Abstract
BCR-ABLp190 oncogene is the result of a reciprocal translocation between chromosomes 9 and 22 and is associated with B-cell acute lymphoblastic leukemia (B-ALL) in humans. Current models expressing the BCR-ABLp190 chimeric gene fail to consistently reproduce the phenotype with which the fusion gene is associated in human pathology, mainly due to the difficulty of being expressed in the appropriate cell type in vivo. We have used here homologous recombination in ES cells to create an in-frame fusion of BCR-ABLp190 that mimics the consequences of the human chromosomal translocation by fusion of BCR-ABL coding sequences into the bcr endogenous gene. The chimeric mice generated with the mutant embryonic stem cells systematically develop B-ALL. Using these chimeric mice, we further show that BCR-ABL oncogene does not require the endogenous bcr product in leukemogenesis. Our results show that BCR-ABLp190 chimeric mice are a new model to study the biology of the BCR-ABL oncogene and indicate the efficacy of this strategy for studying the role of specific chromosome abnormalities in tumor development.
THE HALLMARK OF tumors in general and leukemias in particular is the presence of specific chromosome abnormalities considered to be involved in tumor development. A well-characterized example in the hematopoietic system involves the rearrangements of the BCR and ABL genes in Philadelphia chromosome-positive (Ph) chronic myelogenous leukemia (CML) and acute lymphocytic leukemia (ALL).1-13 It represents a reciprocal translocation between chromosomes 9 and 22, t(9; 22)(q34; q11),14 in which the c-ABL proto-oncogene on chromosome 9 is translocated to the chromosome 22.15 In the resulting BCR-ABL chimeric gene, the N-terminal segment of the c-ABL oncogene is replaced by a BCR-derived sequence,13,16 as observed in the case of v-abl, in which the normal N-terminus of c-ABL is replaced by the viral gag-derived sequence.17 Depending on the precise breakpoint within the BCR gene, fusion proteins of 210 kD (BCR-ABLp210 ) or 190 kD (BCR-ABLp190 ) are produced.1,9,11,12,18-20BCR-ABLp210 and BCR-ABLp190 oncogenes contain identical ABL-derived sequences, respectively, but differ in the number of BCR-encoded amino acid residues.10,19,21 The larger chimeric product, BCR-ABLp210, contains either 902 or 927 BCR-derived amino acid residues and is observed in almost all cases of CML,13,15,16 whereas the smaller form, BCR-ABLp190, includes only the BCR exon 1-encoded sequences fused to the same portion of c-ABL and is mainly associated with B-cell ALL.1,9,17,22,23 Both chimeric proteins possess an enhanced tyrosine kinase activity in comparison with the normal c-ABL product and are thus considered to be implicated in the pathogenesis of Ph-positive human leukemias.11
To better understand the pathogenic processes caused by the BCR-ABL chimeric protein in vivo, it seems to be necessary to establish animal models to investigate the function of BCR-ABL chimeric gene products in vivo. Several different approaches have been used to create the animal models, including virus-mediated gene transfer, the use of transgenic mice, and direct viral injection.3,5-8,10,24-27 These models expressing the BCR-ABL chimeric gene product do not show consistently the same phenotype with which these oncogenes are associated in human pathology, due principally to the difficulty of choosing a promoter to engineer expression in the appropriate cell type in vivo. This problem difficults the study of the biologic function of BCR-ABL chimeric gene product in vivo. Homologous recombination provides an alternative strategy to mimic the consequences of chromosomal translocations by fusion of coding sequences into an endogenous gene.28
In the present study, we have used homologous recombination in embryonic stem (ES) cells to create an in-frame fusion of BCR-ABLp190 oncogene with exon 1 of mouse bcr, resembling the rearrangement that takes place into human BCR-ABLp190–positive cells in which the leukemic cells of patients with Ph-positive leukemia only have one intact BCR allele; the other is rearranged as a result of fusion to ABL. The chimeric mice generated by introducing the mutant ES cells into blastocysts develop consistently B-cell ALL, being an ideal model to study the role of BCR-ABLp190 oncogene in leukemogenesis. Moreover, we have used this strategy to study the contribution of the intact bcr allele to BCR-ABLp190 leukemogenesis, because the presence of a functional bcr product has been proposed as a requirement for malignant transformation by BCR-ABL.29-31 Our results show that BCR-ABLp190 oncogene does not require the endogenous bcr product and indicate the efficacy of this strategy for studying the role of specific chromosome abnormalities in tumor development.
MATERIALS AND METHODS
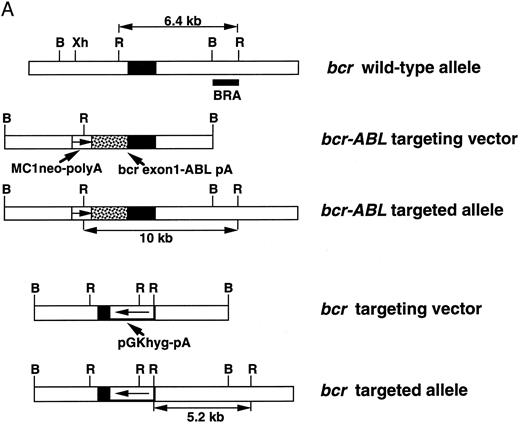
Construction of the targeting vector and generation of chimeric mice. The mouse bcr locus was cloned from a partial Sau3A genomic library made in the λ2001. The genomic fragment containing exon 1 was subcloned in pBluescript. The bcr-ABL targeting vector was constructed by replacing the bcr exon 1 coding sequence by the BCR-ABLp190 cDNA. The MC1-neo-poly(A) cassette was cloned immediately downstream of the bcr-ABL fusion segment. The reading frame of the fusion junction between bcr and BCR-ABL sequences was confirmed by nucleotide sequencing. The bcr targeting vector was made by replacing a 300-bp Eag I-Eag I fragment, spanning part of bcr exon 1, with a 1.7-kb fragment that contained a PGK-hyg-poly(A) cassette. Twenty-five micrograms of linearized vector DNA was used in the transfections of the E14 ES cell line that was maintained as described with leukemic inhibitory factor (LIF) supplementation medium. Selection was commenced 36 hours later, using G418 (GIBCO, Grand Island, NY) at 400 μg/mL or hygromycin B (Calbiochem, La Jolla, CA) at 300 μg/mL. Cells were electroporated at 400 V, 25 μF. Homologous recombination was assessed by filter hybridization using the specific BamHI-EcoRI fragment derived from bcr wild-type allele (Fig 1A). The presence of a single insertion site was confirmed with specific neomycin and hygromycin probes, respectively. Karyotypically normal targeted ES clones were used for the generation of chimeras as essentially described.32 Two independent bcr-ABL/bcr+ and bcr-ABL/bcr− ES clones were plated on feeders 2 days before the injection in Dulbecco's modified Eagle's medium (DMEM) plus 20% fetal calf serum (FCS), 1× nonessential amino acids (GIBCO BRL, Gaithersburg, MD), 0.1 mmol/L β-mercaptoethanol, L-glutamine, and LIF. The ES cells are derived from the 129Sv strain (agouti coat color). Blastocysts were isolated at 3.5 days postcoitum (dpc) from hormone-stimulated females (C57BL strain, with black coat color). Approximately 15 ES cells were injected into each blastocyst and the blastocysts were transferred into the uteri of pseudopregnant recipients 2.5 dpc. When the litters were born, the chimerism was estimated by coat color. As controls, E14 wild-type ES cells and heterozygous bcr +/− ES cells were used. the results presented here were obtained with targeted ES E14 clones, but similar data were observed with targeted ES cells made in the CJ7 line.
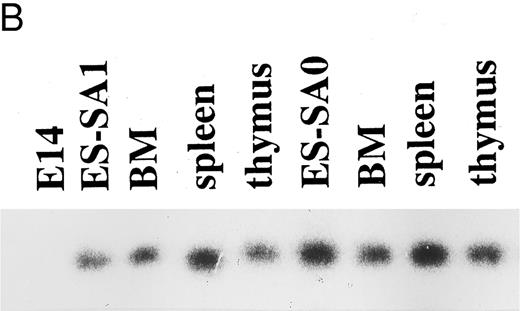
(A) bcr-targeting constructs. The structure of the bcr gene in the proximity of exon 1, the bcr-ABL targeting replacement vector and the predicted structure of the targeted bcr locus after homologous recombination, the bcr targeting vector, and the predicted structure of the bcr targeted allele. Only the relevant restriction sites are shown. B, BamHI; Xh, Xho I; R, EcoRI. Horizontal single arrows indicate the direction of transcription of the MC1-neo-pA and PGK-hyg-pA cassettes. Expected lengths of restriction fragments diagnostic for homologous recombination are indicated by double-headed arrows. The probe used to detect the homologous recombination events (BRA) in Southern filter hybridizations is indicated. (B) Expression of the bcr-ABLp190 fusion gene in ES cells and in chimeric mouse tissues. mRNA was obtained from ES clones with bcr-ABL targeted alleles and from different chimeric mouse tissues. Reverse transcriptase was performed on cDNA made from each RNA preparation using primers specific for BCR-ABL cDNA. Products of the PCR reactions were separated on 1.2% agarose and hybridized with a specific internal oligo as a probe (5′-CCTTCAGCGGCCAGTAGCATCTGACTT-3′). ES-SA1 is a bcr-ABL/bcr+ ES clone and ES-SA0 is a bcr-ABL/bcr− ES clone.
(A) bcr-targeting constructs. The structure of the bcr gene in the proximity of exon 1, the bcr-ABL targeting replacement vector and the predicted structure of the targeted bcr locus after homologous recombination, the bcr targeting vector, and the predicted structure of the bcr targeted allele. Only the relevant restriction sites are shown. B, BamHI; Xh, Xho I; R, EcoRI. Horizontal single arrows indicate the direction of transcription of the MC1-neo-pA and PGK-hyg-pA cassettes. Expected lengths of restriction fragments diagnostic for homologous recombination are indicated by double-headed arrows. The probe used to detect the homologous recombination events (BRA) in Southern filter hybridizations is indicated. (B) Expression of the bcr-ABLp190 fusion gene in ES cells and in chimeric mouse tissues. mRNA was obtained from ES clones with bcr-ABL targeted alleles and from different chimeric mouse tissues. Reverse transcriptase was performed on cDNA made from each RNA preparation using primers specific for BCR-ABL cDNA. Products of the PCR reactions were separated on 1.2% agarose and hybridized with a specific internal oligo as a probe (5′-CCTTCAGCGGCCAGTAGCATCTGACTT-3′). ES-SA1 is a bcr-ABL/bcr+ ES clone and ES-SA0 is a bcr-ABL/bcr− ES clone.
Phenotype analysis. The following phycoerythrin (PE)-conjugated antimouse monoclonal antibodies from Pharmingen (San Diego, CA) were used for cytometry staining: CD45R/B220, Thy-1.1 and Thy-1.2, myeloid markers (Mac 1/CD11b and Gr-1), and SIg. Single-cell suspensions from whole blood samples obtained by routine techniques were incubated with purified antimouse CD32/CD16 (Pharmingen) to block binding via Fc receptors and with an appropriate dilution of the different antibodies at room temperature or 4°C, respectively. Red blood cells were lysed using the lysis solution (Becton Dickinson, Mountain View, CA). The samples were washed twice with phosphate-buffered saline (PBS) and resuspended in PBS. Dead cells in samples were excluded by propidium iodide staining. The samples and the data were analyzed in a FACScan using CellQuest software (Becton Dickinson).
Polymerase chain reaction (PCR) analysis. mRNA was obtained from ES clones with bcr-ABL targeted alleles and from different chimeric mouse tissues. Reverse transcriptase was performed on cDNA made from each RNA preparation treated with RNase free DNase I (HT). The cDNA was subjected to PCR using BCR and ABL specific primers. Reactions were run as described by the manufacturer of Taq polymerase (Perkin-Elmer Cetus, Norwalk, CT) under the following conditions: 1 minute at 95°C, 1 minute at 55°C, and 1 minute at 72°C for 25 cycles, with a final extension of 10 minutes at 72°C.
Histologic analysis. Tissue specimens were fixed in 4% formaldehyde in PBS and embedded in paraffin. Blocks were cut, and slides were processed and stained with hematoxylin-eosin by routine techniques. Slides were examined and photographed.
Western blot analysis. Single-cell suspensions coming from spleen were analyzed by immunoblotting procedures as essentially as described.33 The bcr (AB-2) antihuman BCR monoclonal antibody (Oncogene Science, Uniondale, NY) is directed against an epitope in exon 8.
Ig gene rearrangement. DNA was prepared from various tissues using standard procedures. The DNA was digested with BamHI, and Southern blots were probed with a specific Ig probe.
Cell transfers. Cells from organs of donor mice were suspended, washed, and injected intravenously into tails of 4- to 6-month-old recipient mice (NOD/SCID). Mice were monitored once weekly and were killed for histopathologic studies and collection of tissues for DNA analyses when moribund.
RESULTS AND DISCUSSION
The bcr-ABLp190 oncogene made by homologous recombination produces acute leukemias in chimeric mice.BCR-ABLp190 oncogene was created via homologous recombination by inserting the BCR-ABL cDNA into exon 1 of the mouse bcr locus (Fig 1A). The BCR-ABL cDNA contained a polyA signal sequence downstream of the ABL translation stop codon. The vector also carried the MC1-neomycin resistance gene (MC1-neo-polyA) downstream of the fusion gene to confer resistance to G418 (bcr-ABL targeting construct; Fig 1A). The construct was electroporated into ES E14 cells and correct targeting was obtained. The correctly modified ES cells contained one intact bcr allele and the other is rearranged as a result of fusion to BCR-ABL, recapitulating the rearrangement that takes place into human BCR-ABLp190 leukemic cells (bcr-ABL/bcr+ ES cells). These correctly modified ES cell clones were injected into recipient C57BL/6 blastocysts and chimeric mice produced (bcr-ABL/bcr+). Two independent bcr-ABL/bcr+ ES cell clones with normal karyotypes were injected into C57BL/6 blastocysts.
Introduction of the bcr-ABL targeting vector into E14 ES cells allowed selection in which an bcr-ABL fusion had occurred by homologous recombination. Transcription of the fusion gene occurs from the endogenous bcr promoter. The expression of the bcr-ABL fusion gene was assessed in ES cells and chimeric mouse tissues. RNA obtained from ES cell clones with the targeted allele (bcr-ABL/bcr+) and from the chimeric tissues was used for reverse transcription PCR (Fig 1B). Specific primers corresponding to regions of BCR and ABL sequences yielded PCR products in the ES cell clones with the specific targeted allele and in the tissues of the chimeras, showing that the chimeric bcr-ABLp190 fusion gene is expressed from the targeted allele in bcr-ABL/bcr+ ES cells and chimeric tissues. Normal mouse bone marrow, spleen, and thymus cells treated in parallel did not exhibit any expression. Thus, targeted ES cell derivatives are present in the three major hematologic organs (bone marrow, spleen, and thymus), showing, as it is known,31 that the bcr gene is expressed in many different types of tissues in the mouse.
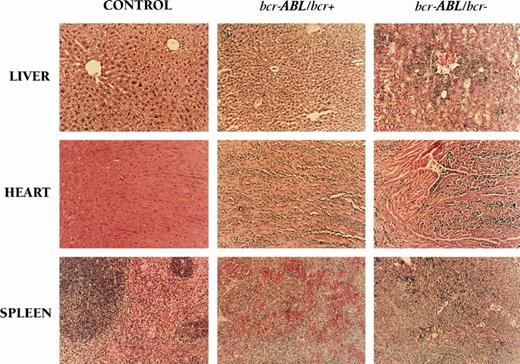
Cohorts of chimeric mice were produced by injection of the modified targeted clone (bcr-ABL/bcr+) into blastocysts to analyze the effect of the bcr-ABLp190 fusion gene. We generated 40 bcr-ABL/bcr+ chimeric mice that gave clear evidence of serious distress from 4 months of age onward. The clinical condition of the chimeric mice deteriorated rapidly and developed into acute leukemia (>30% of the nucleated bone marrow cells were blasts), which was clinically characterized by decreased spontaneous movements in the cage, increased respiratory rates, piloerection, and shivering, and the animals displayed a sustained loss of body weight. This condition persisted and the animals were killed. Macroscopic analysis showed that bcr-ABL/bcr+ chimeric mice were pale with splenomegalia and hepatomegalia with pronounced white spots. The thymi generally appeared normal. This examination is consistent with hematologic disease. However, no tumors of other tissues were found in these chimeric mice, despite widespread activity of the bcr promoter.31 Histologic analysis of these animals showed marked leukemic cell infiltration of hematopoietic (splenic red and whithe pulp) and nonhematopoietic tissues (heart and liver sinusoids) in bcr-ABL/bcr+ chimeric mice (Fig 2). Similar results were obtained with two independently targeted clones and with chimeras derived at different times. There was no relationship between chimerism and disease, because there was not a minimum degree of chimerism required for the development of a leukemic state, and the latent period for the disease did not vary with respect to the degree of chimerism. Although a high proportion of bcr-ABL mice (38 of 40) developed acute leukemia, no disease appeared in chimeras generated with either the unmodified ES-E14 cells or ES-E14 cells in which one bcr allele had been modified. This does not appear simply to reflect the lack of ES cell contribution in these control chimeric mice, because the level of chimerism was as high as 75% to 90% in some mice. Although the bcr gene is expressed in many different types of tissues in the mouse, the lack of bcr alone is not sufficient to perturb hematopoietic development.31 Therefore, these results show the generation of a new mouse model in which the bcr-ABLp190 oncogene expression is directed by the endogenous bcr expression control elements, recapitulating the consequences of the BCR-ABLp190 chromosomal abnormality. As a result of this, the chimeric mice specifically develop acute leukemias, despite widespread activity of the bcr promoter.
Histology of the acute leukemia disease in bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice. Histologic analysis was performed when the mice were moribund. The spleen from bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice that developed an acute leukemia shows the effacement of the normal spleen architecture (compared with control chimeric mice). Similar cells invade visceral organs such as the liver or the heart. Control chimeric mice were generated by injecting wild-type E14 cell clone or ES-E14 cells in which one bcr allele had been modified with the bcr targeting vector into blastocysts. Although the acute leukemia disease in bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice was very similar, the level of leukemic infiltration shown by histologic analysis was not identical (original magnification ×50).
Histology of the acute leukemia disease in bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice. Histologic analysis was performed when the mice were moribund. The spleen from bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice that developed an acute leukemia shows the effacement of the normal spleen architecture (compared with control chimeric mice). Similar cells invade visceral organs such as the liver or the heart. Control chimeric mice were generated by injecting wild-type E14 cell clone or ES-E14 cells in which one bcr allele had been modified with the bcr targeting vector into blastocysts. Although the acute leukemia disease in bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice was very similar, the level of leukemic infiltration shown by histologic analysis was not identical (original magnification ×50).
The bcr-ABLp190 oncogene does not require the endogenous bcr product for leukemogenesis. The leukemic cells of the bcr-ABLp190 chimeric mice recapitulate the rearrangement that takes place into human BCR-ABLp190–positive cells, in which the leukemic cells only have one intact BCR allele (having the other one rearranged as a result of fusion to ABL ). This mouse model is an ideal system to study the biology of the Philadelphia chromosome. In this sense, it has been shown that the endogenous nonrearranged products can influence the effect of the fusion gene as a result of the chromosomal abnormality.29-31,34 There appear to be a number of discrete mechanisms through which BCR contributes to leukemogenesis. One is at the level of transcription: the BCR promoter controls expression of both the remaining nonrearranged BCR allele and the BCR-ABLp190 oncogene in human leukemia. Apart from the structural influence on ABL in BCR-ABL,20,35,36BCR may contribute to leukemia on another level: the presence of a functional BCR protein (p160bcr) has been proposed as a requirement for malignant transformation by BCR-ABL.29-31
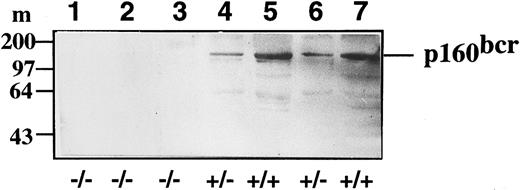
Western blot analysis of bcr expression levels in control (+/+), heterozygote bcr-ABL/bcr+ (+/−), and homozygote bcr-ABL/bcr− (−/−) spleen extracts. The location of p160BCR is as indicated to the left. Lanes 1, 2, and 3 correspond to bcr-ABL/bcr− chimeric mice; lanes 4 and 6 correspond to bcr-ABL/bcr+ mice; and lanes 5 and 7 correspond to control chimeric mice.
Western blot analysis of bcr expression levels in control (+/+), heterozygote bcr-ABL/bcr+ (+/−), and homozygote bcr-ABL/bcr− (−/−) spleen extracts. The location of p160BCR is as indicated to the left. Lanes 1, 2, and 3 correspond to bcr-ABL/bcr− chimeric mice; lanes 4 and 6 correspond to bcr-ABL/bcr+ mice; and lanes 5 and 7 correspond to control chimeric mice.
Our strategy based on homologous recombination to recapitulate the consequences of the Philadelphia oncogene allows us to study the influence of the endogenous nonrearranged bcr allele on tumor cells expressing the bcr-ABLp190 oncogene. To assess the role of the intact BCR product in the Ph1+-leukemogenesis, homologous recombination in ES cells has been used to inactivate the endogenous non-rearranged bcr allele and study the effect of the bcr-ABLp190 oncogene in the context of an inactive bcr allele. A second targeting vector was constructed in which exon 1 of bcr was partially replaced by insertion of the positive hygromycine resistance selection marker (PGK-Hyg-pA; Fig 1A). The construct (bcr targeting vector) was electroporated into bcr-ABL/bcr+ ES cells and correct targeting of the remaining nonrearranged bcr allele, as confirmed by Southern blot analysis, was obtained (bcr-ABL/bcr− ES cells). These correctly modified ES cell clones were injected into recipient C57BL/6 blastocysts and chimeric mice produced (bcr-ABL/bcr−). Two independent bcr-ABL/bcr− ES cell clones with normal karyotypes were injected into C57BL/6 blastocysts. The expression of the bcr-ABLp190 oncogene was determined by reverse transcription PCR in the ES cell clones with the targeted alleles (bcr-ABL/bcr−) and in the chimeric tissues (Fig 1B). bcr-ABLp190 gene expression was detected in both the ES cell clones with the specific targeted alleles and in the tissues of the chimeras (Fig 1B).
Chimeric mice were generated by injection of these modified targeted clones (bcr-ABL/bcr−) into blastocysts to analyze the effect of the bcr-ABLp190 fusion gene in the absence of the endogenous bcr allele. Forty chimeras were generated and 37 of 40 of these bcr-ABL/bcr− chimeric mice developed an acute leukemia disease that was clinically very similar to the disease state of bcr-ABL/bcr+ chimeric mice, which have an nonrearranged bcr allele. The incidence of leukemia was approximately the same in chimeras made from bcr-ABL/bcr+ and bcr-ABL/bcr− ES clones. Histologic analysis of the bcr-ABL/bcr− chimeric animals also showed a marked infiltration of hematopoietic and nonhematopoietic tissues (Fig 2). Similar results were obtained with two independently targeted clones and with chimeras derived at different times.
Therefore, our results show that bcr-ABLp190 oncogene causes leukemia in this mouse model and does not require the endogenous bcr product for malignant transformation, because chimeric mice derived from ES cells that expressed the bcr-ABL fusion product and had a disruption of the remaining nonrearranged bcr allele do develop leukemia and the lack of bcr alone is not sufficient to perturb hematopoietic development.31 To confirm that no bcr gene products were being synthesized in the bcr-ABL/bcr− chimeric mice, Western blot analysis was performed using spleen extracts with a very high infiltration of leukemic cells. bcr-ABL/bcr+ animals expressed levels of bcr approximately half of that of control chimeric mice (Fig 3, compare lanes 4 and 6 with 5 and 7), whereas the bcr-ABL/bcr- mice produced no p160bcr (Fig 3, lanes 1, 2, and 3). These results show that the bcr-ABL/bcr− chimeric mice indeed do not produce bcr.
bcr-ABLp190 chimeras develop B-cell ALL. Although in human pathology the BCR-ABLp190 chimeric product is associate with B-cell ALL (B-ALL),1,9,17,22,23 current mouse models for BCR-ABLp190 have failed to consistently reproduce this pathology,5-8 because in some mice lymphomas will develop, due to the difficulty of choosing a promoter to engineer expression in the appropiate cell type. Therefore, we next defined the acute leukemic disease generated in both bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice, because our bcr-ABL mice mimic the result of the Philadelphia chromosome abnormality in the sense that the transcription of the fusion gene occurs from the endogenous bcr promoter elements.
Detailed analysis of the leukemic cells in the chimeric bcr-ABL mice established the diagnosis as pre-B ALL. Hematoxilin/eosin staining showed that the leukemic cells had a lymphoid morphology. The majority of peripheral blood mononuclear cells from the four leukemic mice examined (2 bcr-ABL/bcr+ and 2 bcr-ABL/bcr−, respectively) had a B-cell precursor immunophenotype (B220+, Sig−; Table 1). A small percentage of cells were positive for Thy-1, myeloid, and SIg (Table 1). This may represent the staining of residual T cells, myeloid, and mature B cells, respectively; although the expression of Thy-1 is a characteristic of early B-precursor cells or pro-B cells that appear to be primary targets for Abelson murine leukemic virus ( MuLV ) transformation in vitro.37 Different tissues from bcr-ABL/bcr+ and bcr-ABL/bcr− leukemic mice were analyzed for Ig heavy chain (IgH) gene rearrangement. Monoclonal patterns were observed in tissues from the mice examined, both bcr-ABL/bcr+ and bcr-ABL/bcr− (Fig 4). In one case, the DNA from one mouse showed no rearranged bands in Southern blots. The tissue samples were heavily infiltrated with leukemic cells. One interpretation of this observation is that the leukemic cells were very primitive or pro-B cells still retaining a germline configuration of IgH genes.37,38 Alternatively, the leukemic cells in these cases might be polyclonal with respect to IgH rearrangements, as described for some cases of lymphoblastic leukemia in humans.39 To test the leukemogenicity of cells from both bcr-ABL/bcr+ and bcr-ABL/bcr− chimeric mice, 1 × 106 cells were injected intravenously into nonirradiated normal NOD/SCID mice. As shown in Table 2, 15 of 16 mice injected developed progressively growing leukemias within 6 to 11 weeks of transplantation. By contrast, none of 20 mice injected with cells from 10 control chimeras developed a leukemia of donor origin. The transplanted cells developed into the same kind of leukemia. Furthermore, the origin of leukemic clones from donor mice was confirmed by PCR analysis showing the presence of the bcr-ABL gene product. The possibility that ecotropic retroviral insertional mutagenesis might be involved in the genesis of the leukemia is ruled out by the observation that neither germline nor acquired clonal ecotropic proviral integrations were detected by Southern blot analyses of the DNAs from leukemia transplants found to have clonal IgH rearrangements (data not shown). These data define the acute leukemia generated in the bcr-ABL mice as a B-cell ALL. Therefore, our mouse model not only resembles the rearrangement that takes place in human BCR-ABLp190 leukemias but also reproduces the same phenotype with which this fusion gene is associated in human pathology. This result validates this mouse model as ideal to study the biology of the BCR-ABLp190 oncogene.
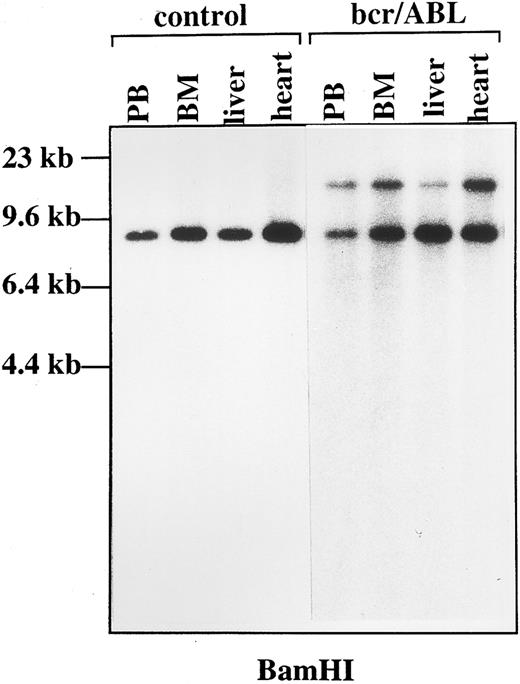
Genomic organization of the Ig heavy chain gene in bcr-ABL chimeric mice. DNA was obtained from control and bcr-ABL chimeric tissues, digested with BamHI, and fractionated into 1% agarose gel. Specific rearrangement was detected by Southern blot analysis with an Ig heavy chain specific probe in bcr-ABL chimeric tissues. PB, peripheral blood; BM, bone marrow.
Genomic organization of the Ig heavy chain gene in bcr-ABL chimeric mice. DNA was obtained from control and bcr-ABL chimeric tissues, digested with BamHI, and fractionated into 1% agarose gel. Specific rearrangement was detected by Southern blot analysis with an Ig heavy chain specific probe in bcr-ABL chimeric tissues. PB, peripheral blood; BM, bone marrow.
CONCLUSIONS
We have used a homologous recombination strategy to generate an in-frame fusion of BCR-ABL to exon 1 of mouse bcr allele, and we find that this causes acute leukemias in the chimeric mice. This strategy has the advantage of using the normal transcriptional control elements of bcr to express the bcr-ABL fusion, recapitulating the effect of the human chromosome translocation. A number of conclusions can be made from our experiments. First, the bcr-ABL fusion gene is important for tumor development and does not require an endogenous bcr product, indicating that bcr-ABL expression is the prime event initiaiting Ph1+-leukemia. Second, the phenotype of these tumors is very close to the phenotype of leukemias seen in patient with BCR-ABLp190 leukemias,9,36 mimicking the effect of the chromosomal translocation. Therefore, these animals will be an ideal model to study the function of BCR-ABLp190 oncogene in malignant transformation40,41 and develop new specific anticancer therapies.42 43
ACKNOWLEDGMENT
The authors are grateful to Dr M. Neuberger for providing the Ig probe, Dr F. Serrano for the NOD/SCID mice, Dr H. Schrewe for the CJ7 ES cell line, and Drs J. Corral and T.H. Rabbitts for assistance.
Supported by Fundación Internacional José Carreras (FIJC-94/INT), European Comission (BMH4-CT96-0375), DGCYT (UE96-0041), and Fundación Cientı́fica of the AECC.
Address reprint requests to I. Sánchez-Garcı́a, MD, PhD, Departamento de Proliferación y Diferenciación Celular, Instituto de Microbiologı́a Bioquı́mica, Edificio Departamental, Avda del Campo Charro s/n, 37007-Salamanca, Spain.