Abstract
Ten patients with adenosine deaminase deficiency (ADA−) have been enrolled in gene therapy clinical trials since the first patient was treated in September 1990. We describe a Japanese ADA− severe combined immune deficiency (SCID) patient who has received periodic infusions of genetically modified autologous T lymphocytes transduced with the human ADA cDNA containing retroviral vector LASN. The percentage of peripheral blood lymphocytes carrying the transduced ADA gene has remained stable at 10% to 20% during the 12 months since the fourth infusion. ADA enzyme activity in the patient's circulating T cells, which was only marginally detected before gene transfer, increased to levels comparable to those of a heterozygous carrier individual and was associated with increased T-lymphocyte counts and improvement of the patient's immune function. The results obtained in this trial are in agreement with previously published observations and support the usefulness of T lymphocyte-directed gene transfer in the treatment of ADA−SCID.
ADENOSINE DEAMINASE (EC3.5.4.4; ADA) is an enzyme in the purine salvage pathway that is critical for the deamination of adenosine and deoxyadenosine and consequent formation of inosine and deoxyinosine, respectively. The deficiency of ADA impairs the function of the human immune system resulting in severe combined immunodeficiency (SCID) characterized by severe T lymphocyte dysfunction and agammaglobulinemia.1-3 The clinical course of inherited ADA deficiency (ADA−) ranges from the rapidly fatal, early onset of classical ADA−SCID to the minimally dysfunctional immune system of patients presenting “partial” ADA deficiency.4,5 A recent review classified ADA deficiency into four types as determined by the age at clinical onset and suggested that these variants are the result of different, specific mutations resulting in various severities of enzyme dysfunction.6
Although the current treatment of choice for ADA−SCID is an HLA-matched bone marrow transplant,7 less than one third of patients have access to an appropriate donor. An alternative is enzyme replacement using polyethene glycol-modified bovine ADA (PEG-ADA). This represents a life saving, but costly, therapeutic option for the patients that do not have an HLA-matched donor.8 9 Although enzyme replacement with PEG-ADA partially reconstitutes the immune function of most patients with ADA−SCID, a few patients have been unresponsive to PEG-ADA.
The determination of the complete sequence of both the ADA cDNA10-12 and the genomic ADA structural gene13has facilitated the molecular analysis of ADA− patients and permitted identification of various genetic mutations in unrelated ADA− patients. Early identification of the mutant gene led ADA−SCID to become the first disorder to be treated by gene therapy. Two ADA−SCID patients who had manifested differing levels of severity of persistent immunodeficiency despite continuous treatment with PEG-ADA thus were enrolled in 1990.14 Since then, 10 patients with ADA−SCID have undergone gene therapy as recently described.14-17 The strategies adopted in these trials have differed and the efficacy of treatment has varied.
We report the molecular analysis of the genetic defect in an ADA−SCID patient enrolled in the first gene therapy protocol in Japan and analyze the clinical results obtained during the first 18 months of this clinical trial.
MATERIALS AND METHODS
Cell culture.
B-lymphoblastoid cell lines (B-LCL) were established from our ADA−SCID patient, his parents and a healthy volunteer by Epstein-Bar Virus (EBV) transformation. B-LCL were maintained in RPMI-1640 medium (GIBCO-BRL, Grand Island, NY) with 10% fetal calf serum (FCS; GIBCO-BRL) and 50 mmol/L β-mercaptoethanol (Sigma Chemical Co, St Louis, MO).
Sequence analysis of patient's ADA cDNA and genomic DNA.
For the analysis of the ADA cDNA sequence, total cellular RNA was isolated from B-LCL using TRIZOL Reagent (GIBCO-BRL). First-strand cDNA was synthesized from 2 μg of total cellular RNA (First strand synthesis kit; Promega, Madison, WI). Full-length ADA cDNA fragments extending from the translation start site codon to 230 base pair (bp) 3′ of the stop codon were amplified by reverse transcriptase-polymerase chain reaction (RT-PCR). Oligonucleotide primers for RT-PCR were as follows: sense primer; CCATGGCCCAGACGCCCGCCTT, antisense primer; ACCATAGCCCATGTGCAAGGGC. Reactions containing 0.5 μL (2.5 U)Taq polymerase (TaKaRa Ex Taq, TaKaRa Shuzo Co, Ltd, Tokyo, Japan) were incubated for 30 cycles of 60 seconds at 92°C, 90 seconds at 58°C, and 180 seconds at 72°C with the extension time at 72°C increased to 10 minutes in the last cycle. Amplified products were isolated from 1.0% agarose gel and then subcloned into pCR II vector (Invitrogen, San Diego, CA). Sequence analysis of double-stranded DNA was performed using Sequenase version II DNA sequencing kit (Amersham Life Science, Arlington Heights, IL) with [35S]dATP (Amersham Life Science) and a series of ADA-specific primers. Amplified products were sequenced through a 6% acrylamide gel (National Diagnostics, Atlanta, GA). To analyze the ADA genomic sequence, high molecular DNA was obtained from B-LCL by standard techniques.18 Primers and PCR conditions for amplification of ADA all exons have also been described previously.19-21 Amplified products were isolated from agarose gel and sequenced directly using the Thermal Cycler DNA sequencing kit (Circum Vent; New England Biolabs Inc, Beverly, MA). ADA cDNA sequences are numbered relative to the start site of translation and genomic DNA according to Wiginton et al.13
Southern blot analysis.
High molecular weight DNA from B-LCL was digested with restriction endonuclease Rsa I, separated in 1.0% agarose gel, and transferred onto a nylon membrane (Biotrace HP; Gelman Sciences, Ann Arbor, MI). Filters were then hybridized to a 32P randomly labeled 444-bp Rsa I-Pst I fragment from the ADA cDNA.
Retroviral-mediated gene transfer into patient's peripheral T cells.
The clinical protocol used here has been described elsewhere.22 Briefly, peripheral T lymphocytes from the patient were obtained by apheresis (CS3000 plus, Baxter Corp, Chicago, IL), isolated by density gradient centrifugation, and then maintained in AIM-V medium (GIBCO-BRL) supplemented with 5% FCS (GIBCO-BRL), 100 U/mL of recombinant human IL-2 (rIL-2, SHIONOGI, Osaka, Japan) and 10 ng/mL of anti-CD3 antibody (Orthoclone OKT3 Injection; Ortho, Raritan, NJ) in gas-permeable culture bags (Nipro Pretobag; Nishyo, Osaka, Japan). After 72 hours, half of the medium was removed and replaced with supernatant containing the LASN retroviral vector23supplemented with interleukin-2 (IL-2) and 10 μg/mL of protamine (Shimizu, Shimizu City, Japan). The LASN supernatant, prepared under Good Manufacturing Practices guidelines, was supplied by Genetic Therapy Inc (Gaithersburg, MD). The transduction procedure was repeated twice following an optimized transduction protocol combining low-temperature (32°C) incubation and centrifugation.24After two rounds of transduction, the virus supernatant was replaced with fresh medium supplemented with IL-2 and the cells were cultured for an additional 6 days. At the 11th day of culture, the cells were harvested and washed extensively with saline containing 0.5% human albumin and then reinfused into the patient.
Analysis of the inserted proviral genome by semi-quantitative PCR.
Sense (GAGGCTGTGAAGAGCGGCAT) and anti-sense (CTCGAAGTGCATGTTTTCCT) primers were designed to match the sequence of the start site of exon 7 and the end of exon 8, respectively. Using these primers, the amplification of DNA samples from vector-containing cells generates two bands; the larger one (250 bp) derived from the endogenous ADA gene containing intron 7 (76 bp) and the smaller one (174 bp) from the LASN provirus. To evaluate the frequency of transduced cells in the patient's peripheral blood, a standard curve was prepared from a serial dilution of in vitro-transduced and G418-selected B-LCL with untransduced cells. The ratio of the amount of amplified ADA cDNA derived from the integrated vector and the amplified genomic sequence was calculated after hybridization with an ADA cDNA probe.
Thin-layer chromatography (TLC) analysis of ADA enzyme activity.
Mononuclear cells were washed twice with phosphate-buffed saline to remove FCS and then suspended in 100 mmol/L Tris, pH 7.4 containing 1% bovine serum albumin. Cell lysates were obtained by 5 rapid freeze-thaw cycles. Cellular debris was removed by centrifugation and the lysates were stored at −80°C until used. ADA enzyme activity was assayed by the measurement of the conversion of [14C] adenosine (Amersham Life Science) to [14C] inosine and [14C] hypoxanthine followed by TLC separation of the reaction products performed as previously described.25 The results were expressed as nanomoles of inosine and hypoxanthine produced per min by 108 cells (nmol/min/108cells).
RESULTS
Clinical course.
The patient is a 5-year-old Japanese male. Symptoms including a chronic productive cough and a purulent nasal discharge began at 8 months of age. At 10 months he developed respiratory distress and was hospitalized for the treatment of severe pneumonia that was unresponsive to antibiotics. On admission at age 10 months, the patient had lymphopenia (absolute lymphocyte count 520/μL), with few mature T and B lymphocytes (CD3, 125/μL; CD4, 62/μL; CD8, 41/μL; CD19, 26/μL) and low serum Ig levels (IgG, 342 mg/dL; IgA, 18 mg/dL; and IgM, 60 mg/dL). Both humoral and cellular immunity were defective, with undetectable isohemagglutinins and absent T-cell proliferative responses to phytohemagglutinin, Concanavalin A, and pokeweed mitogen. Since ADA activity in his red blood cells (RBCs) was undetectable and the deoxyadenosine triphosphate (dATP) level was 506 nmol/mL RBCs (normal <2 nmol/mL), the diagnosis of SCID due to ADA deficiency of the “delayed onset” type6 was established. In the absence of a suitable bone marrow donor, PEG-ADA therapy was initiated at 15 months of age and supplemented with intravenous Ig (IVIG). After treatment with PEG-ADA (37.5 U/kg/wk), the plasma ADA activity in the patient's peripheral blood increased from 0.14 to 53.15 μmol/h/mL and the peripheral blood lymphocyte (PBL) count increased to the range of 1,000 to 2,000/μL. Despite continuous PEG-ADA treatment, however, his Ig levels remained below normal and the lymphopenia recurred during the second year of enzyme replacement. The PBL count decreased to less than 1,000/μL with CD3+ cell counts of 400/μL before the start of gene therapy (PBL, 702/μL; CD3, 400/μL; CD4, 205/μL; CD8, 191/μL; CD19, 57/μL on protocol day 0).
Identification of mutations responsible for ADA deficiency.
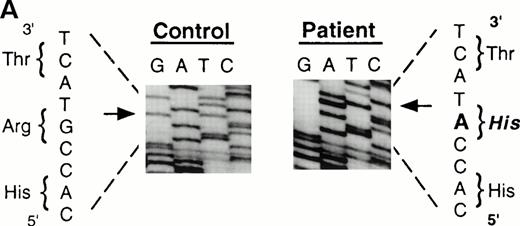
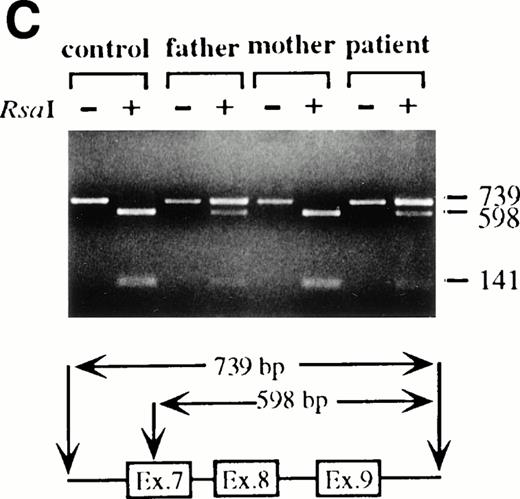
To analyze mutations in our patient, we amplified full-length ADA cDNA from the patient's EBV transformed B-LCL by RT-PCR. Sequence analysis revealed that all of the clones (6/6) carried a G632 to A transition resulting in replacement of the arginine residue by histidine at codon 211 (Fig1A). The mutation eliminates a recognition site for the restriction enzyme Rsa I. We took advantage of this feature to distinguish the mutated allele from the normal allele.19 High molecular weight DNA extracted from the patient's B-LCL was digested with Rsa I, blotted and hybridized to an ADA cDNA probe spanning the region from this mutation site in exon 7 to the end of exon 11 (Fig 1B). Rsa I digestion showed both a normal (3.1 kb) and a larger fragment (4.4 kb) in the patient lane, indicating that the patient was heterozygous for loss of the Rsa I recognition site in exon 7. To determine the parental derivation, amplified genomic fragments spanning intron 6 to intron 9 of the patient and his parents were digested with Rsa I and electrophoresed in 2% agarose gel (Fig 1C). The patient's digestion pattern was identical to that obtained from the analysis of the father's DNA, indicating that this mutation was derived from the paternal allele.
Characterization of the paternal missense mutation. (A) Sequence (sense) of amplified cDNA subclones from a control (left) and the patient (right). The position of the G632 → A transition is indicated by arrows next to the sequence ladder. (B) Identification of heterozygosity for the missense mutation. DNA samples from a control (lane 1) and the patient B-LCL (lane 2) were hybridized to a radiolabeled 444-bp Rsa I-Pst I cDNA probe extending from the mutation site to the end of exon 11 afterRsa I digestion. The normal Rsa I sites are at base pair 27,276 in exon 6, 28,516 in exon 7, and 31,671 in intron 11, predicting a 3.154-kb band hybridized to the probe in normal control, while loss of Rsa I site in exon 7 results in a larger band (4.394 kb). (C) Determination of the paternal mutation. Amplified genomic fragments (739 bp) from intron 6 (base pair 28377) to intron 9 (base pair 29115) were digested with Rsa I and electrophoresed in 2.0% agarose gel, and stained with ethidium bromide. The fragment has one Rsa I recognition site at base pair 28,517 in exon 7, predicting 141- and 598-bp fragments. Loss of the Rsa I site by the mutation results in an undigested fragment.
Characterization of the paternal missense mutation. (A) Sequence (sense) of amplified cDNA subclones from a control (left) and the patient (right). The position of the G632 → A transition is indicated by arrows next to the sequence ladder. (B) Identification of heterozygosity for the missense mutation. DNA samples from a control (lane 1) and the patient B-LCL (lane 2) were hybridized to a radiolabeled 444-bp Rsa I-Pst I cDNA probe extending from the mutation site to the end of exon 11 afterRsa I digestion. The normal Rsa I sites are at base pair 27,276 in exon 6, 28,516 in exon 7, and 31,671 in intron 11, predicting a 3.154-kb band hybridized to the probe in normal control, while loss of Rsa I site in exon 7 results in a larger band (4.394 kb). (C) Determination of the paternal mutation. Amplified genomic fragments (739 bp) from intron 6 (base pair 28377) to intron 9 (base pair 29115) were digested with Rsa I and electrophoresed in 2.0% agarose gel, and stained with ethidium bromide. The fragment has one Rsa I recognition site at base pair 28,517 in exon 7, predicting 141- and 598-bp fragments. Loss of the Rsa I site by the mutation results in an undigested fragment.
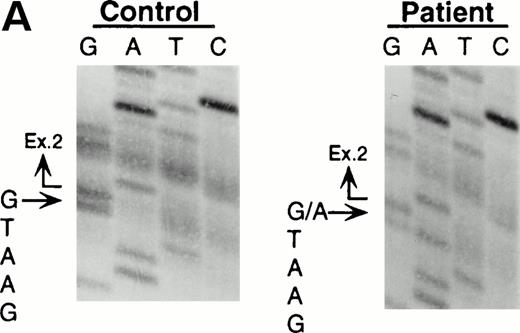
Northern blot analyses showed that the quantity of the ADA message from both the patient and his mother was reduced to approximately half of control (data not shown). All cDNA clones carried the paternal missense mutation, suggesting that the mutation derived from the maternal allele resulted in undetectable mRNA. To characterize this mutation, we analyzed exons 1 to 11 by PCR amplification of genomic DNA and direct sequencing. Sequence analyses of the amplified fragments including exon 2 showed the patient to be heteroallelic for a splice site mutation at the first position of intron 2 (G+1 → A transversion) (Fig 2A). This mutation eliminates a recognition site for the restriction enzyme BspMI.BspMI digestion showed that the patient and his mother were heterozygous for this mutation, while the father showed a normal individual digestion pattern (Fig 2B). Reports of mutation analyses of other patients have shown that a mutation affecting a mRNA splicing mechanism may give rise to a nonfunctional or unstable mRNA.26 27 This mechanism is also supported by the fact that Rsa I digestion showed that all full-length cDNA clones (48/48) from the patient's B-LCL carried the paternal G632to A missense mutation.
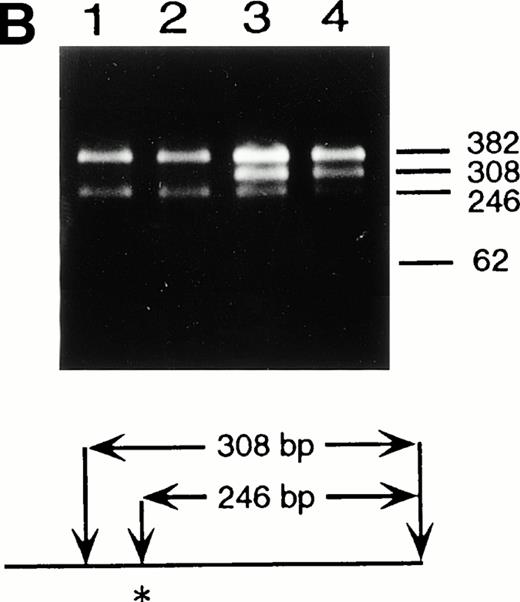
Identification of the maternal mutation at the splice donor site in intron 2. (A) Sequence (sense) of the exon 2/intron 2 junction in amplified genomic DNA. Genomic fragments containing exon 2 were amplified from a control (left) and the patient (right) and sequenced directly. A mutation at the splice donor site in intron 2 (G+1 → A) is indicated by arrows. (B) Detection of the splice site mutation by the BspMI digestion. Amplified genomic fragments (690 bp) from intron 1 (bp 14,901) to intron 2 (base pair 15,590) was digested with BspMI, electrophoresed in 2.0% agarose gel, and stained with ethidium bromide. The fragment has twoBspMI recognition sites at bp 15,282 and 15,344, predicting 62-, 246-, and 382-bp fragments in the control lane. Loss of theBspMI site (base pair 15,282) by the mutation results in the undigested fragment (308 bp). Lane 1, control; lane 2, father; lane 3, mother; and lane 4, patient. BspMI digestion shows the patient and his mother were heterozygous for the splice site mutation.
Identification of the maternal mutation at the splice donor site in intron 2. (A) Sequence (sense) of the exon 2/intron 2 junction in amplified genomic DNA. Genomic fragments containing exon 2 were amplified from a control (left) and the patient (right) and sequenced directly. A mutation at the splice donor site in intron 2 (G+1 → A) is indicated by arrows. (B) Detection of the splice site mutation by the BspMI digestion. Amplified genomic fragments (690 bp) from intron 1 (bp 14,901) to intron 2 (base pair 15,590) was digested with BspMI, electrophoresed in 2.0% agarose gel, and stained with ethidium bromide. The fragment has twoBspMI recognition sites at bp 15,282 and 15,344, predicting 62-, 246-, and 382-bp fragments in the control lane. Loss of theBspMI site (base pair 15,282) by the mutation results in the undigested fragment (308 bp). Lane 1, control; lane 2, father; lane 3, mother; and lane 4, patient. BspMI digestion shows the patient and his mother were heterozygous for the splice site mutation.
Retroviral mediated gene transfer into peripheral T cells.
At the age of 4, the patient was enrolled in a clinical gene therapy trial that repeated the protocol of the first gene therapy experiment at the National Institutes of Health (NIH) in 1990.22 The patient's peripheral mononuclear cells, obtained by apheresis, were stimulated with IL-2 (100 U/mL) and anti-CD3 antibody (OKT3; 10 ng/mL). After 72 hours of stimulation, they were transduced twice during the next 48 hours by exposure to the ADA retroviral vector LASN, expanded 20- to 50-fold in number by culturing for 6 days after the beginning of transduction, and then reinfused into the patient (see Materials and Methods). No selection procedure to enrich for gene-transduced cells was performed. Semiquantitative PCR of the cells in the first and second infusions revealed that the frequency of the vector-carrying cells ranged from 3% to 7% (data not shown).
Clinical course after gene therapy.
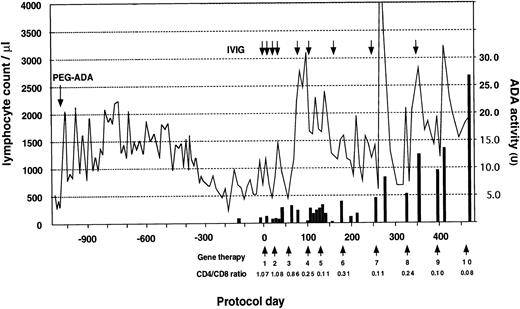
The patient received a total of 10 infusions over the 18-month period (Fig 3). A striking increase in lymphocyte number was observed early in the trial, followed by a gradual return to the basal level. This was followed by a sustained increase after the 8th infusion (protocol day 322) and the patient's PBL count has since remained in the normal range (PBL, 1,980/μL; CD3, 1,822/μL; CD4, 240/μL; CD8, 1,538/μL; CD19, 154/μL on protocol day 429). Progressive inversion of CD4/CD8 ratio has been observed since the 4th infusion due to an increase of the absolute CD8+ cell count. This phenomenon is thought to be the result of preferential proliferation of CD8+ cells during in vitro culture and transduction. ADA enzyme activity, nearly undetectable in the patient's lymphocytes before gene therapy, also increased progressively after the 7th infusion (protocol day 252) and reached 27 U on protocol day 476, which is approximately comparable to that of a heterozygous carrier individual (the patient's mother, 34.8 U).
Clinical course before and after gene therapy. Gene therapy started on August 1, 1995 (protocol day 0) with the patient receiving a total of 10 infusions to date. PEG-ADA therapy was initiated at 15 months of age. The lymphocyte count is indicated by a solid line and CD4/CD8 ratio was measured using PBL before infusion. ADA activity shown by a solid bar is expressed as nanomoles of inosine and hypoxanthine produced per minute by 108 cells. Replacement of IVIG after gene therapy is shown as an arrow. The patient received a Ig replacement (2.5 g) monthly before gene therapy.
Clinical course before and after gene therapy. Gene therapy started on August 1, 1995 (protocol day 0) with the patient receiving a total of 10 infusions to date. PEG-ADA therapy was initiated at 15 months of age. The lymphocyte count is indicated by a solid line and CD4/CD8 ratio was measured using PBL before infusion. ADA activity shown by a solid bar is expressed as nanomoles of inosine and hypoxanthine produced per minute by 108 cells. Replacement of IVIG after gene therapy is shown as an arrow. The patient received a Ig replacement (2.5 g) monthly before gene therapy.
The number of transduced cells in the patient's peripheral blood were assessed by semiquantitative PCR using PBL obtained before each infusion (Fig 4). The frequency of the genetically modified cells increased with the number of infusions of the ADA gene transduced lymphocytes and exceeded 10% of total circulating mononuclear cells just before the 5th infusion (on protocol day 126; Fig 4, lane 4). The frequency measured before each of the 6th through 10th infusions (on protocol days 210 to 462) has remained stable at 10% to 20%.
Semiquantitative PCR analysis to evaluate the frequency of vector-carrying cells in the patient's peripheral blood. Patient's mononuclear cells were obtained before the indicated infusion: before gene therapy (lane 1), 2nd infusion (protocol day [D] 21-lane 2), 4th infusion (D 98-lane 3), 5th infusion (D 126-lane 4), 6th infusion (D 175-lane 5), 8th infusion (D 322-lane 6, and 10th infusion (D 462-lane 7), and assayed for the frequency of vector containing cells by semiquantitative PCR. A standard was prepared by diluting cells containing the LASN vector with nontransduced cells. The ratio was determined by comparing the density of the cDNA derived band to that of the genomic DNA derived band.
Semiquantitative PCR analysis to evaluate the frequency of vector-carrying cells in the patient's peripheral blood. Patient's mononuclear cells were obtained before the indicated infusion: before gene therapy (lane 1), 2nd infusion (protocol day [D] 21-lane 2), 4th infusion (D 98-lane 3), 5th infusion (D 126-lane 4), 6th infusion (D 175-lane 5), 8th infusion (D 322-lane 6, and 10th infusion (D 462-lane 7), and assayed for the frequency of vector containing cells by semiquantitative PCR. A standard was prepared by diluting cells containing the LASN vector with nontransduced cells. The ratio was determined by comparing the density of the cDNA derived band to that of the genomic DNA derived band.
To evaluate the functional consequences of the ADA enzyme activity that had been induced by gene transfer, we compared the patient's immune function before and after the treatment (Table1). Eleven months after beginning gene therapy, the patient's isohemagglutinin titer (IgG) increased from undetectable to 1:16 and delayed-type hypersensitivity (DTH) skin test responses became stronger. The interval between IVIG infusions which were given monthly before gene therapy, was widened and eventually stopped after gene therapy. Despite this, the patient's serum Ig levels gradually increased and have remained normal for more than a half year without additional IVIG treatment (Fig3 and Table 1). These results suggest that the accumulated genetically corrected T lymphocytes in the patient's peripheral blood are associated with improvement of cellular and humoral immune responses and an increase in his circulating lymphocyte count. Although he sometimes became transiently febrile after infusions, the patient showed no serious adverse reactions to the treatments.
DISCUSSION
Advances in molecular biology during the past 3 decades have suggested that gene transfer could provide a new approach to the treatment of inherited diseases as well as acquired disorders such as cancer and acquired immune deficiency syndrome.28 The number of active gene therapy protocols has increased greatly since the first clinical gene therapy trial.29 ADA−SCID is one of the few early candidate disorders suitable for such interventions.30 Accordingly, 10 ADA−SCID patients have been enrolled in gene therapy clinical protocols that employed different strategies, retroviral vector designs, and target cell populations. The results obtained from these trials have recently been reported.14-17
This trial of gene therapy for an ADA−SCID patient in Japan began in August 1995. Over the next 18 months he received a total of 10 infusions of cultured-expanded autologous T cells that had been transduced with the LASN retroviral vector. After an initial period of fluctuating counts, the patient's T cells stabilized in the normal range and this has been sustained for the last half year. The frequency of integrated provirus in the patient's peripheral blood increased to approximately 15% (0.1 to 0.2 proviral copies/cell) by the 4th infusion and has remained stable since that time. The patient's cell associated adenosine deaminase enzyme activity has increased from barely detectable before treatment to values approaching those found in the peripheral mononuclear cells of his heterozygous carrier mother. Delayed hypersensitivity skin tests, a measure of T-cell function, have improved. Isohemagglutinin titers have also increased and his dependence on infusions of normal gammaglobulin has eased. The patient has gained 3 kg in weight during this trial. He is still receiving periodic PEG-ADA replacement and is attending public school with no more infections than his classmates.
The period of observation has simply not been sufficient to assess the full breadth or the duration of this improved clinical status and immune responsiveness. Further, additional studies will be required to reconcile the apparent dissociation between the level of T-cell ADA observed and the proportion of cells containing integrated vector at different time points. Also, the effect of withdrawal of the exogenous PEG-ADA treatment must await more complete characterization of the quality of the patient's immune system and the repertoire of specificities represented in the transduced T-cell population.
Four gene therapy clinical trials including 10 ADA−SCID patients have been performed since the first trial in 1990. Although these trials provided much data that suggested how future gene therapy might be improved by changing retroviral vector design, transduction methods and target cell populations, we found it difficult to compare the efficacy of these various trials because of differences inherent within these basic strategies. Our trial has been performed following the identical protocol and vector preparations and autologous T lymphocyte isolation procedures that were used in the NIH trial. From this perspective, our trial provides an additional opportunity to evaluate the effectiveness of peripheral T lymphocyte-directed gene therapy for ADA−SCID patients. Interestingly, the clinical course of our patient is quite similar to that observed in patient 1 in the NIH trial. Both trials have shown high gene transfer efficiency, remarkable increase of the ADA enzyme activity and eventual improvement of immune function. In contrast, patient 2 in the NIH trial experienced a low gene transfer efficiency and no significant increase in the ADA enzyme activity even though she exhibited some increase in immunological function. Although the factors leading to this difference have not yet been completely identified, a striking difference in the transduction efficiency of peripheral T cells between the three patients may be relevant. Transduction efficiencies before infusion were 3% to 7% for the present case, 1% to 10% for patient 1 and 0.1% to 1% for patient 2 in the NIH trial. An abbreviated proliferative capacity of patient 2 in the NIH trial was also observed. In addition, a contribution of the development of an immune response to the neomycin resistance gene must be considered since the existence of dominant selectable markers of nonhuman origin may result in unwanted immune reactivity that could eliminate or functionally impair transgene-expressing cells.31
The severity of the underlying ADA gene defects could also affect gene transfer. In addition to the mutation analysis reported here, specific ADA gene defects have also been reported for the two NIH patients.20 These three cases can be classified by the severity of their clinical presentation. Both the present case and patient 1 in the NIH trial are of the “delayed onset” type, have splice site mutation defects and have achieved significant levels of “gene-corrected” circulating cells. However, the NIH patient 2 carries compound missense mutations and has manifested low transduction efficiency despite her less severe “late onset” type of presentation at age 5. Although there are insufficient numbers of treated patients to draw firm conclusions at this point, it does appear thus far that the responses of patients with “more severe” gene defects and clinical presentations are at least as responsive as cases with “milder ADA defects.”
It should be noted that the ADA gene transduced T lymphocytes possess a selective advantage over the nontransduced cells due to the latter's high intracellular concentration of deoxyadenosine.32,33 In the ADA− newborn trial using gene-corrected CD34+ cells obtained from the patient's umbilical cord blood,16 LASN vector was detected in the peripheral blood T cells of these patients at a stable frequency of approximately 0.01% during the first 18 months of observation. Then, after a 50% reduction in their weekly dose of PEG-ADA, the proportion of ADA vector-containing T cells in the blood increased to approximately 10% in each case (D.B. Kohn, personal communication, September 1995). In the present case, the dosage schedule of PEG-ADA enzyme has remained constant since the beginning of the trial (18 U/kg/wk on the protocol day 431), during which time the patient's immune function has substantially improved. It might be expected that the proportion of the transduced cells in the patient's PBL will increase as the PEG-ADA dosage is decreased.
To date, three clinical trials have been performed to assess the possibility of treating ADA−SCID patients by correcting hematopoietic progenitor cells.15-17 The results obtained from these trials suggest that cord blood provides a stem cell population more suitable for efficient retroviral-mediated gene transfer than does bone marrow. Taken with the observations made in the NIH trial, our results strongly suggest that the effectiveness of T lymphocyte-directed gene transfer is a viable addition to the treatment programs that should be considered for ADA−SCID patients. After additional courses of treatment and continued observation to determine the breadth and durability of these positive responses, we hope to reduce or eliminate exogenous ADA enzyme supplementation in this patient. Improvements in vector design to permit higher levels of ADA expression and innovative strategies that provide greater efficiency of stem cell gene transduction may make gene therapy the treatment of choice for ADA−SCID patients.
ACKNOWLEDGMENT
We are grateful to Dr M.S. Hershfield for measuring some ADA activities and providing PEG-ADA, Dr A. Wakisaka for semiquantitative PCR, Drs L.M. Muul, C. Carter, and C. Wannebo for the transduction methods, Drs R.A. Knazek, F. Candotti, and D.M. Nelson for their critical review for the manuscript.
Supported by the grant for Scientific Research Expenses for Health and Welfare Programs (Funds of Highly Advances Medical Research), Tokyo, Japan.
Address reprint requests to Yukio Sakiyama, MD, PhD, Department of Pediatrics, Hokkaido University School of Medicine, North 15, West 7, Kita-ku, Sapporo, 060 Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


![Fig. 4. Semiquantitative PCR analysis to evaluate the frequency of vector-carrying cells in the patient's peripheral blood. Patient's mononuclear cells were obtained before the indicated infusion: before gene therapy (lane 1), 2nd infusion (protocol day [D] 21-lane 2), 4th infusion (D 98-lane 3), 5th infusion (D 126-lane 4), 6th infusion (D 175-lane 5), 8th infusion (D 322-lane 6, and 10th infusion (D 462-lane 7), and assayed for the frequency of vector containing cells by semiquantitative PCR. A standard was prepared by diluting cells containing the LASN vector with nontransduced cells. The ratio was determined by comparing the density of the cDNA derived band to that of the genomic DNA derived band.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/1/10.1182_blood.v91.1.30/3/m_blod4010304.jpeg?Expires=1768408493&Signature=y~HCzaj7KGAz9iy5OBUPxOhhIvuEenCicKzZDOPXZKHLbkyN1NoehLB2GzoIr0pidAPhGWUmu5P2PoQ08rAfJx4tKcOQ04Tw0Ss-zM5ydzWiTKDvgO2SDQPbJsLXby0sV4zzNQaKxTfI8YCfED-i1DR8gB3yWcIHGMgxpoNeZQqOqFiK1Su4L9lxoDqwKDODjJiVs0RagUTfcEXCrPW0TMz2fiPWQUSpIGf0q6iKUv2-ahOIWrzAtrMHbn2nFZ0wM9h5TQgQn-0K8tfr6gEIS7RyyDhrj5gci8s6FY~bbqH3UsNJ3oqV01vT3KWYVcWqcPu~AzVf5X4hpp1d9uAAHg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
