Abstract
The initiation of primary hemostasis is mediated by interaction of the platelet glycoprotein Ib (GPIb) surface receptor and its arterial subendothelial von Willebrand factor (vWF) ligand. The intracellular signaling immediately following GPIb receptor occupancy connecting the adhesive event to platelet activation and aggregation has not been well characterized. The 14-3-3 proteins are a 27- to 30-kD ubiquitous protein family with diverse biologic roles, including functional modulation of several prominent signaling proteins. We used the yeast two-hybrid system and confocal microscopy to characterize the recently described interaction between GPIb and platelet 14-3-3ζ, and provide evidence for the potential signaling role of this protein. Two-hybrid interactions suggest that platelet 14-3-3ζ associates with the cytoplasmic domain of GPIb subunits Ibα and Ibβ in transformed yeast cells. The 14-3-3 interaction with GPIbβ may be partly mediated through the latter's phosphorylated serine 166 residue as its mutagenesis results in 20% to 40% reduced interaction. There was 51% to 59% reduced interaction between GPIb and three 14-3-3ζ deletion mutants compared with full-length 14-3-3ζ, suggesting that either theN-terminal dimerization or membrane-binding domains or more than one noncontiguous 14-3-3ζ element may be required for optimal GPIb interaction. Confocal studies of platelets and a megakaryocyte cell line provided additional evidence for interaction of 14-3-3ζ with GPIbα and GPIbβ. We also found that, similar to the signaling mediators phosphatidylinositol 3-kinase and Src, platelet cytoskeletal 14-3-3ζ content is increased following vWF and ristocetin stimulation. We suggest that platelet 14-3-3ζ interacts with GPIbα and Ibβ, that this interaction may be partly mediated through phosphoserine recognition, and that 14-3-3ζ cytoskeletal translocation may serve as a GPIb post–receptor occupancy signaling event.
PLATELET ADHESION TO arterial subendothelium is an integral component of thrombus formation and is mediated by a shear-dependent interaction between the platelet glycoprotein (GP) Ib/V/IX receptor on the platelet surface and its ligand, von Willebrand factor (vWF). Following this interaction, the platelet undergoes characteristic morphologic and biochemical changes associated with its transition from a resting state to an activated state. This includes a conformational change of the platelet surface GPIIb-IIIa integrin that facilitates its interaction with ligands, including plasma fibrinogen, that lead to platelet aggregation.
The platelet GPIb receptor consists of four polypeptide chains with certain features in common.1 First, they are all transmembrane proteins with extracellular and cytoplasmic domains. The GPIbα subunit is associated covalently with the smaller GPIbβ subunit, while GPIX and GPV are noncovalently associated with the complex. Surface expression of GPIb subunits is significantly reduced or absent in the congenital bleeding disorder Bernard-Soulier syndrome, in which mutations involving three subunits have been described.2,3 Each GPIb subunit contains one or more 24–amino acid tandem repeats rich in leucine that have been demonstrated to participate in protein-protein interactions in other cells.1
Tyrosine phosphorylation of multiple platelet proteins is demonstrable upon GPIb stimulation with vWF and ristocetin, but the precise molecular signaling events leading to platelet activation remain to be elucidated. Previous investigation has demonstrated the translocation of platelet Src and phosphatidylinositol 3-kinase to cytoskeletal elements in response to vWF stimulation, suggesting these enzymes play a role in the cytoskeletal reorganization associated with platelet activation following vWF-GPIb interaction.4
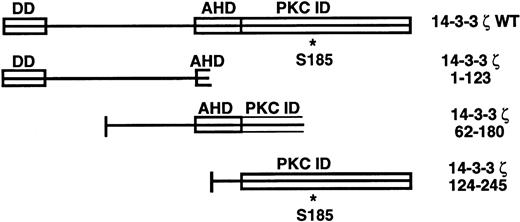
The 14-3-3 proteins are a 27- to 30-kD family originally isolated from the brain, with at least 10 homologous mammalian isoforms now described.5-7 Current knowledge regarding the functional domains of the 14-3-3 family is shown in Fig1. They exist as homodimers and heterodimers in vivo and have been found to exhibit a wide array of biologic functions in many mammalian cell types, plants, and yeast.7-13 The 14-3-3 family has recently been implicated in the regulation of intracellular signaling pathways through interaction with several oncogene and proto-oncogene products. Association with and regulation of proteins with key signaling roles such as Raf-1, protein kinase C, and phosphatidylinositol 3-kinase by 14-3-3 has been described.12,14 15 This group may have their kinase activity altered by this binding depending on the experimental conditions used.
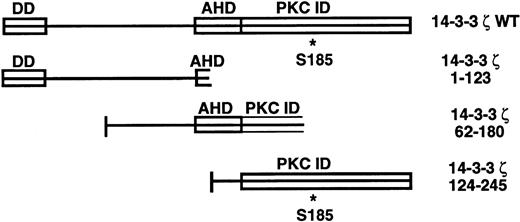
14-3-3ζ structural features and deletion mutants used in yeast 2-hybrid cotransformations with platelet GPIb subunits. Known and putative domains include a 26-residue N-terminal dimerization domain (DD),8,9 a domain with homology to the C terminus of the annexin protein family that acts as a protein kinase C (PKC) inhibitor (AHD),10 and a putative C-terminal PKC-inhibitory domain that a recent mutagenesis investigation was unable to more precisely localize (PKC ID).8 A C-terminal phosphoserine (residue 185*) has also been characterized.11
14-3-3ζ structural features and deletion mutants used in yeast 2-hybrid cotransformations with platelet GPIb subunits. Known and putative domains include a 26-residue N-terminal dimerization domain (DD),8,9 a domain with homology to the C terminus of the annexin protein family that acts as a protein kinase C (PKC) inhibitor (AHD),10 and a putative C-terminal PKC-inhibitory domain that a recent mutagenesis investigation was unable to more precisely localize (PKC ID).8 A C-terminal phosphoserine (residue 185*) has also been characterized.11
Recent affinity chromatography experiments using a monoclonal antibody to GPIbα led to the coelution of a 29-kD protein with GPIb.16 Amino acid sequencing of the protein led to its identification as the ζ isoform of 14-3-3. In view of the association of 14-3-3 with mediators that participate in known early signaling events,14,15,17 and the protein's membrane-binding domain8,18 and phosphoserine site,11 one may speculate that this protein participates in GPIb signaling following vWF stimulation. The current study uses the yeast two-hybrid expression system, confocal microscopy, and platelet compartment immunoblot studies to document and quantify the interaction of 14-3-3ζ with cytoplasmic domains of the GPIb subunits, and begins to characterize its potential role in GPIb signaling.
MATERIALS AND METHODS
Yeast two-hybrid constructs and transformation.
The cytoplasmic domains of platelet GPIbα and GPIbβ were each polymerase chain reaction (PCR)-amplified from platelet cDNA generated by platelet RNA reverse transcription. Platelet RNA was isolated from the therapeutic pheresis material of a patient with essential thrombocythemia. The sense primers each contained EcoRI sites and the antisense primers contained SalI sites to facilitate directional cloning of the restriction enzyme–digested PCR products into the yeast expression vectors pGBT9 and pGAD424 (Clontech Laboratories Inc, Palo Alto, CA). This resulted in the in-frame fusion of each cytoplasmic domain to the 3′ end of either the GAL4 (1-147) DNA-binding domain (pGBT9) or the GAL4 (768-881) activation domain. Similar PCR and cloning procedures led to construction of yeast expression vectors containing full-length platelet 14-3-3ζ.19
Construction of the GPIbβ (ser166ala) substitution mutant was achieved with the site-directed mutagenesis technique of Michael.20 PCR amplification of the Ibβ cytoplasmic domain used the original two wild-type primers along with a third sense primer incorporating the ser166ala mutation and an HpaI site. The latter facilitated colony screening following transformation intoEscherichia coli without altering the amino acid sequence of the resultant protein other than the desired substitution. PCR was performed in the presence of a thermostable DNA ligase (Taq ligase; Sigma Inc, St Louis, MO) to permit 5′ to 3′ ligation of the wild-type sense strand to the first base of the mutagenic sense primer. The 14-3-3 deletion constructs (Fig 1) were prepared similarly to the wild-type constructs using primers with EcoRI (sense) andSalI (antisense) restriction site adaptors. All wild-type and mutant DNA sequences were confirmed by automated sequencing technology.
Transformations of pairwise vector combinations into yeast strain SFY526 (MATa, ura3-52, his3-200, ade2-101, lys2-801, trp1-901, leu2-3, 112, canr, gal4-542, gal80-538, URA3 :: GAL1-lacZ; Clontech) were made as described by Bartel et al.21 After verification of the strain's nutritional phenotype (auxotrophic for tryptophan and leucine), early log-phase (OD600 = 0.2 to 0.3) cultures were grown for 3 hours at 30°C in YPD medium before transformation of 100 μL with 100 ng of each construct. Following agitation in 40% polyethylene glycol/100-mmol/L lithium acetate at 30°C for 30 minutes, addition of 10% dimethyl sulfoxide, and heat shock at 42°C for 15 minutes, the transformants were plated on selective media at 30°C until 2-mm colonies were visible.
Liquid culture and colony filter-lift β-galactosidase assays.
For quantitative β-galactosidase assays, a single yeast colony was innoculated into 5 mL SD medium without tryptophan and leucine and agitated overnight at 30°C, and 2 mL was added to 8 mL YPD liquid and grown until the OD600 was 0.5 to 0.7. Then, 1.5 mL was processed for quantitation of β-galactosidase activity expressed in units using ONPG substrate and taken to represent the strength of interaction between the two fusion proteins.21-24 Colony filter-lifts were performed on transformants by submersion of nitrocellulose transfer membranes lifted from each colony plate into liquid nitrogen (Clontech). This was followed by incubation at 30°C for up to 8 hours over Whatman (Maidstone, UK) filter paper presoaked in Z buffer (60 mmol/L Na2HPO4 · 7H2O, 40 mmol/L NaH2PO4H2O, 10 mmol/L KCl, and 1 mmol/L MgSO4, pH 7.0) with 0.27% β-mercaptoethanol and 1.67% X-gal in 100-mm petri dishes.
Platelet and human erythroleukemia cell confocal microscopy studies.
Human erythroleukemia (HEL) cells were grown at 37°C in a 5% CO2 atmosphere in RPMI 1640 medium (Bio-Whittaker Inc, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum, 2 mmol/L glutamine, 1 mmol/L sodium pyruvate, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 2.5 μg/mL amphotericin B, and 2.05 μg/mL desoxycholate (Fungizone; Life Technologies Inc, Grand Island, NY). To separate the platelets, platelet-rich plasma was centrifuged twice (1,000g for 10 minutes) followed by suspension of the platelet pellet in 35 mL washing buffer (10 mmol/L Tris, pH 7.0, 150 mmol/L NaCl, 1 mmol/L EDTA, and 7 mmol/L theophylline). Following centrifugation at 100g for 10 minutes, the suspended platelets were pelleted at 1,000g for 10 minutes and the concentration was determined in 2 mL Tyrode's buffer.25
Rabbit anti-human GPIbα polyclonal antibody,26 mouse monoclonal antibody Beb1 (anti-human GPIX), and mouse monoclonal antibody C-34 (anti-human Ibα) were used in confocal microscopy studies. The latter two were characterized by our laboratory and others in association with the Fifth International Workshop on Leukocyte Differentiation Antigens.27 Mouse anti-CD62P was obtained from Becton Dickinson Immunocytometry Systems (San Jose, CA). Gi27 (mouse anti-human GPIbβ) was a gift from Dr Sentot Santoso of Universitat Giessen (Giessen, Germany). Rabbit polyclonal anti-human 14-3-3ζ was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Fluorescein-isothiocyanate (FITC)-conjugated goat F(ab′)2 anti-rabbit antibody was obtained from Southern Biotechnology Associates Inc (Birmingham, AL), and CY3-conjugated goat anti-mouse antibody was obtained from Jackson Laboratories (West Grove, PA).
Platelets and HEL cells assessed for 14-3-3 and GPIb colocalization were aliquotted to 30 × 106 platelets/mL or 5 × 106 cells/mL phosphate-buffered saline (PBS) or PBS with 1% BME if subsequently labeled with anti-Ibβ. After 15 minutes, each platelet sample was cytospun onto a slide for the rest of the labeling procedure while HEL cells were centrifuged (130g for 10 minutes) following each of the remaining steps and then resuspended for the next step. The PBS incubation step was followed by suspension of platelets and HEL cells in 90% ethanol in PBS for 45 minutes.28 After washing in PBS, they were incubated for 30 minutes with 1:10 to 1:100 dilutions of two primary antibodies. After washing twice, they were then incubated for 30 minutes with 1:10 to 1:100 dilutions of fluorochrome-conjugated secondary antibodies. Following this, the platelets and HEL cells were washed twice and then suspended in 50 μL PBS. For confocal microscopy, 20 μL antibody-labeled suspended HEL cells were mounted onto a glass slide to which a glass cover slip was applied separated by an adhesive tape spacer to prevent crush artifact. Platelet and HEL cell slides were visualized using an ACAS Ultima Laser Cytometer (Meridian Instruments, Okemos, MI). Slides were scanned in dual-color confocal mode (pinhole = 225 μm for platelets and 400 μm for HEL cells) using a 100× oil-immersion objective (numeric aperture = 1.3), a step size of 0.1 μm/pixel for platelets and 0.2 μm for HEL cells, and a field size of 360 × 360 pixels (platelets) or 180 × 180 or 270 × 270 pixels (HEL cells). FITC fluorescence was detected in PMT1 with a 530/30-nm band-pass filter, and CY3 fluorescence was detected after separation with a dichroic beam splitter (560-nm short-pass filter) in PMT2 with a 580/30-nm band-pass filter. Dual-color digital images were displayed on a DASY 9000 Image analysis workstation (Meridian) using the manufacturer's software, and were saved as tag interchange file format (TIFF) files. Similarly, after using a threshold for background fluorescence, pixel histograms were generated from these images and displayed using the manufacturer's software and printed with a Sony Mavigraph video printer (Sony Corp, Tokyo, Japan).
Platelet 14-3-3 translocation studies.
To study the relative amount of 14-3-3ζ present in platelet compartments during the resting state or following stimulation with known agonists, fresh platelets were first isolated from whole blood as already described. After suspension in Tyrode's buffer, 108 platelets were aggregated for 5 minutes at 37°C following addition of either (1) 10 μg/mL vWF isolated from human cryoprecipitate,29 (2) 1 mg/mL ristocetin (American Biochemical and Pharmaceutical Corp, New York, NY), (3) both 10 μg/mL vWF and 1 mg/mL ristocetin, (4) 1 U/mL thrombin (Sigma), (5) 20 μmol/L adenosine diphosphate (ADP), or (6) 500 μg/mL arachidonic acid (Bio/Data Corp, Hatboro, PA). This was followed by sample incubation in platelet lysis buffer (10 mmol/L EGTA, 2% Triton X-100, 1 mmol/L PMSF, 100 mmol/L benzamidine, 4 mg/mL leupeptin, 100 mmol/L Tris hydrochloride, pH 7.4, and 2 mmol/L sodium orthovanadate) on a rocker at 4°C for 1 hour. Samples were then fractionated into Triton X-100–soluble and –insoluble (cytoskeletal) components by 10,000g centrifugation for 5 minutes. This supernatant was in turn centrifuged at 100,000g for 3 hours at 4°C to separate the membrane skeletal component from platelet cytosol.30Pellet samples, after washing twice in PBS/0.05% Tween 20, along with 50 μL of each supernatant solution were then resuspended in sodium dodecyl sulfate (SDS) protein loading buffer, boiled for 2 minutes, electrophoresed on a 10% SDS-polyacrylamide gel,31electrotransferred to a 0.45-μm nitrocellulose membrane (Millipore Corp, Bedford, MA), and immunoblotted with anti–14-3-3 antibody C-16 or K-19 (Santa Cruz Biotechnology) followed by 32.5 μCi125I-labeled Staphylococcal protein A (New England Nuclear, Boston, MA). Autoradiograph bands were subsequently analyzed quantitatively using area integration on an ImageQuant densitometer (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Yeast two-hybrid assays.
We were interested in characterizing the interaction of the signaling protein 14-3-3ζ with the cytoplasmic domains of the GPIb α and β subunits. Our aims were to determine to which of the GPIb subunits 14-3-3 would bind; to determine what role, if any, was played by a previously documented phosphoserine residue on GPIbβ32; and to map the GPIb binding domain on 14-3-3.
PCR-amplified cDNA fragments encoding the entire cytoplasmic domains of GPIbα and GPIbβ along with a GPIbβ sequence in which serine 166 had been mutated to alanine were fused in-frame to either the DNA-binding domain or activation domain of the yeast plasmids pGBT9 or pGAD424, respectively. cDNAs encoding deletion mutants along with wild-type 14-3-3ζ (Fig 1) were similarly cloned into the same yeast expression vectors. Fusion protein expression of transformed unpaired GPIb and 14-3-3 constructs by themselves did not cause transactivation of the lacZ GAL4 reporter gene present in the yeast host strain SFY526, as determined by colony filter-lift assays. These vectors were next transformed into yeast in pairwise combinations along with three pairwise positive and four negative control transformations as follows: wild-type full-length GAL4 gene in pGBT9 alone; murine p53(72-390)/SV40 large T antigen(84-708); GPIbα/GPIbβ; pGBT9/pGAD424; murine p53(72-390)/pGAD424; pGBT9/SV40 large T antigen(84-708); and human lamin C(66-230)/SV40 large T antigen(84-708)(Clontech). To further exclude nonspecific interactions, each vector was also cotransformed into yeast with other two-hybrid vectors encoding irrelevant proteins not expected to interact with the GPIb or 14-3-3 constructs (Figs 2 and 3).
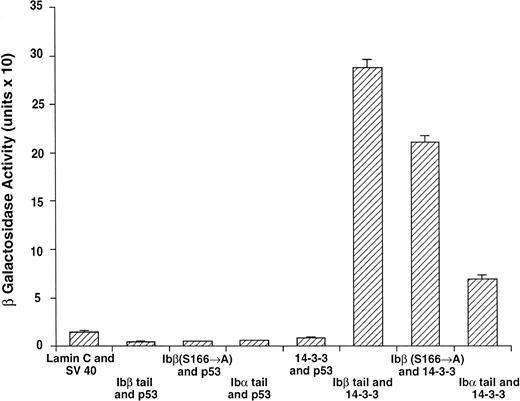
14-3-3ζ binds to the cytoplasmic domains of GPIbα and GPIbβ in yeast. Yeast were cotransformed with fusion proteins composed of GAL 4 modular domains hybridized to either GPIb, 14-3-3ζ, or control proteins. Data bars represent the mean ± SE of 5 separate experiments using analysis of 36 independent colonies and 3 spectrophotometric readings per colony at different concentrations. Five representative negative control transformations among the 8 performed are shown.
14-3-3ζ binds to the cytoplasmic domains of GPIbα and GPIbβ in yeast. Yeast were cotransformed with fusion proteins composed of GAL 4 modular domains hybridized to either GPIb, 14-3-3ζ, or control proteins. Data bars represent the mean ± SE of 5 separate experiments using analysis of 36 independent colonies and 3 spectrophotometric readings per colony at different concentrations. Five representative negative control transformations among the 8 performed are shown.
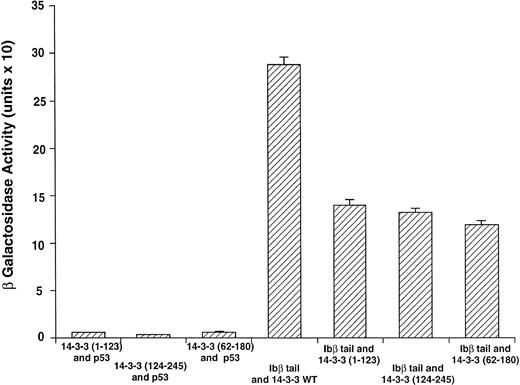
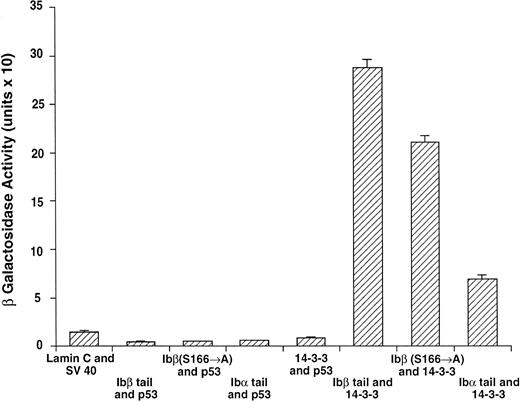
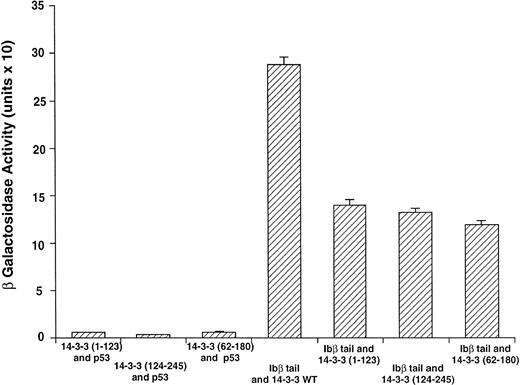
GPIbβ binding to 14-3-3ζ deletion mutants is reduced compared with the interaction with full-length 14-3-3ζ. The binary protein interactions along with the further controls shown in Fig 2 and 3 additional positive and 4 negative control transformations not shown were studied, and quantitative β-galactosidase assays were performed. Deletion mutant activity was reduced 51% to 59% v full-length 14-3-3. Data bars represent the mean ± SE of 5 separate experiments using analysis of 37 independent colonies with 3 spectrophotometric readings per colony at different concentrations.
GPIbβ binding to 14-3-3ζ deletion mutants is reduced compared with the interaction with full-length 14-3-3ζ. The binary protein interactions along with the further controls shown in Fig 2 and 3 additional positive and 4 negative control transformations not shown were studied, and quantitative β-galactosidase assays were performed. Deletion mutant activity was reduced 51% to 59% v full-length 14-3-3. Data bars represent the mean ± SE of 5 separate experiments using analysis of 37 independent colonies with 3 spectrophotometric readings per colony at different concentrations.
Following incubation at 30°C for 3 to 4 days, individual colonies were grown overnight in selective SD medium, and β-galactosidase activity was quantitatively determined using ONPG substrate.21 In this system, the extent of expression of the reporter gene lacZ can be interpreted as an indication of the strength of interaction between the two fusion proteins.21-24 Figure 2 shows the wild-type 14-3-3/GPIb fusion protein quantitative interactions along with five of eight negative control combinations. The findings suggest 14-3-3ζ interacts with both the GPIb α and β cytoplasmic domains, with significantly increased β-gal activity evident in both of these interactions versus the eight negative control combinations (Fig 2 and data not shown). Furthermore, when ser166 on GPIbβ is replaced with alanine, its interaction is reduced 20% to 40% (mean, 27%) with 14-3-3ζ.
Figure 3 shows results for β-galactosidase quantitative assays when the Ibβ cytoplasmic domain is cotransformed into yeast with full-length 14-3-3ζ and the 14-3-3ζ deletion mutants outlined in Fig 1. These findings along with the findings from qualitative X-gal assays (Fig4) suggest that both Ibα and Ibβ interact optimally with full-length 14-3-3 compared with any of the deletion mutants studied. Assays of Ibβ/14-3-3ζ colonies from each of the three 14-3-3ζ deletion mutant transformations showed a reduction in reporter gene activity of 51% to 59% compared with the analysis of Ibβ/14-3-3ζ full-length colonies. Interactions between the cytoplasmic domain of Ibα and the 14-3-3ζ deletion mutants were too low to quantify using an ONPG substrate, and were barely appreciable using the more sensitive X-gal colony filter-lift assay (Fig 4). Thus, we were unable to map the GPIb binding domain on 14-3-3 using the yeast two-hybrid assay, perhaps due to the need for at least two noncontiguous regions of 14-3-3ζ for optimal interaction with GPIb and/or the need for 14-3-3ζ to be dimerized or membrane-associated.
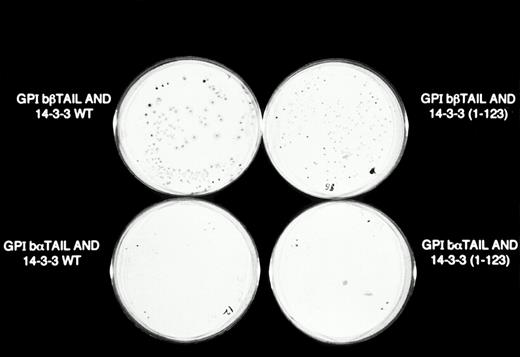
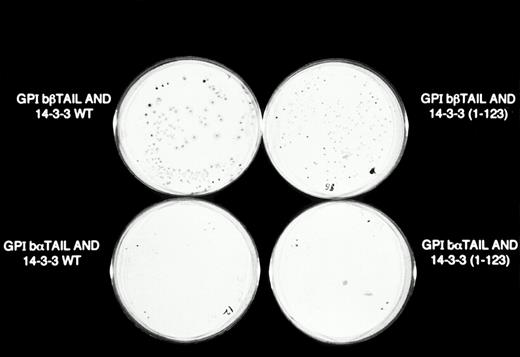
β-Galactosidase colony filter-lift assays of GPIb/14-3-3ζ yeast 2-hybrid transformations. Reporter gene activity is reflected by expression of blue (dark) colonies, most notable with the combination of GPIbβ and full-length 14-3-3ζ, followed by GPIbβ and deletion mutant 14-3-3 (1-123) and GPIbα and full-length 14-3-3ζ. The X-gal substrate used in these assays is considered more sensitive to β-galactosidase activity than the ONPG used in the quantitative assays in Figs 2 and 3.
β-Galactosidase colony filter-lift assays of GPIb/14-3-3ζ yeast 2-hybrid transformations. Reporter gene activity is reflected by expression of blue (dark) colonies, most notable with the combination of GPIbβ and full-length 14-3-3ζ, followed by GPIbβ and deletion mutant 14-3-3 (1-123) and GPIbα and full-length 14-3-3ζ. The X-gal substrate used in these assays is considered more sensitive to β-galactosidase activity than the ONPG used in the quantitative assays in Figs 2 and 3.
Confocal microscopy studies.
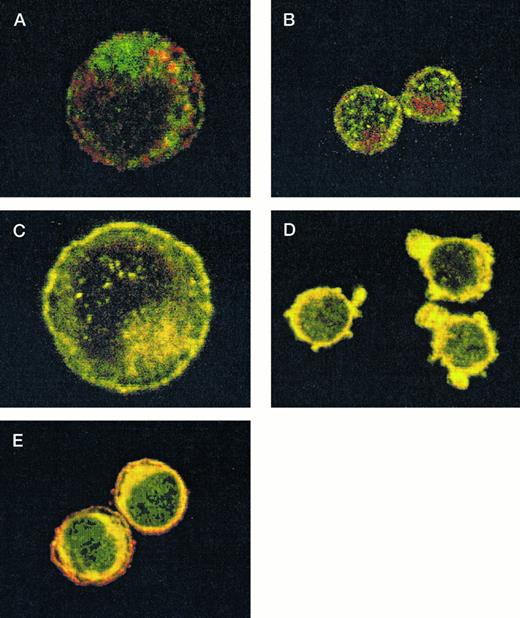
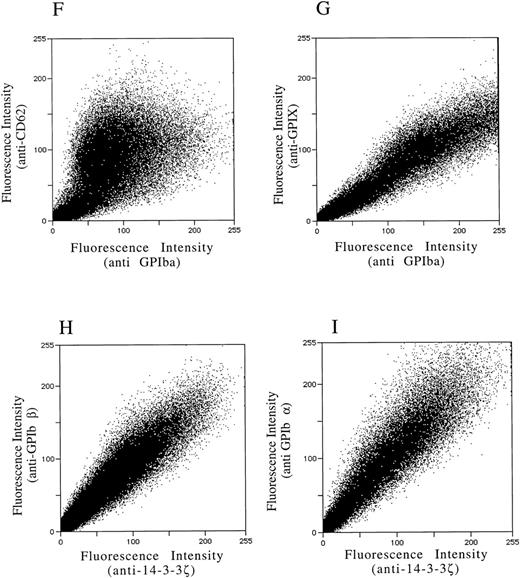
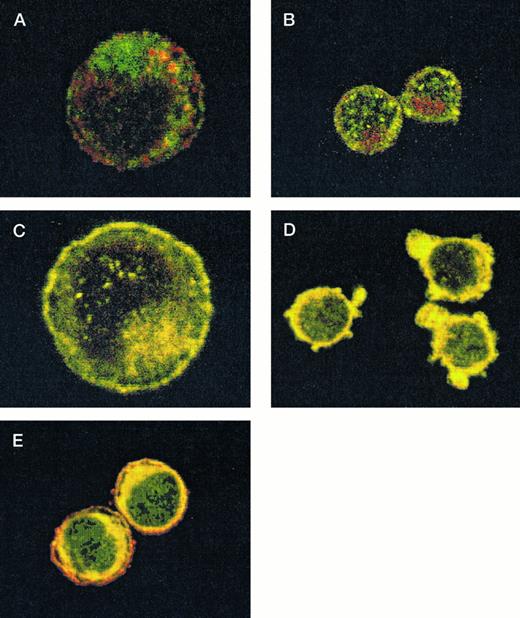
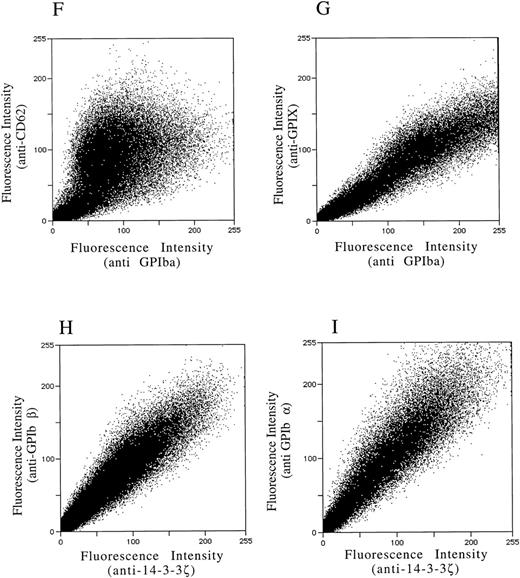
After demonstrating the interaction of 14-3-3ζ with GPIbα and GPIbβ through the yeast two-hybrid system, we were next interested in documenting these relationships in vivo using human platelets and HEL cells, a tumor cell line that manifests several megakaryocyte and platelet antigens including GPIb.33 Following membrane permeabilization, we proceeded to colabel platelets and cells with anti–14-3-3ζ and either anti-human GPIbα or anti-GPIbβ antibodies followed by fluorochrome-conjugated secondary antibody combinations. Samples were also colabeled with primary and secondary antibody combinations to either 14-3-3 and CD62P, Ibα and CD62P (negative controls), or GPIbα and GPIX (positive control). Cytospun slides (platelets) or HEL cells in suspension were then visualized by confocal microscopy. Our objective was to determine if each of the protein pairs colocalized and associated with each other to within the limits of resolution of a single pixel (∼200 nm). Colocalization of FITC (green) and Cy3 (red) is evidenced by a yellow appearance (Fig 5C to E) and by corresponding pixel histograms that demonstrate a linear correlation in antibody expression for each fluorescence intensity level34 (Fig 5G to I). Absence of colocalization is suggested by a red and green appearance (Fig 5A and B) and by a pixel histogram exhibiting a diffuse pattern34 (Fig 5F). These data support the yeast two-hybrid evidence that 14-3-3ζ interacts with GPIbα and GPIbβ present in platelets and HEL cells.
Colocalization of GPIb subunits and 14-3-3ζ in HEL cells (A to E) and platelets (F to I). Permeabilized platelets and HEL cells were colabeled with antibody pairs. HEL cells were labeled with either (A) anti-GPIbα/FITC (green) and anti-CD62P/CY-3 (red) (negative control), (B) anti–14-3-3ζ/FITC and anti-CD62P/CY-3 (negative control), (C) anti-GPIbα/FITC and anti-GPIX/CY-3 (positive control), (D) anti-GPIbβ/CY-3 and anti–14-3-3ζ/FITC, or (E) anti-GPIbα/CY-3 and anti–14-3-3ζ/FITC. Inset: detector 1, FITC fluorescence intensity; detector 2, CY-3 fluorescence intensity. Yellow fluorescence suggests that the 2 antibodies colocalize to within approximately 200 nm. A and C to E field size, 36 × 36 μm; B field size, 54 × 54 μm. Representative platelet pixel histograms for each antibody pair are shown.
(F to I) and reflect colocalization of GPIX/Ibα (G), GPIbβ/14-3-3ζ (H), and GPIbα/14-3-3ζ (I). Note the approximately equal contribution from each fluorochrome-conjugated secondary antibody at different fluorescence intensities in G (positive control), H, and I. This suggests that each computer pixel is detecting an equal amount of both antibodies present, which implies their respective epitopes are present in equal amounts (colocalized) in the 3-dimensional volume scanned by the microscope. This contrasts with F (1 of 2 negative controls), in which a diffuse pattern reflecting noncolocalization is seen. Three experiments with HEL cells and 2 with platelets were performed.
Colocalization of GPIb subunits and 14-3-3ζ in HEL cells (A to E) and platelets (F to I). Permeabilized platelets and HEL cells were colabeled with antibody pairs. HEL cells were labeled with either (A) anti-GPIbα/FITC (green) and anti-CD62P/CY-3 (red) (negative control), (B) anti–14-3-3ζ/FITC and anti-CD62P/CY-3 (negative control), (C) anti-GPIbα/FITC and anti-GPIX/CY-3 (positive control), (D) anti-GPIbβ/CY-3 and anti–14-3-3ζ/FITC, or (E) anti-GPIbα/CY-3 and anti–14-3-3ζ/FITC. Inset: detector 1, FITC fluorescence intensity; detector 2, CY-3 fluorescence intensity. Yellow fluorescence suggests that the 2 antibodies colocalize to within approximately 200 nm. A and C to E field size, 36 × 36 μm; B field size, 54 × 54 μm. Representative platelet pixel histograms for each antibody pair are shown.
(F to I) and reflect colocalization of GPIX/Ibα (G), GPIbβ/14-3-3ζ (H), and GPIbα/14-3-3ζ (I). Note the approximately equal contribution from each fluorochrome-conjugated secondary antibody at different fluorescence intensities in G (positive control), H, and I. This suggests that each computer pixel is detecting an equal amount of both antibodies present, which implies their respective epitopes are present in equal amounts (colocalized) in the 3-dimensional volume scanned by the microscope. This contrasts with F (1 of 2 negative controls), in which a diffuse pattern reflecting noncolocalization is seen. Three experiments with HEL cells and 2 with platelets were performed.
To provide statistical evidence for these conclusions, we calculated the degree to which colocalization occurred in each of the colabeled samples. This was determined by calculating the coefficient of variation ([CV] the standard deviation divided by the mean) for the ratio of red to green pixels for individual cells. These CVs were averaged (4 to 15 cells per sample) for each image and analyzed for differences between them using a two-sample one-tailed t test assuming unequal variances. Thus, for example, in one experiment the colocalization between GPIbα/GPIX was found not to significantly differ versus GPIbβ/14-3-3 (P = .23), while that between GPIbβ/14-3-3 and CD62P/14-3-3 did (P = .05). Lower average HEL cell CVs from three experiments confirmed the increased colocalization of GPIbα and GPIbβ with 14-3-3, compared with significantly less colocalization exhibited by GPIbα and 14-3-3 with CD62P (data not shown). Hence, the confocal photomicrographs, pixel histograms, and statistical analysis suggest that colocalization between 14-3-3ζ and both GPIbα and GPIbβ cytoplasmic domains occurs in platelets and HEL cells as suggested by the yeast two-hybrid experiments.
Platelet 14-3-3ζ translocation studies.
Having used the yeast two-hybrid system and confocal microscopy studies to obtain structural data concerning the interaction of 14-3-3ζ and platelet GPIb, we were interested in determining whether 14-3-3ζ translocated between the platelet cytosol and either the platelet cytoskeleton or membrane skeleton in response to stimulation with known agonists.30 Translocation to the cytoskeletal component in response to surface receptor occupancy is a property of many signaling proteins; these events typically lead to cytoskeletal reorganization, which in turn promotes events such as shape change, locomotion, and activation.4 35
Following isolation from whole blood, platelets were stimulated with either vWF alone, ristocetin alone (controls), both vWF and ristocetin, thrombin, ADP, or arachidonic acid. They were then subjected to aggregometry, lysed, and separated by low- and high-speed centrifugation into cytosolic, membrane skeletal, and cytoskeletal components, which were in turn subjected to SDS-polyacrylamide gel electrophoresis and immunoblotted with one of two anti–14-3-3ζ antibodies. Immunoblot band densitometry determinations from these experiments showed a 4.9-fold increase of cytoskeletal 14-3-3ζ in vWF/ristocetin-stimulated platelets compared with unstimulated platelets, and a 2.9-fold increase compared with thrombin-stimulated platelets (Table 1). Additional studies showed a similar or reduced amount of cytoskeletal 14-3-3ζ in response to ADP and arachidonic acid stimulation, respectively, compared with thrombin stimulation (data not shown).
DISCUSSION
Intracellular signaling that takes place immediately following GPIb receptor occupancy connecting the adhesive event to platelet activation and aggregation has not been well characterized. In the present study, we used yeast two-hybrid experiments and confocal microscopy to further characterize the interaction of platelet GPIb with an isoform of the 14-3-3 protein family. While their exact function remains elusive, the 14-3-3 protein family shows a diverse array of associations and biologic activities related to signaling and cell-cycle progression.7 12
We have shown that 14-3-3ζ will interact in vivo with both the GP Ibα and Ibβ subunit cytoplasmic domains; we are unable to determine the preference, if any, of 14-3-3 for either subunit with these methods. This interaction is modestly reduced when serine 166 on GPIbβ is replaced with alanine. We have also demonstrated that GPIb interacts optimally with full-length 14-3-3ζ, while showing an approximately equally reduced interaction with each of three deletion mutants. Confocal microscopy studies of platelets and HEL cells have provided additional characterization of the 14-3-3 interaction with GPIb receptor subunits. In this respect, the positive confocal data support the yeast two-hybrid findings but do not conclusively prove them, because of the imaging system's limits of resolution. Negative confocal data, on the other hand, would have refuted the findings. Finally, platelet 14-3-3 translocation studies in stimulated and resting platelets have provided evidence that 14-3-3ζ may translocate from the cytosolic component to the cytoskeleton in response to vWF and ristocetin. A similar agonist-induced cytoskeletal relocalization of platelet molecules with signaling roles such as Src, Syk, and FAK has been observed.30 35 Our methods were unable to detect any appreciable amount of agonist-induced translocation to or from the membrane skeleton.
Using affinity chromatography, a 29-kD protein found to be 14-3-3ζ was recently coeluted with GPIb-IX by Du et al,16 who later showed that the 14-3-3 binding site on GPIbα resides within a 15 residue C-terminal sequence.36 Binding was not contingent upon the presence of actin-binding protein and was abolished by removing the C-terminal five residues of GPIbα, suggesting their critical role in the association.16,36 Our results also suggest that 14-3-3 will associate with the cytoplasmic domain of GPIbα in vivo along with GPIbβ (Figs 2, 4, and 5). Du's group provided some evidence that 14-3-3 may not interact with GPIbβ.36 Our study differs from theirs with respect to the methods used to address this question (yeast two-hybrid studies and confocal microscopy v peptide inhibition and synthetic peptide binding studies). In addition, this group's synthetic peptides and anti-GPIbβ immunogens did not incorporate the cytoplasmic domain GPIbβ phosphoserine residue. Recent investigation has provided evidence that phosphoserine recognition may play an important role in 14-3-3 interaction with other proteins.37 38
Since GPIbα contains the receptor's ligand binding site and interacts with the membrane skeleton and GPIbβ contains a potentially signal-transducing phosphoserine residue in its intracellular domain, it is not unreasonable to speculate that 14-3-3ζ may indeed interact with both subunits. In this respect, dimerization of 14-3-3 not only may serve to potentially link receptors and signaling proteins but may also bridge important functional elements within a multiple subunit complex like GPIb.39
The reduction in interaction between 14-3-3 and GPIbβ when serine 166 is replaced by alanine is supported by the observation that phosphoserine recognition may play an important role in the former's interaction with other proteins.37,38,40,41 It is thus reasonable to speculate that the platelet GPIb interaction with 14-3-3 may be mediated, in part, by the phosphoserine-containing cytoplasmic domain of GPIbβ. Other groups have suggested that 14-3-3 may recognize clusters of serine residues in an amphipathic helical structure appropriate for its ligand-binding groove recently characterized through crystallography studies.36,42,43 Such a serine-rich cluster exists at the C-terminal region (residues 596 to 610) of GPIbα where interaction with 14-3-3 has been recently characterized.36 Our yeast two-hybrid and confocal microscopy data provide evidence for both of the 14-3-3 recognition motif theories by demonstrating 14-3-3 interactions with GPIbα and GPIbβ.
Our yeast two-hybrid experiments suggest that GPIb interacts optimally with full-length 14-3-3ζ, with 51% to 59% reduced interaction demonstrable between GPIb and deletion mutants of 14-3-3ζ (Figs 1 and3). Using these techniques, we were therefore unable to map a specific GPIb binding site on 14-3-3ζ. These results may be attributable to the dimerization domain involving the N-terminal 26–amino acid residues, suggesting that 14-3-3 may need to homodimerize or heterodimerize for optimal activity.8,9,13,44,45 The 14-3-3 proteins have been found at high concentrations on synaptic plasma membranes, and another study suggests the N-terminal region is responsible for this membrane binding.8,18 Optimal GPIb–14-3-3 interaction may thus be facilitated by binding of the latter to the platelet membrane. A third possible contributing cause to these results rests with the fact that GPIb–14-3-3 interaction may be mediated by at least two distinct and noncontiguous regions of 14-3-3. In this respect, the C-terminal phosphoserine 185 of 14-3-3 is surrounded by a consensus sequence for cyclin-dependent kinases, and amino acids 171 to 213 are required for interaction with phosphorylated tryptophan hydroxylase, the rate-limiting enzyme of serotonin synthesis.11 46 Therefore, for example, if both theN-terminal dimerization domain and C-terminal residues 171 to 213 were required for optimal interaction, only the full-length 14-3-3 species would be capable of maximal interaction with GPIb, as our findings suggest.
The translocation of 14-3-3ζ to the platelet cytoskeleton upon vWF and ristocetin stimulation is suggestive evidence that this protein participates in GPIb signaling (Table 1). Similar fractionation procedures have been used to demonstrate the cytoskeletal translocation of membrane skeleton and selective signaling molecules, such as the tyrosine kinases Src and FAK, upon platelet surface receptor binding.4,30,35,47 vWF stimulation and aggregation of platelets is capable of inducing cytoskeletal translocation and activation of both the lipid kinase phosphatidylinositol 3-kinase and Src independently of other agonists.4 These events were not observed in Bernard-Soulier platelets. Recent investigation has demonstrated an association of 14-3-3τ and phosphatidylinositol 3-kinase with additional evidence showing that it may act as a negative regulator of this enzyme by directly binding to its catalytic subunit.15 It could be hypothesized that 14-3-3 may interact with platelet GPIb under resting conditions and thereby exert a similar negative regulatory effect on phosphatidylinositol 3-kinase activity.
ACKNOWLEDGMENT
We are grateful to Chao-Yang Li and Kin Ritchie for helpful discussion, to Susan Danner and Tanya Hill for excellent technical assistance, and to Sally Swedine for help with the graphics.
Supported by Grants No. HL39947 and ES07033 from the National Institutes of Health, a Merit Review Award from the Department of Veterans Affairs, and the Medical Research Council of Canada.
Presented in part at the 1996 38th Annual Meeting of the American Society of Hematology, Orlando, FL (December 7-10, 1996).
Address reprint requests to Gerald J. Roth, MD, Medical Service, Seattle VA Medical Center (111), 1660 S Columbian Way, Seattle, WA 98108.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.