Abstract
Mitochondrial iron overload in acquired idiopathic sideroblastic anemia (AISA) may be attributable to mutations of mitochondrial DNA (mtDNA), because these can cause respiratory chain dysfunction, thereby impairing reduction of ferric iron (Fe3+) to ferrous iron (Fe2+). The reduced form of iron is essential to the last step of mitochondrial heme biosynthesis. It is not yet understood to which part of the respiratory chain the reduction of ferric iron is linked. In two patients with AISA we identified point mutations of mtDNA affecting the same transmembrane helix within subunit I of cytochrome c oxidase (COX I; ie, complex IV of the respiratory chain). The mutations were detected by restriction fragment length polymorphism analysis and temperature gradient gel electrophoresis. One of the mutations involves a T → C transition in nucleotide position 6742, causing an amino acid change from methionine to threonine. The other mutation is a T → C transition at nt 6721, changing isoleucine to threonine. Both amino acids are highly conserved in a wide range of species. Both mutations are heteroplasmic, ie, they establish a mixture of normal and mutated mitochondrial genomes, which is typical of disorders of mtDNA. The mutations were present in bone marrow and whole blood samples, in isolated platelets, and in granulocytes, but appeared to be absent from T and B lymphocytes purified by immunomagnetic bead separation. They were not detected in buccal mucosa cells obtained by mouthwashes and in cultured skin fibroblasts examined in one of the patients. In both patients, this pattern of involvement suggests that the mtDNA mutation occurred in a self-renewing bone marrow stem cell with myeloid determination. Identification of two point mutations with very similar location suggests that cytochrome c oxidase plays an important role in the pathogenesis of AISA. COX may be the physiologic site of iron reduction and transport through the inner mitochondrial membrane.
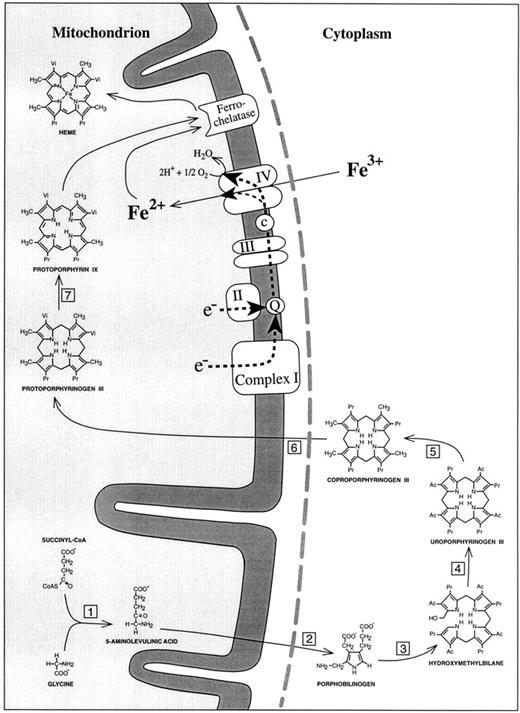
SIDEROBLASTIC ANEMIA is characterized by inadequate formation of heme and excessive accumulation of iron in erythroblast mitochondria. Whereas recent molecular biology approaches identified abnormalities of erythroid δ-aminolevulinic acid synthase in the hereditary, X-linked type of sideroblastic anemia,1-7 the pathogenesis of the most prevalent form of sideroblastic anemia, acquired idiopathic sideroblastic anemia (AISA), has remained an enigma. For many years, investigators tried to clarify the disorder by searching for enzyme defects of heme biosynthesis (Fig 1). A major shortcoming of this approach was its inability to explain why the penultimate product of the heme synthetic pathway, protoporphyrin IX, is elevated rather than decreased in most AISA patients.8 In our opinion, this finding provides a strong argument for a possible defect in the last step of heme synthesis, namely insertion of iron into protoporphyrin IX. This step is catalysed by ferrochelatase. However, this enzyme is unlikely to be responsible, because increased red blood cell (RBC) protoporphyrin concentrations in sideroblastic anemia are not correlated with low ferrochelatase activities.9 With normal ferrochelatase activity, and with plentiful iron and protoporphyrin IX, iron is still not inserted into the heme precursor. The likely explanation is that iron is not in the right chemical form. Iron deposits in sideroblastic mitochondria contain iron in the trivalent ferric (Fe3+) state,10 whereas ferrochelatase can only use ferrous iron (Fe2+) for heme synthesis.11 Because Fe2+ is not stable under aerobic conditions, it is necessary for erythropoietic cells to have an enzyme system that can maintain a supply of Fe2+ as a substrate for ferrochelatase. About 20 years ago, reduction of Fe3+ was shown to be accomplished by the respiratory chain as the source of reducing equivalents.12 13 At complete inhibition of mitochondrial respiration, heme synthesis becomes negligible. However, it is not known to which part of the respiratory chain the reduction of ferric iron is linked.
Schematic representation of the heme biosynthetic pathway and its connection with the electron transport chain of the inner mitochondrial membrane. (1) 5-Aminolevulinic acid synthase; (2) 5-aminolevulinic acid dehydratase; (3) porphobilinogen deaminase; (4) uroporphyrinogen III synthase; (5) uroporphyrinogen III decarboxylase; (6) coproporphyrinogen III oxidase; (7) protoporphyrinogen III oxidase. Complex I, NADH dehydrogenase; complex II, succinate dehydrogenase; complex III, b/c1-complex = ubiquinol cytochrome c reductase; complex IV, cytochrome c oxidase; Q, coenzyme Q = ubiquinone; c, cytochrome c. The broken line (grey) represents the outer mitochondrial membrane. In accordance with the point mutations identified in mtDNA-encoded subunit I of cytochrome c oxidase, the reduction of Fe3+ to Fe2+ has been placed into complex IV of the respiratory chain.
Schematic representation of the heme biosynthetic pathway and its connection with the electron transport chain of the inner mitochondrial membrane. (1) 5-Aminolevulinic acid synthase; (2) 5-aminolevulinic acid dehydratase; (3) porphobilinogen deaminase; (4) uroporphyrinogen III synthase; (5) uroporphyrinogen III decarboxylase; (6) coproporphyrinogen III oxidase; (7) protoporphyrinogen III oxidase. Complex I, NADH dehydrogenase; complex II, succinate dehydrogenase; complex III, b/c1-complex = ubiquinol cytochrome c reductase; complex IV, cytochrome c oxidase; Q, coenzyme Q = ubiquinone; c, cytochrome c. The broken line (grey) represents the outer mitochondrial membrane. In accordance with the point mutations identified in mtDNA-encoded subunit I of cytochrome c oxidase, the reduction of Fe3+ to Fe2+ has been placed into complex IV of the respiratory chain.
We recently hypothesized that, in sideroblastic anemia, one or several of the enzyme complexes of the respiratory chain are disturbed in such a way that efficient reduction of iron no longer occurs.14 Under these circumstances, the imported iron cannot be used for heme synthesis and will accumulate in the mitochondrial matrix. The proposed malfunction could be due to mutations of nuclear DNA or mitochondrial DNA (mtDNA), because both genomes contribute to the assembly of respiratory chain complexes. Human mtDNA is a 16,569-bp circular molecule that codes for the genes of 13 essential polypeptide subunits of the respiratory chain, together with the 12S and 16S ribosomal RNAs and the 22 mitochondrial transfer RNAs that are required for the intramitochondrial translation of the protein-coding units. In recent years, a variety of disorders with predominantly neurologic symptoms have been linked to mutations in mtDNA.15 16
Our focus on mtDNA is partly explained by the fact that Pearson's syndrome, a rare congenital disorder, points to a relation between mtDNA mutations and sideroblastic anemia. This condition presents in infancy with a variety of symptoms, including refractory sideroblastic anemia, thrombocytopenia, neutropenia, pancreatic exocrine dysfunction, and lactic acidosis.17 The bone marrow shows dysplastic changes including ring sideroblasts and prominent vacuolization of precursor cells.18,19 The cause of Pearson's syndrome was discovered in 1989, when Rötig et al20 found an mtDNA deletion in a patient with the disorder. Investigation of further patients confirmed that large deletions of mtDNA are a constant feature of this disease.21 However, the relation between mtDNA deletions and sideroblastic anemia was not understood. According to our pathogenetic model,14 deficient iron reduction, attributable to respiratory chain dysfunction, provides the missing link.
Having excluded large deletions of mtDNA as a cause of AISA,22 we started looking for short deletions and point mutations. Recently, we reported a patient who had a heteroplasmic point mutation of mitochondrial tRNALeu(CUN) in his bone marrow cells.23 However, this mutation did not help to localize the crucial site of iron reduction and transport through the mitochondrial inner membrane, because mitochondrial tRNA mutations cause a general impairment of mitochondrial protein synthesis and can thus affect all respiratory chain complexes possessing components encoded by mtDNA. More informative in this respect are the two point mutations presented here.
MATERIALS AND METHODS
Case Reports
Patient no. 1. A 58-year-old woman was first evaluated for moderate macrocytic anemia in 1992. The hemoglobin (Hb) level was 11 g/dL, an RBC count of 2.94 × 106/μL, a hematocrit level of 33%, a mean corpuscular volume (MCV) of 112 fL, a mean corpuscular Hb (MCH) level of 37.8 pg, and a mean corpuscular Hb content (MCHC) of 33.7 g/dL. The leukocyte count was 8.5 × 103/μL, and the white blood cell (WBC) differential was 50% neutrophils, 2% bands, 2% eosinophils, 1% basophils, 5% monocytes, and 40% lymphocytes. The platelet count was 596,000/μL. Bone marrow biopsy showed a hypercellular marrow with dysplastic changes in all three myeloid cell lines. On iron staining, 40% of erythroblasts were ring sideroblasts. There was no excess of myeloblasts. A diagnosis of refractory anemia with ring sideroblasts was made (RARS; according to the French-American-British [FAB] classification). Cytogenetic analysis showed a normal karyotype; in particular, there was no evidence of a 5q− abnormality. The patient had psoriatic arthritis since 1979, but was never treated with methotrexate or cyclophosphamide. On follow-up examinations, bone marrow biopsies showed that ring sideroblasts were greater than 50%; there were no significant changes otherwise. No chromosomal abnormalities were found. Until 1996, the course of the disease was uneventful, with Hb values fluctuating between 8.5 and 10 g/dL. In May 1996, the patient received her first blood transfusion because her Hb level had decreased to 8 g/dL. Meanwhile, transfusion of 2 U of packed RBCs is required every 3 to 4 weeks. In the bone marrow, there has been no evidence of progression toward a more advanced type of myelodysplastic syndrome (MDS).
Patient no. 2. A 68-year-old man presented with exertional dyspnea and fatigue in December 1989. The Hb level was 7.8 g/dL, the MCV was 77.7 fL, the MCH was 32.2 pg, and the serum ferritin level was 664 μg/mL. The leukocyte count was 3.8 × 103 μL, and the WBC differential was 27% neutrophils, 1% bands, 3% eosinophils, 3% monocytes, and 66% lymphocytes. The platelet count was 338,000/μL. Bone marrow biopsy showed hyperplasia and dysplasia of the erythroid series, with megaloblastoid changes and nuclear lobulation and fragmentation. Iron staining showed 70% to 80% ring sideroblasts. Multinucleated megakaryocytes were found. Granulocytopoiesis appeared inconspicuous, and there was no excess of blasts. A diagnosis of RARS was made. In addition, the patient was found to have a multinodular goiter and to be hyperthyroid. He was successfully treated with radioactive iodine. In March 1990, a control bone marrow biopsy showed no significant change, but the patient became transfusion dependent. In September 1991, anemia worsened, bone marrow biopsy showed greater than 90% ring sideroblasts, and cytogenetic examination was abnormal for the first time [in 4 of 20 mitoses, a 5q− abnormality was found, ie, del(5) (q13-14)].
The patient was entered into a pilot study of all-trans retinoic acid24 and responded with an increase in Hb values up to 11.8 g/dL. Ring sideroblasts in the bone marrow decreased to 45% (July 1992). The ferritin level was 385 ng/mL. In January, 1993, anemia worsened again, a bone marrow biopsy showed 70% ring sideroblasts, and karyotype analysis confirmed the 5q− anomaly. The patient received a second trial of ATRA until June 1993. He responded again, became independent of transfusions, and showed an Hb level of 10.2 g/dL. The disease remained stable from 1993 to 1995, when the patient turned up for a follow-up examination: Hb level of 11 g/dL, RBC count of 4.52 × 106 /μL, hematocrit level of 34.6%, reticulocytes level of 13.4%, and MCV of 76.7 fL. The platelet count was 227,000/μL. Bone marrow biopsy showed dysplastic changes in all three cell lines, including some megakaryocytes with nonlobulated nuclei. Ring sideroblasts were 90%. Karyotype analysis showed 5q−. Despite the high proportion of ring sideroblasts, Hb levels have remained stable.
Preparation of Cell Fractions
The patients were part of a series of 20 patients with MDS whose bone marrow samples were subjected to restriction fragment length polymorphism (RFLP) screening for mtDNA mutations. Among those 20 patients, 12 had sideroblastic anemia. Fifty hematologically normal individuals (cardiac surgery patients) were specifically screened for the mutations of cytochrome c oxidase (COX) at nucleotide positions 6721 and 6742 that we identified in AISA patients; bone marrow samples from the controls had been obtained for a previous study on mtDNA deletions.25 In the two patients under discussion, mtDNA was extracted from an unfractionated bone marrow sample and from peripheral blood cells of different lineages. Platelet-rich plasma was obtained from citrated blood samples by centrifugation at 100g for 15 minutes at room temperature. Contaminating blood cells were removed by centrifugation at 200g for 10 minutes. Platelets were pelleted from the supernatant by centrifugation at 1,000g for 10 minutes and resuspended in phosphate-buffered saline. Granulocytes were enriched by depletion of peripheral blood mononuclear cells through density gradient centrifugation with Lymphoprep 1.077 (Nycomed, Oslo, Norway). A 200 μL sample from the granulocyte/erythrocyte pellet was used for DNA extraction. Erythrocytes do not contain mtDNA,26 because mitochondria are eliminated during the reticulocyte stage.27 Therefore, mtDNA isolated from the granulocyte/erythrocyte pellet mainly derives from mature peripheral blood granulocytes, apart from a small contamination by reticulocyte mtDNA. For isolation of B and T lymphocytes from EDTA-anticoagulated blood, immunomagnetic beads (Dynabeads M-450 Pan-B [CD19] and Dynabeads M-450 Pan-T [CD2], respectively) were used according to the recommendations of the manufacturer (Dynal, Oslo, Norway). With antibody-coated beads still bound to the cell surface, lymphocytes were entered into the cell lysis step of the DNA extraction procedure. After cell lysis, beads were removed with a magnet. Buccal mucosa cells were pelleted from a vigorous mouthwash performed with 20 mL normal saline and depleted of contaminating granulocytes by density gradient centrifugation with Nycoprep 1.150 (Nycomed). A skin biopsy (right buttock) was obtained from patient no. 2, and fibroblast cultures were established by outgrowth from this biopsy according to a standard method.28 Cells were grown to confluency in Dulbecco's modified Eagle's Medium (Flow, Meckenheim, Germany) supplemented with sodium ascorbate (50 μg/mL), glutamine (300 μg/mL), penicillin (400 μg/mL), streptomycin (50 μg/mL), and 10% fetal calf serum (FCS) on tissue culture dishes in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Colony Assay of Hematopoietic Cells
Mononuclear cells were obtained from peripheral blood (PB-MNC) through density gradient centrifugation with Lymphoprep 1.077 (Nycomed), washed three times in RPMI 1640 medium, and plated in tissue culture-grade 35-mm Petri dishes (2 × 105 cells per dish) in a volume of 1 mL MethoCult H4433 (StemCell Technologies, Vancouver, British Columbia, Canada), containing 0.9% methylcellulose, 5% serum (containing phytohemagglutinin-leukocyte conditioned medium [PHA-LCM]), 30% FCS, 10% bovine serum albumin, 13% Iscove's medium, erythropoietin (150 U/mL), 2-mercaptoethanol (0.1 mol/L), and L-glutamine (200 mmol/L). After 14 days of incubation at 37°C and 5% CO2 , colonies and clusters were examined using an inverted microscope. For polymerase chain reaction (PCR) amplification of a particular COX I segment of mtDNA, individual colonies were picked with a micropipette and entered directly into PCR reaction mixtures (before adding specific primers and polymerase).
DNA Isolation
Total DNA was isolated from the various cell samples using the QIAamp Kit (Qiagen, Hilden, Germany), which is based on cell lysis in the presence of guanidine hydrochloride and subsequent binding of DNA to an anion-exchange resin in a spin-column. The procedure ensures extraction of cytoplasmic (eg, mitochondrial) DNA in addition to nuclear DNA.
mtDNA Amplification
mtDNA was amplified in 17 overlapping segments using the PCR. The exact position of segments according to the standard Cambridge sequence29 and the respective primer sequences were as previously reported.23 DNA was amplified using 35 cycles of denaturation (1 minute at 94°C), annealing (30 seconds at 55°C to 71°C, depending on the pair of primers used), and extension (60 seconds at 72°C). The 100 μL PCR reactions contained 10 mmol/L KCl, 20 mmol/L Tris-HCl (pH 8.8), 10 mmol/L (NH4 )2SO4 , 4 mmol/L MgS04 , 400 μmol/L of each dNTP, 50 pmol of each primer, and 1 U of Vent DNA polymerase (New England Biolabs, Dreieich, Germany). Double-stranded PCR products were purified with a commercial DNA purification system (Wizard PCR Preps; Promega, Heidelberg, Germany), separated in 1% agarose gels, and visualized by ethidium bromide staining.
RFLP Analysis
Each amplified mtDNA segment was entered into 20 different restriction enzyme digestions using Alu I, BamHI, Cfo I, Dde I, EcoRI, Hae II, Hae III, HincII, HindIII, HinfI, Hpa I, Msp I, Pvu II, Rsa I, Sac I, Sca I, Stu I, Taq I, Tru9I, and Xba I. Endonucleases were purchased from Boehringer Mannheim (Mannheim, Germany). All restriction enzyme reactions were performed according to manufacturer's recommendations, using approximately 100 to 200 ng of PCR-generated DNA for digestion with 5 U of enzyme. Restriction fragments were separated by electrophoresis in 3.5% Visigel matrix (Stratagene, Heidelberg, Germany) and visualized by ethidium bromide staining and UV illumination.
Heteroduplex Analysis by Temperature Gradient Gel Electrophoresis (TGGE)
Point mutations in double-stranded DNA can be detected by denaturing-gradient gel electrophoresis (DGGE)30 or TGGE,31 32 because mutations alter the denaturation pattern of double-stranded DNA at the site of the mutation and in its neighborhood. The sensitivity of TGGE analysis is greatly enhanced through heteroduplex generation. During a denaturation/renaturation cycle, hybrids are formed between wild-type DNA fragments and homologous fragments carrying the mutation. The mismatch in the double helix at the site of the mutation leads to significant lowering of the midpoint melting temperature of the heteroduplex fragments. When exposed to increasing temperatures during electrophoresis (the TGGE procedure), heteroduplex hybrids show partial denaturation earlier than homoduplexes do. Because early partial denaturation causes early retardation in the gel, heteroduplexes can be resolved from homoduplex molecules by TGGE.
In the two patients reported, intraindividual heteroduplex analysis was performed, ie, TGGE was used to examine whether heteroduplexes formed after PCR amplification of a mtDNA fragment of interest. This only happens when there is coexistence of wild-type and mutated mtDNA in the patient's sample, which is also called heteroplasmy. Among other segments of mtDNA, a 420-bp segment carrying the COX I mutation was amplified by PCR using primers A (nt 6504-6523), 5′-GTCCTAGCTGCTGGCATCAC-3′, and B (nt 6923-6904), 5′-TCAGAGCACTGCAGCAGATC-3′. PCR was followed by quantitative denaturation of the PCR product (heating in 4 mol/L urea at 95°C for 5 minutes) and subsequent renaturation (incubation at 50°C for 20 minutes). Samples were then loaded onto thin polyacrylamide gels (5%) covalently bound to polyethylene gel support films. After electrophoresis, bands of heteroduplex- and homoduplex-DNA were visualized by silver staining. TGGE equipment was purchased from Qiagen.
Sequencing
For sequencing of the region of interest, mtDNA from bone marrow was amplified using a modified primer A (A-EcoRI) and primer B. Primer A-EcoRI (5′-GGCG ↓ AATTC GTCCTAGCTGCTGGCATCAC-3′) was designed to introduce a restriction site for EcoRI. PCR was performed as described above, with an annealing temperature of 59°C and an extension time of 30 seconds, yielding a 429-bp fragment. The PCR product was purified with QIAamp spin columns (Qiagen) and digested with EcoRI and Pst I to yield a shortened fragment 416 bp in length. The digested DNA was extracted with a mixture of phenol and chloroform (70:30), brought to a 0.3 mol/L concentration of sodium acetate, and precipitated with 2.5 vol of ethanol. Using T4 DNA-Ligase, the 416-bp fragment was ligated into the sequencing vector pUC19,33 which had been digested with the same two enzymes (EcoRI and Pst I). After transformation into Escherichia coli strain DHαF′, white colonies were selected on Luria broth (LB) medium containing 150 mg/L ampicillin and X-Gal.34 Plasmid DNA from transformants was prepared using Tip20 columns (Qiagen) and double-stranded DNA was sequenced with the Sequenase kit (US Biochemicals, Cleveland, OH) using 35S-dATP, according to the method of Sanger et al.35
RESULTS
In two patients with AISA we identified point mutations of mtDNA in the region encoding subunit I of cytochrome c oxidase (nt 5904-7444). These mutations, in the following referred to as COX I mutations, are located at nt 6721 and nt 6742, affecting amino acids M273 and I280, respectively, of the human sequence.
Patient No. 1
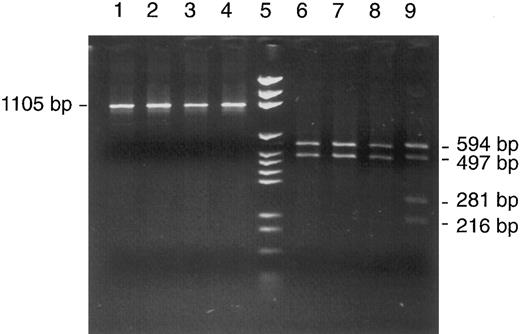
A heteroplasmic mutation was detected by RFLP analysis of unfractionated bone marrow cells when Rsa I digestion of mtDNA segment no. 7 (nt 6504-7608) showed an aberrant pattern with two unexpected bands representing fragments of 281 and 216 bp length (Fig 2, lane 9). Their appearance implied the presence of a new Rsa I site, because the two regular sites in this segment (nt 6999 and 7013) only explained fragments of 594 and 497 bp and a very short fragment of 14 bp that was not visualized in the gel. The relative intensities of bands were estimated by gel photography with an electronic camera and data analysis with suitable software (Enhanced analysis system, EASY plus; Herolab GmbH, Wiesloch, Germany). The system sums up the light intensities of all pixels belonging to a specific band (with automatic background subtraction). The intensities were 182, 113, and 66, respectively, for the 497-bp band representing normal molecules and the 281-bp and 216-bp bands representing cleavage products of mutated mtDNA molecules. After correcting for molecular weight, this translated into 49% mutated mtDNA in the patient's bone marrow.
Detection of a heteroplasmic mtDNA mutation in a bone marrow aspirate from patient no. 1. RFLP analysis of mtDNA segment 7 (nt 6504-7608) with Cfo I in four patients (lanes 1 through 4) and with Rsa I in the same four patients (lanes 6 through 9) showed an abnormal restriction pattern in patient no. 1 (lane 9); for explanation, see text. Lane 5, DNA size marker (pGEM).
Detection of a heteroplasmic mtDNA mutation in a bone marrow aspirate from patient no. 1. RFLP analysis of mtDNA segment 7 (nt 6504-7608) with Cfo I in four patients (lanes 1 through 4) and with Rsa I in the same four patients (lanes 6 through 9) showed an abnormal restriction pattern in patient no. 1 (lane 9); for explanation, see text. Lane 5, DNA size marker (pGEM).
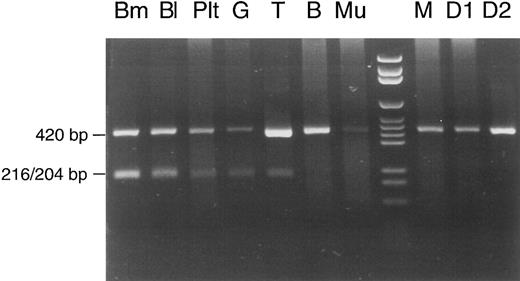
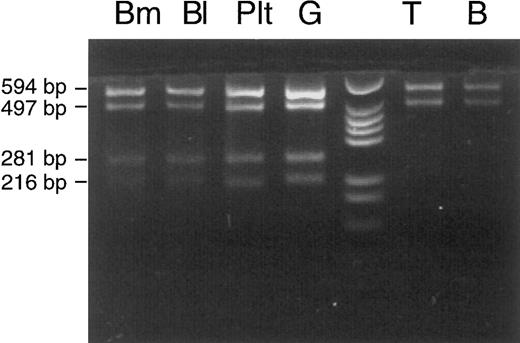
The same proportion (50% mutated mtDNA) appeared when the calculation was based on data of another experiment that was performed for analysis of different cell lineages (Fig 3). This experiment involved shortening of segment no. 7. A 420-bp PCR product, defined by primers A (nt 6504-6523) 5′-GTCCTAGCTGCTGGCATCAC-3′ and B* (nt 6923-6904) 5′-TCAGAGCACTGCAGCAGATC-3′, contained the mutation but no regular Rsa I site. It therefore remained undigested in controls (the patient's mother and two daughters) and in the patient's buccal mucosa cells and lymphocytes (the faint pathologic band in T lymphocytes was inconsistent and may have been due to contamination with phagocytes during immunomagnetic bead isolation of lymphocytes). In contrast, 50% of the PCR product was cut into two fragments of 216 and 204 bp when DNA from bone marrow, whole blood, or granulocytes was examined. For unknown reasons, platelets showed a lower percentage of mutated mtDNA (20%). Lymphocytes from another peripheral blood sample were also isolated with immunomagnetic beads and used for a repeat analysis (Fig 4). This time, the Rsa I restriction pattern of the full-length mtDNA segment no. 7 was clearly normal with T and B lymphocytes (no faint pathologic bands). This pattern of lineage involvement, and the absence of the mutation in the patient's mother and daughters, excluded an inherited mutation. We suggest that the mutation arose in a myeloid stem cell of the colony-forming unit granulocyte, erythroid, monocyte, megakaryocyte (CFU-GEMM) type, although we cannot exclude with certainty the involvement of a pluripotent hematopoietic stem cell. The aberrant pattern was not detectable in 10 other patients with sideroblastic anemia, 8 patients with other types of MDS, and 50 hematologically normal controls (data not shown).
Cell lineage involvement of the COX I mutation in patient no. 1. Total DNA was extracted from different types of cells: Bm, bone marrow; Bl, peripheral blood; Plt, platelets; G, erythrocyte/granulocyte pellet; T, T lymphocytes; B, B lymphocytes; Mu, buccal mucosa cells; lane 8, DNA size marker; M, peripheral blood from the mother of patient no. 1; D1 and D2, peripheral blood from the first and second daughter of patient no. 1. See text for explanation of the restriction pattern.
Cell lineage involvement of the COX I mutation in patient no. 1. Total DNA was extracted from different types of cells: Bm, bone marrow; Bl, peripheral blood; Plt, platelets; G, erythrocyte/granulocyte pellet; T, T lymphocytes; B, B lymphocytes; Mu, buccal mucosa cells; lane 8, DNA size marker; M, peripheral blood from the mother of patient no. 1; D1 and D2, peripheral blood from the first and second daughter of patient no. 1. See text for explanation of the restriction pattern.
Cell lineage involvement of the COX I mutation in patient no. 1. Immunomagnetic bead isolation of T and B lymphocytes was repeated with another peripheral blood sample, and a normal Rsa I restriction pattern of mtDNA segment 7 (nt 6504-7608) was obtained for T and B lymphocytes. For abbreviations, see Fig 3.
Cell lineage involvement of the COX I mutation in patient no. 1. Immunomagnetic bead isolation of T and B lymphocytes was repeated with another peripheral blood sample, and a normal Rsa I restriction pattern of mtDNA segment 7 (nt 6504-7608) was obtained for T and B lymphocytes. For abbreviations, see Fig 3.
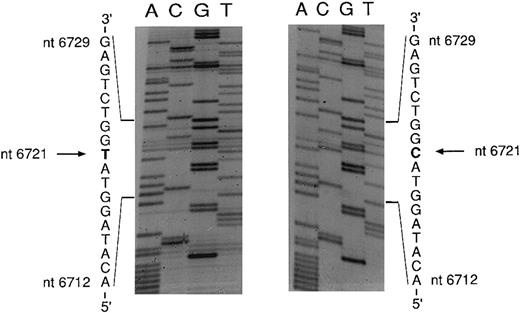
An mtDNA fragment containing the mutation was cloned in E coli. Five clones were sequenced; three yielded the wild-type sequence and two showed a point mutation characterized by a T → C transition at nt 6721 (Fig 5). The mutation changes an ATG codon to ACG, thus causing an amino acid change from methionine to threonine. Complementary strands of mtDNA were also sequenced and showed the expected A → G transition at nt 6721 (data not shown).
Sequencing of the COX I mutation in patient no. 1. mtDNA light strand; normal sequence on the left, mutated sequence on the right. The pathologic clone showed a T → C transition at nt 6721.
Sequencing of the COX I mutation in patient no. 1. mtDNA light strand; normal sequence on the left, mutated sequence on the right. The pathologic clone showed a T → C transition at nt 6721.
On RFLP analysis in patient no. 1, we also detected a hitherto unknown homoplasmic Taq I polymorphism at nucleotide position 8958 (Fig 6). Cloning and sequencing (data not shown) showed a C → T transition, changing the Taq I recognition sequence TCGA to TTGA. This polymorphism, located within the gene for ATPase subunit 6, is not associated with an amino acid change. As shown in Fig 6, the polymorphism was present in the blood cells of patient no. 1 and her two daughters. This inherited homoplasmic genetic marker stands in contrast to the acquired heteroplasmic mutation at nt 6721, which occurred in the patient's hematopoietic system and was detectable neither in cells of extramedullary origin nor in the patient's relatives.
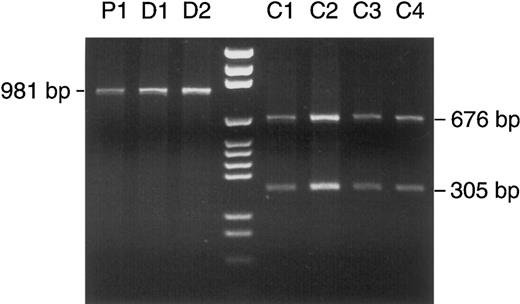
Detection of a homoplasmic polymorphism in patient no. 1 and her two daughters. PCR-amplified mtDNA segment no. 9 (nt 8282-9262) was treated with Taq I. According to the Cambridge sequence, Taq I has a restriction site at nt 8957, which divides the segment of 981 bp into two fragments of 676 and 305 bp. In patient no. 1 (lane P1) and her two daughters (D1 and D2), complete (homoplasmic) loss of the restriction site left the PCR product intact after Taq I digestion, whereas the expected cleavage occurred in four other patients (C1 through 4). Lane 4, DNA size marker (pGEM).
Detection of a homoplasmic polymorphism in patient no. 1 and her two daughters. PCR-amplified mtDNA segment no. 9 (nt 8282-9262) was treated with Taq I. According to the Cambridge sequence, Taq I has a restriction site at nt 8957, which divides the segment of 981 bp into two fragments of 676 and 305 bp. In patient no. 1 (lane P1) and her two daughters (D1 and D2), complete (homoplasmic) loss of the restriction site left the PCR product intact after Taq I digestion, whereas the expected cleavage occurred in four other patients (C1 through 4). Lane 4, DNA size marker (pGEM).
Patient No. 2
When the mutation at nt 6721 had been detected in patient no. 1, the other patients in our series were screened for mutations in the COX I region by TGGE analysis. As a result, a further COX I mutation was found in patient no. 2 that had not been detected by RFLP analysis. Figure 7 shows a typical heteroduplex finding in perpendicular TGGE, and Fig 8 shows a parallel TGGE with heteroduplex bands of patients no. 1 and 2 in comparison with normal findings. DNA sequencing showed that the mutation in patient no. 2 was a T → C transition at nt 6742 (Fig 9). This mutation changes an amino acid from isoleucine (ATT) to threonine (ACT). It also creates a restriction site for Bsr I (ACTGGN ↓), which permitted screening of different cell lineages in this patient, too. Figure 10 clearly shows that skin fibroblasts and buccal mucosa cells were not affected by the mutation, whereas myeloid cells showed the abnormal restriction pattern. Analysis of band intensities showed a proportion of 52% mutated mtDNA in the bone marrow, 59% in a peripheral blood sample, and 71% in platelets. Faint pathologic bands in B and T lymphocytes could again be due to contamination with phagocytic cells during immunomagnetic bead isolation of lymphocytes or represent a low percentage of mutated mtDNA in lymphocytes. As in patient no. 1, the data suggest that the mutation was restricted to the hematopoietic system.
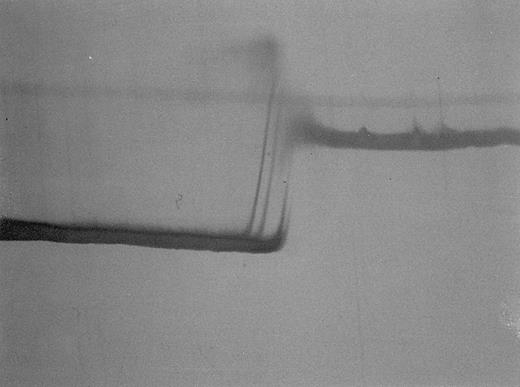
Perpendicular TGGE in patient no. 2. MtDNA fragment, nt 6504-6923. 5% polyacrylamide gel, silver staining. The typical heteroduplex pattern (two clearly separated heteroduplex bands on the left, two homoduplex bands not clearly separated on the right) indicates the presence of two homologous mtDNA species (wild-type and mutated), which after a cycle of denaturation and renaturation produce hybrids differing in their melting behavior.
Perpendicular TGGE in patient no. 2. MtDNA fragment, nt 6504-6923. 5% polyacrylamide gel, silver staining. The typical heteroduplex pattern (two clearly separated heteroduplex bands on the left, two homoduplex bands not clearly separated on the right) indicates the presence of two homologous mtDNA species (wild-type and mutated), which after a cycle of denaturation and renaturation produce hybrids differing in their melting behavior.
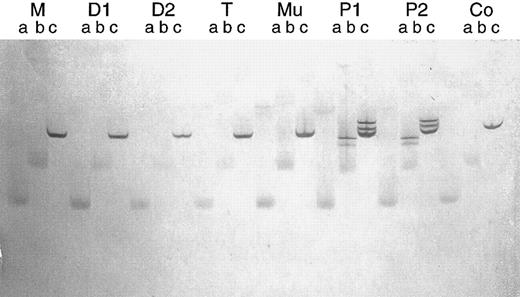
Parallel TGGE. Same mtDNA fragment as in Fig 7. After denaturation and renaturation, each PCR product was brought onto the gel in triplicate at time intervals of 15 minutes (lanes a, b, and c, respectively). The running time was too long for the samples designated a and b, so that bands became diffuse. Clear bands can be seen in lanes c. M, D1, and D2, normal bands in the mother and two daughters of patient no. 1, respectively; T and Mu, normal bands in T lymphocytes and buccal mucosa cells of patient no. 1, respectively; P1 and P2, heteroduplex bands in patients no. 1 and 2, respectively; Co, normal bands in an additional control person.
Parallel TGGE. Same mtDNA fragment as in Fig 7. After denaturation and renaturation, each PCR product was brought onto the gel in triplicate at time intervals of 15 minutes (lanes a, b, and c, respectively). The running time was too long for the samples designated a and b, so that bands became diffuse. Clear bands can be seen in lanes c. M, D1, and D2, normal bands in the mother and two daughters of patient no. 1, respectively; T and Mu, normal bands in T lymphocytes and buccal mucosa cells of patient no. 1, respectively; P1 and P2, heteroduplex bands in patients no. 1 and 2, respectively; Co, normal bands in an additional control person.
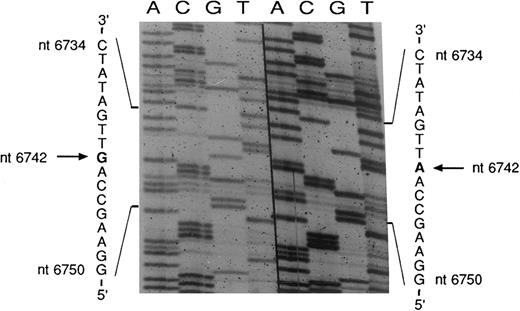
Sequencing of the COX I mutation in patient no. 2. mtDNA heavy strand; normal sequence on the right, mutated sequence on the left. The pathologic clone showed an A → G transition at nt 6742. The complementary light strand was also sequenced and showed the expected T → C transition (data not shown).
Sequencing of the COX I mutation in patient no. 2. mtDNA heavy strand; normal sequence on the right, mutated sequence on the left. The pathologic clone showed an A → G transition at nt 6742. The complementary light strand was also sequenced and showed the expected T → C transition (data not shown).
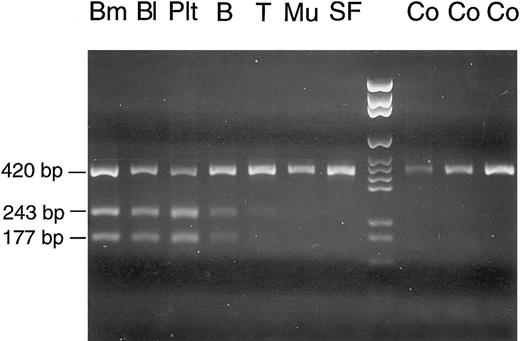
Cell lineage involvement of the COX I mutation in patient no. 2. Total DNA was extracted from different types of cells: Bm, bone marrow; Bl, peripheral blood; Plt, platelets; B, B lymphocytes; T, T lymphocytes; Mu, buccal mucosa cells; SF, skin fibroblasts; Co, control bone marrow samples; lane 8, DNA size marker. In affected cell lineages, the COX I mutation creates a restriction site for Bsr I in the mtDNA segment nt 6504-6923, so that a considerable proportion of the 420-bp PCR product is cleaved into fragments of 243 and 177 bp.
Cell lineage involvement of the COX I mutation in patient no. 2. Total DNA was extracted from different types of cells: Bm, bone marrow; Bl, peripheral blood; Plt, platelets; B, B lymphocytes; T, T lymphocytes; Mu, buccal mucosa cells; SF, skin fibroblasts; Co, control bone marrow samples; lane 8, DNA size marker. In affected cell lineages, the COX I mutation creates a restriction site for Bsr I in the mtDNA segment nt 6504-6923, so that a considerable proportion of the 420-bp PCR product is cleaved into fragments of 243 and 177 bp.
Interspecies Comparison
The positions of both mutations are in close proximity to each other within subunit I of cytochrome oxidase and affect amino acids M273 and I280 of the human sequence. During evolution, methionine and isoleucine in these positions have been strongly conserved, as shown by interspecies comparison (Table 1). Furthermore, high resolution screening of human populations involving hundreds of individuals has never shown restriction site polymorphisms at nt 6721 or 6742.36
mtDNA From Clonal Cultures
Whereas erythropoietic cell growth in culture was completely lacking in patient no. 1, erythropoietic clusters and some smaller colonies were observed in patient no. 2. Growth of colony-forming units-granulocyte (CFU-G) and colony-forming units-monocyte (CFU-M) appeared unimpaired in both patients. PCR amplification and RFLP analysis of the mtDNA segment containing the COX I mutation showed that all clusters and colonies that we picked (erythropoietic, granulocytic, and monocytic) showed the same pathologic restriction pattern as the mtDNA derived from uncultured blood or bone marrow cells of the respective patient (data not shown). The percentage of mutated mtDNA varied between different colonies, but the average percentage was similar to that observed in whole blood or bone marrow samples. In patient no. 2, there was no significant difference in the percentage of mutated mtDNA between erythropoietic clusters and granulocytic colonies.
DISCUSSION
The finding of mtDNA mutations affecting cytochrome c oxidase supports our hypothesis that AISA is a disorder involving mitochondrial respiratory chain dysfunction.14 Cytochrome c oxidase (COX) is a good candidate for playing a major role in mitochondrial iron metabolism. A strong hint in this direction comes from the copper connection,37 which refers to the close relation between copper and iron in hematopoiesis, including the role of copper in mitochondrial iron metabolism. COX is the only respiratory chain complex that contains copper, namely CuA1 and CuA2 in subunit II and CuB in subunit I.38,39 Copper is an essential part of the metal redox centers of this enzyme complex. Intriguingly, copper deficiency can cause sideroblastic anemia, with usually moderate numbers of ring sideroblasts.40-47 Williams et al48 found that mitochondria isolated from copper-deficient animals were deficient in COX activity and failed to synthesize heme from Fe3+ and protoporphyrin at the normal rate. The rate of heme synthesis correlated with the COX activity. Through experiments with respiratory chain inhibitors, these investigators confirmed that an active and intact electron transport chain is required for the reduction of Fe3+ on the inner mitochondrial membrane and for a supply of Fe2+ as substrate for ferrochelatase. Williams et al48 suggested that electrons may be donated to Fe3+ from cytochrome c oxidase. Because both COX I mutations that we identified are located in transmembrane helix no. VII in the immediate vicinity of the heme a3 -CuB center,38 it is tempting to speculate that this metal redox center is involved in supplying electrons to Fe3+.
The copper connection has recently been strengthened by the finding that high-affinity transmembrane uptake of iron in Saccharomyces cerevisiae requires copper. This is explained by the involvement of a copper-containing oxidoreductase, FET3, in iron uptake.49 50 Yeast mutants defective in the fet3 gene are deficient in high-affinity Fe2+ transport, and copper depletion of wild-type S cerevisiae also results in a selective reversible decrease in Fe2+ transporter activity. The FET3 protein exhibits extensive similarity to the familiy of blue multicopper oxidoreductases, an enzyme superfamily that also includes mitochondrial cytochrome c oxidase.
Both COX I mutations fulfill the criteria that are generally applied to exclude nonpathogenic polymorphisms of mtDNA, namely (1) the observed nucleotide change is absent in unaffected individuals; (2) it occurs in an evolutionary conserved region; and (3) the mutation displays heteroplasmy. As far as the first criterion is concerned, data are available from extensive population genetic studies involving more than 1,800 individuals from various ethnic backgrounds.36,51-61 To our knowledge, these studies never identified nucleotide changes in positions 6721 or 6742 of the mitochondrial genome. The second criterion is not reliable, because mutations of conserved nucleotides can occur without having a direct effect on the clinical phenotype.62 The third criterion, heteroplasmy, requires special consideration. When an mtDNA mutation arises, it creates an intracellular mixture of mutant and normal mtDNA molecules called heteroplasmy. Heteroplasmy is considered an indicator for pathogenic mutations because it is typical of diseases associated with defects of mtDNA, but generally absent in normal individuals.63 64 Accordingly, the COX I mutations in patients no. 1 and 2 are heteroplasmic, whereas the inherited nonpathogenic polymorphism at nt 8958 in patient no. 1, her mother, and her two daughters is homoplasmic. However, it must be realized that heteroplasmy of an acquired mtDNA mutation in the hematopoietic system is not quite the same as heteroplasmy in postmitotic tissues such as muscle or nerve. The matter is more complicated in the bone marrow because the dynamics of hematopoietic cell proliferation must be taken into account. Thus, the degree of heteroplasmy that we observed with the two COX mutations may reflect a balance between (1) a putative replicative advantage of mutated mtDNA over wild-type mtDNA, (2) the proliferative advantage of the cells belonging to the pathologic clone of sideroblastic anemia, and (3) a proliferative disadvantage of cells containing a particularly high proportion of mutated mtDNA.
A very interesting aspect of heteroplasmy is its potential to explain a peculiar feature of AISA, namely the heterogeneity in the degree of mitochondrial iron accumulation. Not all erythroblasts in AISA are ring sideroblasts, and not all ring sideroblasts show the same degree of mitochondrial iron overload. This heterogeneity has also been demonstrated within single erythroid colonies cultured in vitro.65-67 Whereas such intraclonal heterogeneity is hard to explain in terms of mutations involving nuclear genes, mtDNA mutations could easily account for the phenomenon. Each cell contains hundreds of mitochondria and thousands of copies of the mitochondrial genome. Therefore, cells can harbor mixtures of mutant and normal mtDNAs (heteroplasmy), and each time a heteroplasmic cell divides, the mutant and normal mtDNAs are randomly segregated into the daughter cells.15 In each cell, the severity of the resulting respiratory chain defect depends on the proportion of mutated mtDNA. A heterogeneous population consisting of normal and grossly abnormal as well as intermediate cells can thus be the consequence of an mtDNA mutation.
It is conceivable that the COX mutations that we identified mainly disturb the process of mitochondrial iron reduction, without producing significant impairment of other respiratory chain functions. Although such a defect would interfere with the major metabolic activity of erythroblasts (ie, heme synthesis), causing considerable damage through secondary effects of iron loading, its consequences may be minimal for other cell lineages. In accordance with a defect primarily disturbing erythropoiesis, we found that erythropoietic colony growth (burst-forming unit-erythroid [BFU-E] and colony-forming unit-erythroid [CFU-E]) was virtually absent or severely impaired, whereas growth of CFU-GM was not significantly reduced, despite the fact that cells in granulocyte and macrophage colonies carried the same mtDNA mutation.
In general, mtDNA mutations associated with mitochondrial diseases arise in the female germline and are present in various tissues, albeit at widely differing percentages. To our knowledge, the two COX mutations described (and a mitochondrial tRNA mutation that we recently reported23 ) are the first examples of disease-associated mtDNA mutations that are specifically related to a particular organ, ie, the hematopoietic system. As a prerequisite for this unusual constellation, a mtDNA mutation must arise in a hematopoietic stem cell, which in turn must populate the bone marrow in a dominant fashion. The clonal expansion that is necessary to spread the mtDNA mutation in the bone marrow cannot be effectuated by the mtDNA mutation per se, because severe mutations of mtDNA are known to produce a growth handicap rather than a growth advantage for proliferating cells.25,68 69 A hematopoietic stem cell affected with a pathogenic mtDNA mutation must therefore acquire an additional nuclear mutation conferring growth advantage to become capable of establishing a clonal bone marrow disease such as sideroblastic anemia. In patient no. 2, the 5q− anomaly may have been the transforming event.
The mtDNA mutations reported here fit into the mitochondrial-DNA mutation hypothesis of cell ageing,70,71 which is based on (1) an approximately 10 to 20 times higher frequency of mutations in mitochondrial as compared with nuclear DNA; (2) the compactness of the mitochondrial genome (no introns), increasing the probability that mutations will affect functionally important regions; (3) the lack of sufficient repair mechanisms; and (4) the increased production of oxygen radicals in mitochondria with increasing age, promoting chemical modification of mitochondrial proteins and mtDNA. Apparently, hematopoietic stem cells must be included among cells suffering age-related changes to their mitochondrial genome.25
Supported by Deutsche Forschungsgemeinschaft (Grant No. Ga 527/1-1), Leukämie-Liga e.V., and Ministerium für Wissenschaft und Forschung des Landes Nordrhein-Westfalen.
Presented in part at the Joint Annual Meeting of the German and Austrian Societies for Hematology and Oncology, October 4-7, 1996, Düsseldorf, Germany.
Address reprint requests to Norbert Gattermann, MD, PhD, Klinik für Hämatologie, Onkologie und Klinische Immunologie, Heinrich-Heine-Universität, Moorenstraβe 5, D-40225 Düsseldorf, Germany.