2 and II3 have inherited the P1 and M1 haplotypes intact, individual II1 has inherited P1 and a maternal recombinant chromosome, as identified by the haplotype M1/M2. The site of recombination is located within the 361-bp interval between positions −911 and −550 relative to the β-globin gene cap site.
The β haplotypes for each member of the family were derived from the results of 14 RFLPs (HindII-ε,HindIII-Gγ,HindIII-Aγ, HindII-Ψβ,HindII-3′ Ψβ, AvaII-β,BamHI-β, Taq I-5′δ, Pst I-3′δ,HinfI-5′β at positions −1823 and −990, RsaI-5′β, HgiAI-β, and HinfI-3′β (see Fig 2). There are four β haplotypes in this family, the phase of the RFLPs was established using the two virtually homozygous offspring, II2 and II3. Both II2 and II3 have inherited the β-thalassemia alleles from the father (I1) and the mother (I2) associated with haplotypes P1 and M1, respectively. II1, who is also a compound heterozygote for β-thalassemia, has inherited an intact paternal chromosome, P1. However, while II1 has inherited the maternal β-thalassemia mutation, the β-thalassemia allele in II1 is associated with the 5′ haplotype of M2 and the 3′ haplotype of M1. On the basis of RFLP analysis, the M2 chromosome is intact 5′ up to the Taq I-5′δ site, and the M1 chromosome is intact 3′ up to the Rsa I-5′β site, but the origin of the variant sites in the 10.4-kb segment between these two sites, including the Pst I-3′δ andHinfI-5′β, is not clear.
Direct sequence analysis of the area between the Taq I-5′δ and Rsa I-5′β site was undertaken to search for additional sequence polymorphisms. The analysis identified several polymorphisms; the T/C polymorphism at position −1866, G/A polymorphism at position −1069, deletion of a single C at position −911 (a novel polymorphism), and A/C polymorphism at position −704 from the cap site of the β-globin gene (see Fig 2). Analysis of the (TG)n repeat24 was not attempted as it lay outside the area of interest.
The direct sequence analysis established that the proband, II1, had inherited an intact maternal M2 chromosome from HindII-ε to the C deletion at position −911 (5′ to 3′), and an intact M1 chromosome from BamHI-β to the Rsa I-5′β site at position −550 relative to the β-globin gene cap site (3′ to 5′) (Fig 3). The segment bordered by these two sites is 361 bp; no informative polymorphisms were found within this segment.
DNA sequence immediately 5′ of the β-globin gene in the Greek Cypriot family. The individuals II1, I1, I2, II2, and II3 (as shown in Fig 2) are represented by lanes 1, 2, 3, 4, and 5. The termination reactions are loaded in consecutive lanes in a block, ddGTP (G), ddATP (A), ddTTP (T), and ddCTP (C). (Panel A) The 5′ breakpoint indicated by the 1-bp deletion (arrowed) at position −911; (Panel B) the 3′ breakpoint indicated by the arrow (Rsa I site) at position −550.
DNA sequence immediately 5′ of the β-globin gene in the Greek Cypriot family. The individuals II1, I1, I2, II2, and II3 (as shown in Fig 2) are represented by lanes 1, 2, 3, 4, and 5. The termination reactions are loaded in consecutive lanes in a block, ddGTP (G), ddATP (A), ddTTP (T), and ddCTP (C). (Panel A) The 5′ breakpoint indicated by the 1-bp deletion (arrowed) at position −911; (Panel B) the 3′ breakpoint indicated by the arrow (Rsa I site) at position −550.
Asian Indian family.
The β haplotypes of both parents and the three offspring were generated from 13 RFLPs: HindII-ε,HindIII-Gγ,HindIII-Aγ, HindII-Ψβ,HindII-3′ Ψβ, Taq I-5′β,HinfI-5′β at position −1823, HinfI-5′β at position −990, Rsa I-5′β, HgiI-β,AvaII-β, HinfI-3′β, and BamHI-β (see Fig 4). In this family, the inheritance of the β-alleles is not associated with thalassemia. Phase of the RFLPs was established in the children based on the phase in the mother, I2, who was homozygous for most of the RFLPs. Two of the children, II2 and II3, have inherited intact chromosomes, P2 and M2, from their father (I1) and mother (I2), respectively. The proband, II1, has inherited an intact chromosome (M1) from the mother but the paternally inherited chromosome comprised of the 5′ end of P1 (up toHind-3′Ψβ) and the 3′ end of P2 (up toHinfI-3′β). The origin of the chromosome in the 15-kb segment between the HindII-3′Ψβ andHinfI-3′β sites is not clear from RFLP analysis because the sites within it are uninformative both in the father and proband.
The Asian Indian family showing segregation of the β haplotypes and informative recombination events. Individual II1 has inherited a paternal recombinant chromosome, as identified by the haplotype P1/P2. The site of recombination is located within a region of ∼1,100 bp between the (AT)xTy repeats at position −542 and the G/T polymorphism at position +568. (AT)xTy at position −542 is represented by A: (AT)7T7 and B: (AT)9T5.
The Asian Indian family showing segregation of the β haplotypes and informative recombination events. Individual II1 has inherited a paternal recombinant chromosome, as identified by the haplotype P1/P2. The site of recombination is located within a region of ∼1,100 bp between the (AT)xTy repeats at position −542 and the G/T polymorphism at position +568. (AT)xTy at position −542 is represented by A: (AT)7T7 and B: (AT)9T5.
Sequence analysis of the 5′ of the β-globin gene showed several polymorphisms, including the T/C polymorphism at position −1866, (ATTTT)n at position −1411, G/A polymorphism at position −1069, A/C polymorphism at position −704, (AT)xTy at positions −542 to −522, and G/T polymorphism in intron 2 (at position +568 from the β-globin gene cap site).
The sequence analysis established that II1 had inherited the 5′ P1 chromosome intact up to the (AT)xTy region and that the 3′ P2 chromosome was inherited intact up to the G/T polymorphism in intron 2 of the β-globin gene (Fig5).
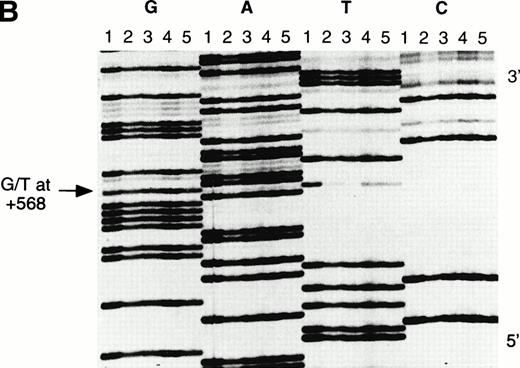
DNA sequence of the 5′ region of the β-globin gene in the Asian Indian family. The individuals I1, I2, II1, II2, and II3 (as in Fig 4) are represented by lanes 1, 2, 3, 4, and 5, respectively. The termination reactions are loaded in consecutive lanes in a block, ddGTP (G), ddATP (A), ddTTP (T), and ddCTP (C). (a) The 5′ breakpoint of the crossover region is located in the region of the (AT)xTy repeats; (b) the 3′ breakpoint is located at the G/T polymorphic site at position +568 (arrowed).
DNA sequence of the 5′ region of the β-globin gene in the Asian Indian family. The individuals I1, I2, II1, II2, and II3 (as in Fig 4) are represented by lanes 1, 2, 3, 4, and 5, respectively. The termination reactions are loaded in consecutive lanes in a block, ddGTP (G), ddATP (A), ddTTP (T), and ddCTP (C). (a) The 5′ breakpoint of the crossover region is located in the region of the (AT)xTy repeats; (b) the 3′ breakpoint is located at the G/T polymorphic site at position +568 (arrowed).
Amish family.
Previous study on the third family, of Amish descent, by Gerhard et al,16 had established that a single recombination event occurred in the paternal chromosome of II2 between the TaqI-5′δ and BamHI-3′β sites, within a region of 15 kb (see Fig 6), which was confirmed by our RFLP analysis of the same sites. Our analysis of additional RFLPs suggested that the putative recombination occurred within a 2.5-kb segment between the Rsa I-5′β and the HinfI-3′β sites. There are three haplotypes in this family; the parents I1 and I2 share a common β haplotype (M1 and P1).
The Amish family showing segregation of the β haplotypes and informative recombination events. Individual II2 has inherited a paternal recombinant chromosome as identified by the haplotype P1/P2. The site of recombination is located within a region of ∼1,100 bp between the (AT)xTy repeats at position −542 and the G/T polymorphic site at position +568. (AT)xTy is represented by A: (AT)7T7 and B: (AT) 9T5.
The Amish family showing segregation of the β haplotypes and informative recombination events. Individual II2 has inherited a paternal recombinant chromosome as identified by the haplotype P1/P2. The site of recombination is located within a region of ∼1,100 bp between the (AT)xTy repeats at position −542 and the G/T polymorphic site at position +568. (AT)xTy is represented by A: (AT)7T7 and B: (AT) 9T5.
Direct sequence analysis proved that the family was informative at the same sequence polymorphisms as in the Indian family with similar recombination breakpoints, ie, the (AT)xTy site at positions −542 to −522 and the G/T polymorphism at position +568.
There are four possible explanations for the recombinant chromosomes in the three families: (1) a meiotic recombination event has occurred within the β-globin cluster; (2) false parentage; (3) point mutations at multiple sites; or (4) gene conversion. False parentage had been previously excluded in the Amish family.16 In the Greek-Cypriot family, Southern blot hybridization of HinfI digest with a pool of seven hypervariable minisatellite probes showed 10 bands in the father, 14 in the mother, 13 in II1, 9 in II2, and 11 in II3. Of the 13 bands in I1, 6 were shared with father and 7 with mother. Of the 9 bands in II2, 5 were shared with father and 4 with mother. Of the 11 bands in II3, 6 were shared with father and 5 with mother. Similarly, in the Asian Indian family, 12 bands could be scored in the father, 10 in the mother, 11 in II1, 10 in II2, and 12 in II3. In II1, 5 of the 11 bands were shared with father, 6 with mother; in II2, 5 of the 10 bands were shared with the father and 5 with the mother, while in II3, 6 of the 12 bands were shared with father and the other 6 with mother. In all cases, all the alleles present in the offspring could be traced to either of the parents, indicating that the genetic relationships have been correctly assigned with no evidence of false paternity or maternity. The third explanation is unlikely because it requires several independent point mutations at the different RFLP sites and, in the case of the Greek family, including the C-T mutation at codon 39 of the β-globin gene to create a β-thalassemia mutation identical to the one in the mother. The fourth explanation is not ruled out but seems unlikely as it would involve gene conversion of a large stretch of DNA (∼22 kb) in the Amish and Asian Indian families. In the Greek Cypriot family a conversion of ∼1 kb of M2 by M1, from 3′ of the A/C at position −704 to 3′ of the thalassemia mutation at codon 39, could explain the composite maternal chromosome in II1.
The most likely explanation is that a meiotic recombination has occurred within the β-globin gene cluster, between the C deletion at position −911 and the Rsa I site at position −550 in the Greek family, and between the (AT)xTy repeats at position −542 and the G/T site at position +518 in the Asian Indian and Amish families.
DISCUSSION
β Haplotypes generated from RFLPs have led to the identification of a recombination event in the β-globin gene cluster in two families, bringing the total reported to six. We then used DNA sequence analysis to delineate the region of crossover to 361 bp in one family and 1,100 bp in the other family. The region of crossover was also sequenced in a previously reported family and this corresponded to the same region of 1,100 bp. These represent the most precisely defined region of recombinations within the human β-globin gene cluster.
The 9-kb region immediately 5′ to the β-globin gene has been proposed as a recombination ‘hotspot’ with recombination rates of 3 to 30 times higher than those of the surrounding regions. The ‘hotspot,’ as identified by Chakravarti et al,8 extends from theTaq I-5′δ site to the HgiAI-β site (Fig 1). Although the crossover region falls within this ‘hotspot’ in one family (Greek Cypriot), the 3′ boundary of the crossover region in the other two families extends ∼500 bp 3′ beyond the ‘hotspot.’ However, in the absence of any further polymorphisms within the 1,100-bp segment, it is quite possible that the cross-over region in the Amish and Asian Indian families falls within the boundaries of the 9 kb ‘hotspot.’
Previous studies have suggested several potential signal sequences that could enhance recombination within the ‘hotspot.’25 These included the (ATTT)n, (TG)n, and (AT)xTy repeats. In all three families, the crossover regions are located 3′ of the (TG)n24and (ATTTT)n repeats which lie ∼1,800 bp and ∼500 bp, respectively, upstream of position −911 (the 5′ crossover breakpoint in the Greek family) which makes these repeats as unlikely candidates initiating recombination, at least in these three families. Interestingly, a previous study of sequence-specific meiotic recombination in Saccharomyces cerevisiae comparing three adjacent restriction fragments immediately 5′ of the human β-globin gene showed that deletion of the (TG.CA)n sequences at the −2678 to −2648 interval does not significantly reduce the high frequency of genetic recombination in this region.26
It is of note that in all three families, the crossover region lies within the small well-defined 2-kb fragment (between positions −1461 and +476) which contains the replication origin of the β-globin cluster.19,27 This initiation region (IR) contains a 16-bp consensus sequence (5′-GGNNGAGGGAGARRRR-3′ at positions −63 to −48) for the Pur protein, which has been shown to have preferential binding for single-stranded DNA.20 On this basis it has been suggested that Pur would be expected to function as a helix-destabilizing protein, especially at the position of the Pur element, a role implicated in initiation of replication and recombination. The Pur element is located within the 1,100-bp crossover region. Although the Purelement lies outside the crossover region in the Greek family, it is still possible that it is involved in the initiation of recombination/replication as initial duplex opening is not restricted to a single site but can occur throughout within each initiation zone.28 Evidence to support this possibility includes, firstly: in many mammalian loci where the Pur element has been identified, it is located at least 1 kb away from the actual start site of replication.20 Secondly, it has been observed in primate cells that the binding of replication initiator protein dnaA, for oric of Escherichia coli,destabilizes the DNA helix to promote duplex opening at a region removed from the original recognition site.29Frequently, this region is a reiterated AT-sequence,30 such as the (AT)xTy repeats, which forms the 5′ breakpoint of the 1,100-bp crossover regions. Furthermore, the 3′ breakpoint of the 361-bp crossover region also lies within an AT-rich sequence.
Another consensus DNA sequence implicated in the initiation of DNA replication and recombination is the 21-bp 5′-WAWTTDDWWWDHWGWHMAWTT-3′ where M = A or C, W = A or T, D = A or G or T, and H = A or C or T.21 This consensus is found, with two base mismatches, within the ‘hotspot’ at position −3550 upstream of the β-globin gene. However, it is located ∼1 kb outside the IR region and certainly outside the crossover regions in the three families. The 1,100-bp crossover region also contains a chi (χ) sequence (5′-GCTGGTGG-3′) at ∼300 bp 5′ of the G/T polymorphic site in exon 2 of the β-globin gene. The χ sites are known to be hotspots for homologous recombination in E coli, increasing recombination by 5- to 10-fold.25,31 32 However, the exact role of these elements in recombination remains unclear.
Although all the three crossover regions fall within the broad 9-kb ‘hotspot,’ sequence analysis indicates that the crossover events occurred at distinct sites within this area. It is possible that this 9-kb hotspot contains several smaller areas with increased rates of recombination that are nonrandomly distributed. This suggests a clustering of recombination breakpoints such as has been observed in the MHC complex where the crossover breakpoints occurred within several defined areas associated with an increased frequency of recombination, all located within the larger ‘hotspot.’3 Despite sequence analysis, only an estimate of the minimal crossover regions can be obtained, the limiting factor being the presence of informative polymorphisms. In one family this region is 361 bp and 1,100 bp in the other two families. In contrast, sequence analysis of a crossover site was defined to a 138-bp segment in a recombination event in the HLA class II region.33
Although only six families with recombination events within the β-globin cluster have been reported to date, in all six the crossovers occurred within the region immediately 5′ to the β-globin gene, which suggests that the frequency of recombination in this area is higher than those in the rest of the β complex. These observations support the results of a recent study of allelic sequence diversity at the human β-globin locus which concluded that recombination and gene conversion in the 5′ region of the β-globin gene has contributed to the β-haplotype diversity, which was higher than expected on the basis of the observed nucleotide polymorphisms.34 However, to fully appreciate the frequency of recombination, ie, to establish if this is truly a ‘hotspot’ or just a ‘warm patch’ in cold surroundings, a systematic study of the β-globin cluster compared with its flanking regions on chromosome 11p involving sperm DNA typing or a larger number of families (such as the other CEPH pedigrees) is needed.
ACKNOWLEDGMENT
We thank Dr John Old for the gift of some of the PCR primers; Prof Sir D.J. Weatherall for encouragement and support; and Liz Rose and Milly Graver for preparation of the manuscript.
P.J.H. was a Nuffield Dominions Fellow; R.A.S. was supported by the Medical Research Fund, Oxford; and J.R.K. is supported in part by a grant (SBR-96 32509) from the National Science Foundation (USA). We thank the MRC, UK, for support.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Swee Lay Thein, MD, Medical Research Council Molecular Haematology Unit, Institute of Molecular Medicine, John Radcliffe Hospital, Headington, Oxford, OX3 9DS UK; e-mail: swee.thein@imm.ox.ac.uk.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal