Abstract
Interferon consensus sequence binding protein (ICSBP) was first identified as a transcription factor of the interferon (IFN) regulatory factor family (IRF) which regulates expression of IFN-dependent genes by binding to DNA at specific sites, IFN-stimulated responsive elements. Analysis of ICSBP-deficient mice showed hematologic alterations similar to chronic myelogenous leukemia (CML) in humans and suggested a novel role for ICSBP in regulating proliferation and differentiation of hematopoietic progenitor cells. Here we show that ICSBP-mRNA expression is impaired in human myeloid leukemias: 27 of 34 CML patients (79%) and 21 of 32 patients with acute myeloid leukemia (AML) (66%) showed very low or absent transcript numbers of ICSBP. In contrast, only 2 of 33 normal volunteers (6%) showed low transcription of ICSBP(P < .0001 both for CML and AML values). The lack of expression was not associated with lack of lymphatic cells, which normally have been shown to express ICSBP at the highest level. More detailed analysis showed an absence of ICSBP-mRNA also in sorted B cells derived from CML patients. To analyze whetherICSBP may be induced in leukemic cells, ex vivoexperiments using a known inducer of ICSBP, IFN-γ, were performed. Ex vivo treatment of primary CML cells using IFN-γ resulted in induction of ICSBP transcripts. Furthermore, samples of CML patients during IFN-α treatment were analyzed. In 11 of 12 CML patients ICSBP-mRNA was inducible upon in vivo treatment with IFN-α, but decreased with progression of CML. Stable transfection of K-562 cell line with ICSBP led to no difference in bcr-abl expression in vitro, although two patients showed an inverse correlation between bcr-abl andICSBP in vivo. These data suggest that lack of ICSBPmay have an important role also in human myeloid leukemogenesis.
INTERFERONS (IFNs), divided into type I (IFN-α and -β) and type II (IFN-γ), are cytokines regulating antiviral activity, immune responses, and cell growth in mammals through IFN-regulated genes. The expression of IFN-inducible genes is regulated by IFN regulatory factors (IRFs), which bind to DNA sites containing IFN-stimulated responsive elements (ISRE).1-3IFN consensus sequence binding protein (ICSBP)4,5is one member of the growing family of the IRFs.6-10Although some are expressed in many different tissues, ICSBP is preferentially expressed in cells of hematopoietic origin. Specifically, highest ICSBP expression is detected in mature B cells, while resting T cells and mature macrophages harbor relatively low expression.11
It has been shown that mice with deleted IRF-1 andIRF-2 do not reveal any gross abnormalities.12 In human leukemias, deletion of IRF-1 has been reported,13 but it has been questioned whetherIRF-1 was indeed the target gene of the deletion of chromosome 5q, which is common in myeloid disorders.14 Furthermore, expression of IRF-1 and IRF-2 has been investigated in chronic myelogenous leukemia (CML) and no abnormality was detected.15 In contrast to these findings, mice with a null mutation of ICSBP exhibit two prominent features: (1) enhanced susceptibility to viral infections; and (2) granulocytic leukemia with enlargement of lymph nodes, liver and spleen, similar to CML in humans.16 Strikingly, there seemed to be a ‘dose-effect’ of ICSBP in that ICSBP−/− homozygous mice were more prone to blastic transformation of the respective myeloid proliferation compared to ICSBP+/−heterozygous mice.16 However, in contrast to CML in humans, where the rearrangement of the c-abl gene into thebcr-gene is observed in more than 90% of the patients,17,18 no gross genomic alteration of the c-abl gene was detected in the leukemic cells of theICSBP-null mice.16 These data pointed toICSBP as a potential tumor-suppressor gene and a role ofICSBP in leukemogenesis.
To address the question whether ICSBP may play a role also in human leukemic transformation, we investigated the transcriptional level in human leukemias and normal hematopoietic tissues. Therefore, we used a sensitive semi-quantitative polymerase chain reaction (PCR) assay.
MATERIALS AND METHODS
Patient samples.
Patient samples were taken from a single institution (Virchow Klinikum, HU Berlin, Germany). Ten to 20 mL heparinized peripheral blood (20 U/mL) were drawn after informed consent. All acute myeloid leukemias (AMLs) exhibited blast counts above 75%, and acute lymphoblastic leukemias had blast counts above 50%.
Cells and cell lines.
Cell lines U-937, Jurkat, and K-562 were obtained from the ATCC (American Type Culture Collection; Rockville, MD) and BV-173 from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany). Cell lines HL-60, Daudi and Raji were kindly provided by Dr F. Schriever (Virchow Klinikum) and cell lines MOLT-4, EHEB, and DHL4 were gifts from Dr I.G. Schmidt-Wolf (Virchow Klinikum). All cell lines were maintained at 5% CO2 in RPMI 1640 medium with 1% glutamine (GIBCO-BRL Eggenstein, Germany) supplemented with 10% fetal calf serum (FCS, GIBCO-BRL), 1% penicillin/streptomycin (Biochrom KG, Berlin, Germany). Mononuclear cells were separated from peripheral blood by centrifugation over a Ficoll (Biochrom KG) gradient and were stimulated with 1,000 U/mL IFN-α (Intron A; Essex Pharma GmbH, München, Germany) or IFN-γ (Boehringer Mannheim, Mannheim, Germany) for 6, 24, and 48 hours, respectively.
Separation of CD19+ B cells.
B cells were separated from peripheral blood of normal healthy volunteers and CML-patients using MACS CD19 MultiSort Kit (Miltenyi Biotec GmbH, Sunnyvale, CA) as recommended by the manufacturer. Purity of some B-cell fractions was verified to be approximately 90% using CD19-FITC antibodies (Dako Diagnostika GmbH, Hamburg, Germany) and FACScan analysis (Becton Dickinson, Heidelberg, Germany).
RNA isolation and cDNA synthesis.
RNA was extracted from heparinized peripheral blood using the RNAzol-kit (Paesel, Frankfurt, Germany) as recommended by the manufacturer. One microgram of total RNA was heat denaturated at 90°C for 5 minutes in the presence of 100 pmol random hexamers (Pharmacia, Freiburg, Germany) and cooled on ice for 2 minutes. The RNA was reverse transcribed in 20 μL final volume of 1X PCR buffer (Perkin Elmer, Weiterstadt, Germany), 625 nmol/L of each dNTP (Boehringer Mannheim), 10 mmol/L dithiothreitol (GIBCO-BRL), 40 U RNasin (Promega, Madison, WI), and 140 U SuperScript reverse transcriptase (GIBCO-BRL). The reaction mixture was incubated for 10 minutes at room temperature (25°C), 40 minutes at 42°C, and 5 minutes at 95°C.
ICSBP-mRNA expression analysis by PCR.
PCR was performed using 50 ng single-stranded cDNA in 25 μL 1X PCR buffer, 200 μmol/L dNTP, 500 nmol/L of each primer, and 0.625 U AmpliTaq DNA Polymerase (Perkin Elmer) under following cycling conditions: 94°C for 2 minutes for denaturation; then 94°C for 1 minute, 55°C (for β-actin) or 61°C (for ICSBP) for 1 minute, 72°C for 1 minute for 21 (for β-actin) or 27 cycles (for ICSBP), followed by 90°C for 1 minute and 60°C for 10 minutes. The sequences of the primers are as follows: β-actinsense primer, 5′-CCTTCCTGGGCATGGA GTCCT-3′; β-actin reverse primer, 5′-AATCTCATCTTGTTTTCTGCG-3′, which results in a 407-bp PCR-product;ICSBP sense primer, 5′-CAGTGGCTGATCGAGCAGATTGA-3′;ICSBP reverse primer, 5′-ATTCACGCAGCCAGCAGTTGCCA-3′, which results in a 360-bp PCR product. The products were electrophoresed on a 3% agarose gel. Gels were stained with ethidium bromide and photographed. The gel-photos were scanned and integrated optical densities (IntOD) were calculated using the ONE-Dscan 1.0 software (Scanalytics, Billerica, MA). The ratio ‘IntODICSBP/IntOD β-actin’ was then calculated. Analysis of normal control samples suggested a value of 0.200 as cut-off-level.
Other reference genes, porphobilinogendeaminase (pbgd) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), showed comparable results (data not shown).
Detection of the bcr-abl translocation and mRNA expression analysis by PCR.
The PCR for detection of the bcr-abl translocation was performed as described elsewhere.19 Quantitativebcr-abl-PCR was performed using a competitive differential quantitative PCR-assay which enabled us to determine the relative amplification-equivalence points (EP) of bcr-abl and a reference gene, pbgd, and to calculate the ratio ‘EP bcr-abl/EP pbgd.’20
Construction of ICSBP expression vectors.
An expression-plasmid for hICSBP was constructed by cloning a PCR-product of 1,372 bp into pCR3.1 (Invitrogen, NV Leek, The Netherlands). PCR was performed using 50 ng single-stranded cDNA in 25 μL 1X PCR buffer, 200 μmol/L dNTP, 500 nmol/L of each primer, and 0.625 U Expand Long PCR System (Boehringer Mannheim) under following cycling conditions: 94°C for 2 minutes for denaturation then 94°C for 1 minute, 62°C for 1 minute 68°C for 1 minute for 34 cycles, followed by 90°C for 1 minute and 60°C for 10 minutes. The sequences of the primers are as follows: sense 5′-GCGGCGAGACGGCGGCAGGA-3′; reverse 5′-GGCCACTGTAACAGGGAGATGGA-3′. The primers are based on the recently corrected sequence deposited at GenBank/EMBL Data Bank (accession no. M91196).
Transfection and cloning of stable transfectans.
K-562 cells (1.2 × 107) were transfected with 20 μg of control pCR3.1 (without insert) and pCR3.1 containing the coding sequence for hICSBP by electroporation with a Gene Pulser II (BioRad, München, Germany). Cells were selected with 0.5 mg/mL geneticin (G-418; GIBCO-BRL) for 2 to 3 weeks and then cloned by limiting dilution. Ten clones propagated from each transfection group were screened by reverse transcriptase (RT)-PCR for ICSBP-mRNA expression. Stable transfectans were maintained in culture medium with 0.2 mg/mL geneticin. Three different clones each were used for stimulation experiments with 1,000 U/mL IFN-α (Intron A; Essex Pharma) or IFN-γ (Boehringer Mannheim).
DNA sequencing.
ICSBP-PCR products and ICSBP expression vectors were verified by automated sequencing with the ABI Prism DNA Sequencer 377 (Perkin Elmer) using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer) as recommended by the manufacturer.
Statistical analysis.
Differences in the ICSBP expression of various leukemias were calculated by Fisher's exact test using Statistica 5.0 software (StatSoft, Tulsa, OK).
RESULTS
Determination of ICSBP-transcript levels in human samples.
We used a sensitive and semi-quantitative RT-PCR-approach to study transcription levels of ICSBP in human leukemias, as well as in normal hematopoietic tissues. Data obtained using RT-PCR were confirmed for several samples with the RNase protection assay (data not shown). Due to the restricted availability of RNA amounts from leukemia patients, RT-PCR was used for expression analysis in our further work.
To analyze the number of ICSBP transcripts, the relative expression of ICSBP was compared to a reference gene, β-actin. For both of these genes, PCR protocols were standardized such that the cycle number for each of these genes ensured that PCR-amplification was in its exponential phase. The optimal cycle number for ICSBP was determined to be 28 cycles, and 22 cycles for β-actin.
In each PCR, three positive controls were run as controls. The data obtained showed reproducible results (coefficient of variation of 16 experiments for three different controls: CV = 12.7%, 9.9%, and 8.1%, respectively).
We also performed control experiments with two other reference genes (pbgd and GAPDH) which showed comparable results with the data obtained with β-actin(data not shown). Therefore, all other experiments were performed using β-actin as reference gene.
In addition, we diluted mononuclear cells from a normal healthy volunteer with ICSBP-nonexpressing K-562 cells to determine the accuracy of the semi-quantitative RT-PCR. As expected, no linear correlation was found (Fig 1). However, the resulting near exponential curve fitted well for a semi-quantitative assay.
ICSBP-transcript numbers in a dilution series of mononuclear cells from a normal healthy volunteer with cells fromICSBP-nonexpressing cell line K-562. ICSBP-mRNA levels were detected by a semi-quantitative RT-PCR assay. Five different dilutions were done and the percentages of normal cells (ICSBP-expressing cells) were shown (0%, 10%, 50%, 90%, and 100%). The mean of three experiments is displayed (±SEM).
ICSBP-transcript numbers in a dilution series of mononuclear cells from a normal healthy volunteer with cells fromICSBP-nonexpressing cell line K-562. ICSBP-mRNA levels were detected by a semi-quantitative RT-PCR assay. Five different dilutions were done and the percentages of normal cells (ICSBP-expressing cells) were shown (0%, 10%, 50%, 90%, and 100%). The mean of three experiments is displayed (±SEM).
Furthermore, we tested different cell lines with known expression levels of ICSBP, which had been previously determined using standard Northern blotting techniques.5 The results of the RT-PCR approach were in keeping with these results: HighICSBP-transcript levels were detected in cell lines Daudi and U-937, whereas K-562, MOLT-4, and HL-60 exhibited no or lowICSBP transcripts (data not shown).
Expression patterns of ICSBP in human leukemias.
To study expression patterns in primary human leukemias, we first determined the level of ICSBP transcription in normal, healthy individuals. In 2 of 33 (6%) normal individuals, a relative expression below 0.2 (arbitrary value) was detected (Fig2 and Table 1).Because ICSBP−/− mice harbor a CML-like disease, we next investigated blood samples from CML patients in chronic phase, all of them bcr-abl-positive. Here, very low transcript numbers of the ICSBP gene were found in most patients (Fig 2 and Table 1). Twenty-seven of 34 (79%) CML patients showed no or significantly impaired ICSBP-transcript levels (P < .0001, Fisher's exact test).
ICSBP-transcript numbers in peripheral blood from normal healthy individuals and patients with different kinds of leukemias. (A) Lack of ICSBP-transcript numbers in CML-patients was detected by a semi-quantitative RT-PCR assay. Comparison of three healthy normal blood donors (NB; lanes 1 through 3) with four CML patients at diagnosis (CML; lanes 4 through 7) and three hydroxyurea-treated CML patients (CML, lanes 8 through 10). The expression level is displayed as integrated optical density (IntOD). (B) As (A), except that ICSBP-transcript numbers of 33 normal healthy blood donors, 34 CML, 32 AML, 11 CLL, 11 ALL, and 6 CMMoL patients were compared.
ICSBP-transcript numbers in peripheral blood from normal healthy individuals and patients with different kinds of leukemias. (A) Lack of ICSBP-transcript numbers in CML-patients was detected by a semi-quantitative RT-PCR assay. Comparison of three healthy normal blood donors (NB; lanes 1 through 3) with four CML patients at diagnosis (CML; lanes 4 through 7) and three hydroxyurea-treated CML patients (CML, lanes 8 through 10). The expression level is displayed as integrated optical density (IntOD). (B) As (A), except that ICSBP-transcript numbers of 33 normal healthy blood donors, 34 CML, 32 AML, 11 CLL, 11 ALL, and 6 CMMoL patients were compared.
ICSBP-Transcript Numbers in Peripheral Blood From Normal Healthy Individuals and Patients With Different Kinds of Leukemias
| Diagnosis . | Amount of Samples . | Mean (CI 95%)* . | % Samples With Impaired ICSBP Expression† . | Fisher's Exact Test . |
|---|---|---|---|---|
| NB | 33 | 0.406 (0.350-0.462) | 6 | — |
| CML‡ | 34 | 0.113 (0.054-0.171) | 79 | P < .0001 |
| AML§ | 33 | 0.225 (0.121-0.330) | 66 | P < .0001 |
| CLL | 11 | 0.725 (0.628-0.822) | 0 | P = .5581 |
| ALL∥ | 11 | 0.322 (0.176-0.468) | 36 | P = .0269 |
| CMMoL | 6 | 0.491 (0.100-0.881) | 17 | P = .4008 |
| Diagnosis . | Amount of Samples . | Mean (CI 95%)* . | % Samples With Impaired ICSBP Expression† . | Fisher's Exact Test . |
|---|---|---|---|---|
| NB | 33 | 0.406 (0.350-0.462) | 6 | — |
| CML‡ | 34 | 0.113 (0.054-0.171) | 79 | P < .0001 |
| AML§ | 33 | 0.225 (0.121-0.330) | 66 | P < .0001 |
| CLL | 11 | 0.725 (0.628-0.822) | 0 | P = .5581 |
| ALL∥ | 11 | 0.322 (0.176-0.468) | 36 | P = .0269 |
| CMMoL | 6 | 0.491 (0.100-0.881) | 17 | P = .4008 |
Abbreviations: NB, normal healthy blood donors; CI, confidence interval.
Relative ICSBP expression is displayed as arbitrary value, calculated as ratio of the integrated optical densities (IntOD) ofICSBP- to β-actin- transcript numbers.
The cut-off for normal ICSBP expression was fixed at the arbitrary value of 0.200.
All CML patients were untreated, treated with hydroxyurea, or exhibit relapses of CML after bone marrow transplantation.
All AML patients exhibited blast counts above 75%.
∥All ALL patients exhibited blast counts above 50%.
For further investigation of myeloid leukemias, samples from patients with AMLs, all bcr-abl-negative, were analyzed. Only samples with a blast count above 75% were investigated. In 21 of 32 (66%) of these AML samples, ICSBP-transcript levels were low or absent (P < .0001). Some AML patients harbored high to very high levels of ICSBP-mRNA. Interestingly, these were samples derived from patients with AML-M4 and M5, ie, monocytic differentiating leukemias. In keeping with this, most patients with chronic myelomonocytic leukemia (CMMoL), a disorder classified as myelodysplasia and characterized through monocytic differentiation, exhibited high ICSBP levels (Fig 2 and Table 1).
We also analyzed lymphatic neoplasias; none of 11 patients with chronic lymphocytic leukemia (CLL) and 4 of 11 (36%) patients with acute lymphoblastic leukemia (ALL) harbored low values ofICSBP-transcript numbers (Fig 2 and Table 1). Out of these 11 ALL patients, all with blast counts above 75%, 3 exhibited highICSBP values. These samples were drawn from patients who suffered from c-ALL, a B-cell–derived disorder. All of our samples from CLL patients, in approximately 95% also a disease of B cells, showed high levels of ICSBP transcripts (Fig 2). In keeping with this, all tested B-cell lines (DHL-4, Raji, BV-173, EHEB, Daudi) exhibited high, whereas T-cell lines (Jurkat, MOLT-4) showed low,ICSBP values (data not shown).5
ICSBP expression can be induced by IFNs both in vitro and in vivo.
ICSBP is inducible in vitro with IFN-γ.4 5 Given that ICSBP levels were impaired in CML samples, we first addressed the question whether ICSBPmay be inducible in vitro inp210bcr-abl-positive CML cells. Therefore, leukemic cells of a patient with bcr-abl-positive CML were obtained by centrifugation over a Ficoll gradient and incubated in vitrofor 6, 24, and 48 hours using different IFNs (Fig3). ICSBP-mRNA levels were low at the beginning of the in vitro incubation and increased 3- to 10-fold during incubation with IFN-γ, but not with IFN-α. These data suggested that ICSBP may be downregulated in myeloid leukemias, because in vitro cultivation with IFN-γ led to a significant increase of ICSBP message.
ICSBP-transcript numbers in mononuclear cells of a CML patient during in vitro stimulation with IFN-α and IFN-γ. An initial control was obtained (lane 1); samples were obtained after 6 hours (lanes 2, 5, and 8), after 24 hours (lanes 3, 6, and 9) and after 48 hours (lanes 4, 7, and 10). No increase ofICSBP-transcript numbers was seen without IFN (lanes 2 through 4) or with IFN-α treatment (1,000 U/mL; lanes 5 through 7). Only during IFN-γ treatment (1,000 U/mL) ICSBP-transcript numbers increased significantly (lanes 8 through 10). The mean of three experiments is displayed (±SEM).
ICSBP-transcript numbers in mononuclear cells of a CML patient during in vitro stimulation with IFN-α and IFN-γ. An initial control was obtained (lane 1); samples were obtained after 6 hours (lanes 2, 5, and 8), after 24 hours (lanes 3, 6, and 9) and after 48 hours (lanes 4, 7, and 10). No increase ofICSBP-transcript numbers was seen without IFN (lanes 2 through 4) or with IFN-α treatment (1,000 U/mL; lanes 5 through 7). Only during IFN-γ treatment (1,000 U/mL) ICSBP-transcript numbers increased significantly (lanes 8 through 10). The mean of three experiments is displayed (±SEM).
Since IFN-γ induced ICSBP in normal and malignant cells in vitro, we asked whether ICSBP may also be induced after in vivo treatment with IFN-α, where a survival benefit has been shown in large multicenter trials.21-23 Therefore, 12 patients were investigated during follow-up. Indeed, ICSBP-transcript numbers were significantly increased in 11 of them (91.7%) while patients were receiving IFN-α, but also decreased with progression of CML (Figs 4and 5, Table2).
ICSBP-transcript numbers, increasing upon in vivo treatment with IFN-α, correlated with impairment ofbcr-abl-transcript (•), leukocyte (□), and lymphocyte (▪) numbers. The sample at diagnosis exhibits few ICSBPtranscripts, even with only 1% blast cells (lane 1). Samples during IFN-α treatment showed an increase of ICSBP (lane 2: after 3 weeks, bone marrow; lane 3: 6 weeks; lane 4: 36 weeks), and a decrease of ICSBP after IFN-α was withdrawn (lane 5: 68 weeks). The mean of three experiments is displayed. All analyzed metaphases during the patient's course were 100% Philadelphia-chromosome positive. The relative equivalence points (EP) of bcr-abl and the reference gene, pbgd, were determined by quantitative PCR.
ICSBP-transcript numbers, increasing upon in vivo treatment with IFN-α, correlated with impairment ofbcr-abl-transcript (•), leukocyte (□), and lymphocyte (▪) numbers. The sample at diagnosis exhibits few ICSBPtranscripts, even with only 1% blast cells (lane 1). Samples during IFN-α treatment showed an increase of ICSBP (lane 2: after 3 weeks, bone marrow; lane 3: 6 weeks; lane 4: 36 weeks), and a decrease of ICSBP after IFN-α was withdrawn (lane 5: 68 weeks). The mean of three experiments is displayed. All analyzed metaphases during the patient's course were 100% Philadelphia-chromosome positive. The relative equivalence points (EP) of bcr-abl and the reference gene, pbgd, were determined by quantitative PCR.
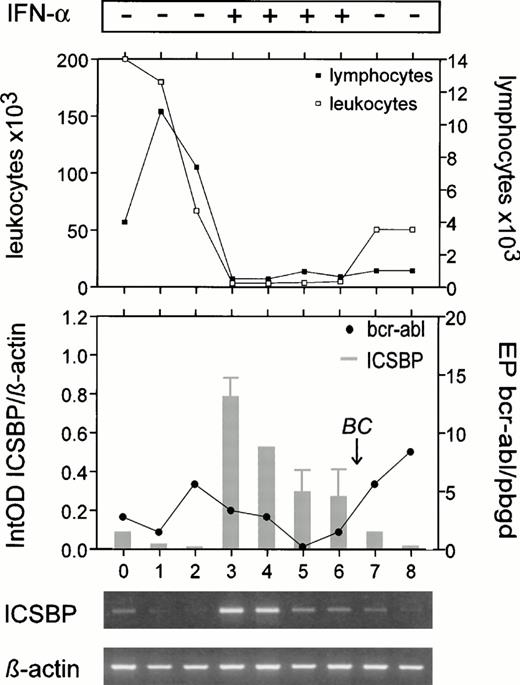
Increase of ICSBP-transcript numbers in a CML patient during in vivo IFN-α treatment, inversely correlated with bcr-abl-transcript (•), leukocyte (□), and lymphocyte (▪) numbers. At diagnosis (lane 0), and during treatment with hydroxyurea (lane 1: after 0.5 weeks; lane 2: 1 week) few ICSBPtranscripts were visible, although in this period the patient exhibited only 0% to 3% blast cells. During early IFN-α treatmentICSBP-transcript numbers increased (lane 3: 15 weeks; lane 4: 15 weeks), but decreased with progression to blast crisis (lane 5: 22 weeks; lane 6: 31 weeks; lane 7: 37 weeks, blast crisis with 60% blast cells; lane 8: 37 weeks, blast crisis, bone marrow). The mean of three experiments is displayed (±SEM). The patient exhibited an additional cytogenetic aberration, a monosomie 7, during blast crisis. BC, blast crisis.
Increase of ICSBP-transcript numbers in a CML patient during in vivo IFN-α treatment, inversely correlated with bcr-abl-transcript (•), leukocyte (□), and lymphocyte (▪) numbers. At diagnosis (lane 0), and during treatment with hydroxyurea (lane 1: after 0.5 weeks; lane 2: 1 week) few ICSBPtranscripts were visible, although in this period the patient exhibited only 0% to 3% blast cells. During early IFN-α treatmentICSBP-transcript numbers increased (lane 3: 15 weeks; lane 4: 15 weeks), but decreased with progression to blast crisis (lane 5: 22 weeks; lane 6: 31 weeks; lane 7: 37 weeks, blast crisis with 60% blast cells; lane 8: 37 weeks, blast crisis, bone marrow). The mean of three experiments is displayed (±SEM). The patient exhibited an additional cytogenetic aberration, a monosomie 7, during blast crisis. BC, blast crisis.
ICSBP-Transcript Numbers From CML Patients Treated With IFN-α During Follow-up
| Patient No. . | ICSBP-Transcript Levels . | ||
|---|---|---|---|
| At Diagnosis . | Early IFN-α Treatment . | Late IFN-α Treatment* . | |
| 1 | − | + | − |
| 2 | − | + | −/+ |
| 3 | − | + | ND |
| 4 | − | −/+ | −/+ |
| 5 | − | + | − |
| 6 | − | − | − |
| 7 | − | + | ND |
| 8 | − | + | −/+ |
| 9 | − | + | −/+ |
| 10 | − | + | − |
| 11 | − | −/+ | ND |
| 12 | − | + | − |
| Patient No. . | ICSBP-Transcript Levels . | ||
|---|---|---|---|
| At Diagnosis . | Early IFN-α Treatment . | Late IFN-α Treatment* . | |
| 1 | − | + | − |
| 2 | − | + | −/+ |
| 3 | − | + | ND |
| 4 | − | −/+ | −/+ |
| 5 | − | + | − |
| 6 | − | − | − |
| 7 | − | + | ND |
| 8 | − | + | −/+ |
| 9 | − | + | −/+ |
| 10 | − | + | − |
| 11 | − | −/+ | ND |
| 12 | − | + | − |
Abbreviations: ND, not determined; −, no/low ICSBPexpression; −/+, moderate ICSBP expression; +, highICSBP expression.
Progression of CML.
In particular, in one patient, ICSBP levels were low at the beginning (0% to 3% blast cells), increased during IFN-α therapy, and decreased during progression to blast crisis (60% blast cells) (Fig 5). These results suggest that ICSBP expression is low in chronic and blastic phase of CML; however, it may be induced in cells of chronic phase by IFN-α.
The detected changes in ICSBP-mRNA levels are not due to variations in the blood differential.
To rule out the possibility that an effect of the peripheral blood differential, especially lymphocyte numbers, accounted for the lack ofICSBP transcripts in samples from CML patients at diagnosis and under hydroxyurea, we compared blood counts from different CML patients (Table 3). We found that in three of six selected patients in chronic phase (Table 3, patients 2, 4, and 6), peripheral blood counts and percentage of lymphocytes were near normal. Still, ICSBP-transcript levels were significantly impaired or absent, arguing against the possibility that lack of ICSBPtranscripts in CML was a result of changes in the blood differential. In keeping with this notion was the comparison to CML patients under IFN-α: Three of five selected patients (Table 3, patients 7, 9, and 11) with high ICSBP-transcript numbers had comparable leukocyte and lymphocyte counts as other patients with low ICSBP levels and not receiving IFN-α.
Comparison of Blood Differentials and ICSBP-Transcript Numbers in Selected CML Patients Without or Under IFN-α Treatment
| Patient No. . | IFN-α Therapy . | Leukocytes (×103/μL) . | Lymphocytes (%) . | Blasts (%) . | ICSBP-mRNA Level* . |
|---|---|---|---|---|---|
| 1 | − | 137.0 | 0 | 15 | 0.000 |
| 2 | − | 2.7 | 13 | 0 | 0.071 |
| 3 | − | 16.6 | 11 | 1 | 0.150 |
| 4 | − | 4.6 | 10 | 0 | 0.010 |
| 5 | − | 453.0 | 2 | 4 | 0.000 |
| 6 | − | 7.8 | 17 | 1 | 0.170 |
| 7 | + | 3.4 | 15 | 0 | 0.718 |
| 8 | + | 2.3 | 49 | 0 | 0.510 |
| 9 | + | 8.8 | 8 | 0 | 0.710 |
| 10 | + | 5.7 | 18 | 0 | 0.416 |
| 11 | + | 6.8 | 15 | 0 | 0.569 |
| Patient No. . | IFN-α Therapy . | Leukocytes (×103/μL) . | Lymphocytes (%) . | Blasts (%) . | ICSBP-mRNA Level* . |
|---|---|---|---|---|---|
| 1 | − | 137.0 | 0 | 15 | 0.000 |
| 2 | − | 2.7 | 13 | 0 | 0.071 |
| 3 | − | 16.6 | 11 | 1 | 0.150 |
| 4 | − | 4.6 | 10 | 0 | 0.010 |
| 5 | − | 453.0 | 2 | 4 | 0.000 |
| 6 | − | 7.8 | 17 | 1 | 0.170 |
| 7 | + | 3.4 | 15 | 0 | 0.718 |
| 8 | + | 2.3 | 49 | 0 | 0.510 |
| 9 | + | 8.8 | 8 | 0 | 0.710 |
| 10 | + | 5.7 | 18 | 0 | 0.416 |
| 11 | + | 6.8 | 15 | 0 | 0.569 |
Abbreviations: −, no IFN-α treatment; +, under IFN-α therapy.
Relative ICSBP expression is displayed as arbitrary value, calculated as ratio of the integrated optical densities (IntOD) ofICSBP- to β-actin-transcript numbers.
In addition, we compared ICSBP-transcript numbers with the amount of leukocytes and lymphocytes in two patients (Figs 4 and 5) during follow-up. The number of lymphocytes decreased in both patients while the ICSBP message was upregulated. In one of these patients the relative amount of lymphocytes remained the same whileICSBP-transcript numbers decreased during a given period (lanes 3 through 6, Fig 5). The other patient never showed a cytogenetic response during his clinical course, ie, all metaphases analyzed were always 100% Philadelphia-positive (Fig 4), suggesting that an increase of ‘normal,’ Philadelphia-negative cells was not responsible for the observed increase of ICSBP transcripts in this patient.24
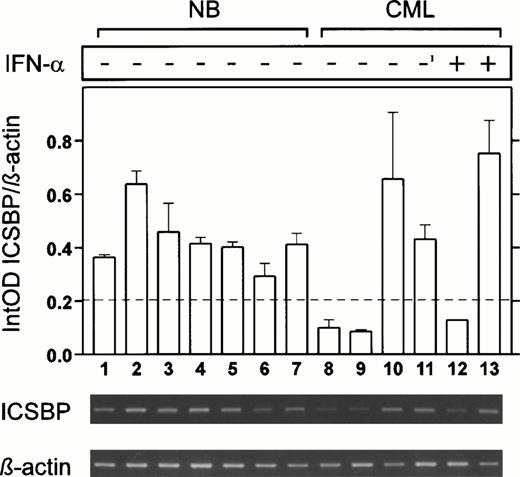
To investigate alteration of ICSBP expression in CML in more detail, we analyzed ICSBP-transcript numbers in CD19+ B cells, known to express ICSBP at the highest level (Fig 6). Seven samples from normal healthy individuals showed high ICSBP-mRNA levels whereas two of three samples from CML patients without IFN-α therapy exhibited low ICSBP-transcript numbers (Fig 6, lanes 1 through 10). Another sample, taken from a CML patient previously treated with IFN-α, had a high ICSBP level as did one of two samples from CML patients currently under IFN-α (Fig 6, lanes 11 through 13). These data correlated favorably to our results with unsorted peripheral blood cells.
ICSBP-transcript numbers in CD19+ B cells from normal healthy individuals and CML patients. High transcript levels were detected in cells from normal individuals (lanes 1 through 7), low levels in two of three CML patients without IFN-α (lanes 8 through 10) whereas another patient with high ICSBP levels had received IFN-α before (lane 11; IFN-α was withdrawn 6 months before sample was taken). One of two CML patients still under IFN-α exhibited low, the other high ICSBP levels (lanes 12 and 13). The mean of three experiments is displayed (±SEM).
ICSBP-transcript numbers in CD19+ B cells from normal healthy individuals and CML patients. High transcript levels were detected in cells from normal individuals (lanes 1 through 7), low levels in two of three CML patients without IFN-α (lanes 8 through 10) whereas another patient with high ICSBP levels had received IFN-α before (lane 11; IFN-α was withdrawn 6 months before sample was taken). One of two CML patients still under IFN-α exhibited low, the other high ICSBP levels (lanes 12 and 13). The mean of three experiments is displayed (±SEM).
All together these results made it unlikely that the observed lack ofICSBP transcripts and the upregulation during IFN-α therapy in CML patients was due to changes in the blood differential, and seemed mainly to be a phenomenon of aberrant ICSBP expression in B lymphocytes.
No regulating effect of ICSBP- on bcr-abl-transcript numbers was detected in vitro.
Since levels of bcr-abl transcripts have been correlated with progression of the disease, we asked if there may be a inverse association between bcr-abl and ICSBP expression. In two CML patients we compared ICSBP-expression patterns with the quantitative determination of the bcr-abl mRNA.20We found an inverse correlation between transcript levels ofICSBP and bcr-abl (Figs 4 and 5).
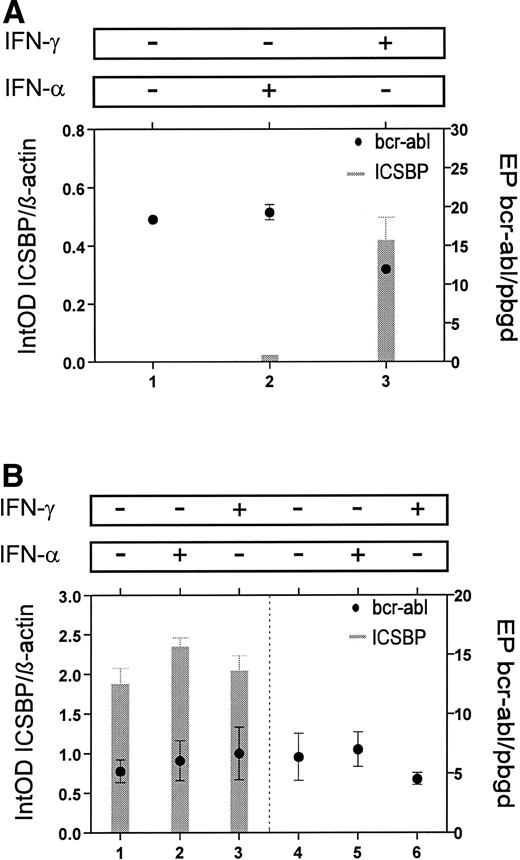
To evaluate the possible effect of ICSBP on bcr-ablexpression, we analyzed the relationship between ICSBP- andbcr-abl-transcript numbers in mononuclear cells from a patient with CML. Untreated cells and cells, which were treated in vitro with IFN-α or IFN-γ for 24 hours, showed no significant differences in bcr-abl expression, though an increase ofICSBP transcripts after incubation with IFN-γ was detected (Fig 7A). To analyze a possible interaction of ICSBP and p210bcr-abl in more detail, we stably transfected the bcr-abl-positive cell line K-562 with anICSBP-expression vector. The K-562 cells stably transfected with ICSBP were found to express ICSBP at high levels (Fig 7B). In these cells no change of bcr-abl-transcript numbers was detected compared to ICSBP-nonexpressing K-562 cells. Also, after incubation for 24 hours with IFN-α or IFN-γ, K-562 cells exhibited, as expected, nearly the same levels ofbcr-abl-transcripts (Fig 7B).
Comparison of ICSBP- andbcr-abl-transcript levels in vitro. (A) Mononuclear cells from a patient with CML were incubated for 24 hours with no cytokine (lane 1), IFN-α (lane 2), or IFN-γ (lane 3). In IFN-γ–treated cells ICSBP-mRNA levels increased, but no change was seen with bcr-abl-mRNA levels. (B) Stable transfectans of ICSBP-expressing (lanes 1 through 3) and nonexpressing K-562 cells (lanes 4 through 6) are compared. Incubation for 24 hours with no cytokine (lanes 1 and 4), IFN-α (lanes 2 and 5), or IFN-γ (lanes 3 and 6) showed no differences inbcr-abl-mRNA levels. Experiments were performed with three different transfected clones each and the mean is displayed (±SEM).
Comparison of ICSBP- andbcr-abl-transcript levels in vitro. (A) Mononuclear cells from a patient with CML were incubated for 24 hours with no cytokine (lane 1), IFN-α (lane 2), or IFN-γ (lane 3). In IFN-γ–treated cells ICSBP-mRNA levels increased, but no change was seen with bcr-abl-mRNA levels. (B) Stable transfectans of ICSBP-expressing (lanes 1 through 3) and nonexpressing K-562 cells (lanes 4 through 6) are compared. Incubation for 24 hours with no cytokine (lanes 1 and 4), IFN-α (lanes 2 and 5), or IFN-γ (lanes 3 and 6) showed no differences inbcr-abl-mRNA levels. Experiments were performed with three different transfected clones each and the mean is displayed (±SEM).
Although an inverse correlation was seen in vivo, our results suggest no direct regulating effect of ICSBP on bcr-ablexpression, but an effect of bcr-abl on ICSBPexpression could not be ruled out and has to be further investigated.
DISCUSSION
Recently, ICSBP−/− mice have been generated. These mice showed no difference in size, behavior, and reproductive ability as compared with normal littermates. However, in 100% of these mice, a hematologic neoplasia resembling CML in humans was observed.16 The most prominent features of these mice were hepatosplenomegaly and enlargement of lymph nodes with infiltrations of mature granulocytes, metamyelocytes, and bands. Blood films of these mice showed leukocytosis with enlargement of immature cells and, in keeping with CML in humans, transformation to blastic phase was observed. Interestingly, there seemed to be a ‘dose-response’ in that homozygous mice were more prone to develop blast crisis as compared with heterozygous mice.16 These data prompted us to investigate the role ICSBP may play in human leukemia.
We used a semi-quantitative RT-PCR approach to determineICSBP-transcript numbers in different kinds of leukemia samples, because no monoclonal antibody was available to study humanICSBP-protein expression. The data presented here showed thatICSBP transcripts are absent or significantly lower expressed in peripheral blood cells in the vast majority of patients with myeloid leukemias. In addition, in some patients with acute lymphoblastic leukemia, mostly T-ALL, ICSBP transcripts were also found at lower levels, but not in B-cell–derived disorders like c-ALL or CLL. These data raise the possibility that loss of ICSBP expression may play a significant role in human myeloid leukemogenesis. We do not believe that the lack of ICSBP transcripts was simply due to expansion of more immature progenitors in the investigated leukemias, because in some of the investigated CML samples a nearly normal blood differential due to treatment with hydroxyurea was observed, yetICSBP levels were still absent, in contrast to samples from CML patients under IFN-α, which had comparable blood counts but highICSBP-transcript numbers. In keeping with this, analysis of CD19+ B cells, normally highly expressing ICSBP,showed absence of ICSBP transcripts in CML samples.
Furthermore, ICSBP was inducible in the leukemic cells, bothin vitro and in vivo. These data suggest thatICSBP is not genomically lost but rather downregulated in myeloid leukemias, the mechanism of which remains to be determined. Downregulation leading to gene-‘silencing’ has been described in human cancer cells for MTS-1, a gene coding for an inhibitor of cyclin-dependent kinases (p16INK-4/CDKN2).25 26
Although patients with CML may respond beneficially to treatment with IFNs, treatment with IFN-γ, in contrast to IFN-α, remains anecdotal.27-30 In our clinic CML patients were only treated with IFN-α, so we were unable to assess the effect of IFN-γ, a known inducer of ICSBP,4 5 onICSBP expression in vivo. In several clinical samples, although many CML patients showed low levels of ICSBPtranscripts at diagnosis, IFN-α therapy led to an increase inICSBP message. These high levels were also detected in some CD19+ B cells from CML patients under or after IFN-α treatment. The induction of ICSBP by IFN-α has not been described in vitro, in keeping with data obtained in this study. One explanation for the missing induction of ICSBPduring IFN-α incubation in vitro may be that the increase ofICSBP transcripts in CML patients during IFN-α therapy cannot be found within a short period of time, and may take at least 1 to 3 months. Further, the observed upregulation of ICSBP during IFN-α treatment in vivo may also be an indirect effect mediated by other factors or cells not present in the in vitroincubation.
When the patients progressed to accelerated/blastic phase,ICSBP-mRNA levels decreased significantly, even during treatment with IFN-α. Thus, this raises the possibility that low or absent levels of ICSBP transcripts may be associated with progression of myeloid leukemias, and upregulation may be of some unknown benefit. The importance of some kind of ICSBP-‘dose-response’ for myeloid cell differentiation may also be supported through data from ICSBP knock-out mice, in that mice with a complete loss of ICSBP progressed to blastic phase in 33% within 50 weeks of observation, whereas heterozygous mice showed acute leukemia in only 9%.16
The data of this study make it unlikely that loss of ICSBP is necessary for the development of all myeloid leukemias. However, the majority of myeloid leukemias displayed decreased or absent expression of ICSBP transcripts. The mechanism by which loss ofICSBP may induce leukemias is unclear at present. IFNs play an important role in the negative regulation of human hematopoiesis. In keeping with this, the beneficial role of these cytokines in human myeloproliferative diseases has been shown in recent years. There may be direct repressing effects of ICSBP on downstream, positive effector genes. Data obtained in two patients of this study showed that upregulation of ICSBP-mRNA was inversely correlated with downregulation of bcr-abl transcripts. However, data from theICSBP-transfected K-562 cells make a direct effect onbcr-abl unlikely. Thus, the possible interaction betweenICSBP and transforming genes need to be determined, especially a possible effect of bcr-abl on ICSBP expression. In addition, the ‘homing’ of myeloid cells could be negatively influenced by the lack of ICSBP, ie, through the loss of expression of certain adhesion molecules. Interestingly, IFN-α therapy may induce adhesion molecules, and this, at least in part, may explain its beneficial effect in human CML.31 On the other hand, IFN-α induces negative cytokines in the human bone marrow stromal cells, and this has been implicated in the therapeutic efficacy of these drugs in treating CML patients.32 33
In summary, ICSBP loss induces myeloid leukemias in mice, and this report describes that ICSBP-mRNA is very low or absent in the majority of human myeloid leukemias. Together these data raise the possibility that ICSBP may be a tumor-suppressor, but the exact role ICSBP plays in hematopoiesis remains to be determined.
ACKNOWLEDGMENT
We thank C. Fieger and S. König for help with electroporation procedures, Dr C. Brendel for FACScan assistance, Dr C. Busemann for providing patient data, J. Laser for technical assistance, and Dr K. Seeger for GAPDH primers. We are grateful to Dr F. Schriever and Dr I.G. Schmidt-Wolf for providing cell lines, and to Prof Dr H. Kleinkauf for helpful discussions.
Supported by grants from the ‘Deutsche Forschungsgemeinschaft’ (Ne310/6-3 to A.N. and SFB 465 to I.H.).
Address reprint requests to Andreas Neubauer, MD, Medizinische Klinik I, Universitätsklinik Carl Gustav Carus, Fetscherstraße 74, 01307 Dresden, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 17.34 solely to indicate this fact.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal