In this issue of Blood, Ma et al1 report on the loss of G protein–coupled receptor, class C, group 5, member D (GPRC5D) expression post–anti-GPRC5D chimeric antigen receptor T-cell (CAR-T) therapy in multiple myeloma (MM), and unravel a potentially reversible silencing mechanism driven by hypermethylation of its regulatory genomic loci.
GPRC5D targeting CAR-T cells and bispecific T-cell engagers (TCEs) have demonstrated deep responses in relapsed refractory MM, even among patients previously exposed to B-cell maturation antigen (BCMA)-targeting therapies.2-4 However, the majority of recipients of anti-GPRC5D CAR-T or TCE therapy eventually relapse.
The reported mechanisms of acquired resistance to anti-GPRC5D immunotherapies to date consistently point to antigen escape as the predominant driver, particularly after anti-GPRC5D TCEs. Several reports have demonstrated that relapse after talquetamab is associated with GPRC5D biallelic deletions, monoallelic deletion coupled with single nucleotide variations or indels, as well as epigenetic silencing of GPRC5D enhancer and promoter regions.5-7 For acquired resistance to anti-GPRC5D CAR-T cells, 6 of 10 patients relapsing post-MCARH109 demonstrated absent GPRC5D expression by immunohistochemistry,4 including 1 confirmed case of biallelic GPRC5D deletion by whole-exome sequencing.8
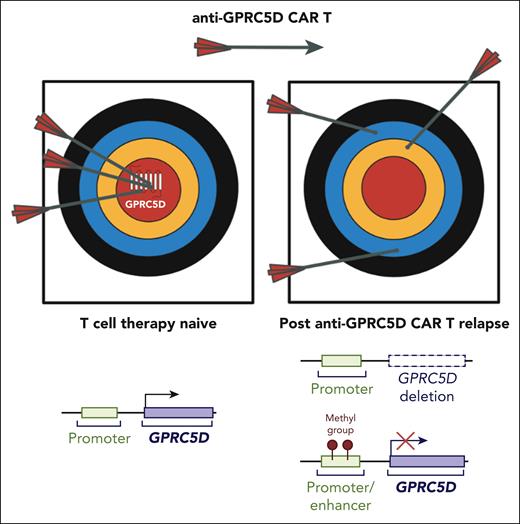
Ma et al leveraged whole-genome sequencing and whole-genome bisulfite sequencing of tumor samples from 10 patients with MM progressing on anti-GPRC5D CAR-T therapy to investigate the genomic and epigenetic events leading to anti-GPRC5D immune evasion. In all 10 cases, the sorted CD138+ MM cells demonstrated downregulation (n = 2) or complete loss of GPRC5D expression (n = 8), reinforcing the notion that GPRC5D antigen loss manifests as a class effect after GPRC5D targeting therapies, regardless of whether TCE or CAR-T modalities are used. Distinct genomic and epigenetic processes were observed in the post–CAR-T relapse GPRC5D-negative tumors, which included (1) biallelic deletion of GPRC5D, (2) biallelic deletion involving GPRC5D promoter and enhancer sites, as well as (3) hypermethylation in GPRC5D regulatory regions (see figure).
Mechanisms of GPRC5D loss mediating resistance to anti-GPRC5D CAR T therapy. In the T-cell therapy–naive setting, GPRC5D is highly expressed on multiple myeloma (MM) tumor cells, enabling effective targeting by anti-GPRC5D CAR T cells (arrows hitting the bullseye). After relapse, tumor cells evade immune recognition through loss of GPRC5D, making anti-GPRC5D CAR T therapy ineffective. Mechanisms of GPRC5D antigen escape include genomic deletion of the GPRC5D locus and epigenetic silencing via promoter/enhancer methylation. CAR, chimeric antigen receptor. Figure generated using BioRender.
Mechanisms of GPRC5D loss mediating resistance to anti-GPRC5D CAR T therapy. In the T-cell therapy–naive setting, GPRC5D is highly expressed on multiple myeloma (MM) tumor cells, enabling effective targeting by anti-GPRC5D CAR T cells (arrows hitting the bullseye). After relapse, tumor cells evade immune recognition through loss of GPRC5D, making anti-GPRC5D CAR T therapy ineffective. Mechanisms of GPRC5D antigen escape include genomic deletion of the GPRC5D locus and epigenetic silencing via promoter/enhancer methylation. CAR, chimeric antigen receptor. Figure generated using BioRender.
Notably, MM tumors from 5 of the 7 patients who relapsed post–anti-GPRC5D CAR-T therapy without GPRC5D structural variations, single nucleotide variations, or indels exhibited a genome-wide hypermethylation signature, involving key differentially methylated CpG sites within the GPRC5D transcriptional regulatory regions, in contrast to samples from anti-GPRC5D therapy-naive patients. This inverse correlation between hypermethylation of GPRC5D regulatory sites and GPRC5D expression was further corroborated in established MM cell lines. Importantly, functional assays revealed that treatment of these MM cell lines with azacytidine-induced GPRC5D transcript and protein expression, providing additional support that hypermethylation in the identified GPRC5D regulatory regions contributes to GPRC5D downregulation. Given the strong association between epigenetic imprinting and GPRC5D loss, the authors proposed a novel therapeutic strategy using demethylating agents to restore GPRC5D expression and circumvent antigen downregulation.
Based on the current report and the cumulative data thus far, it is becoming increasingly apparent that the rates of target antigen loss after anti-GPRC5D vs anti-BCMA CAR-T therapy differ significantly. In the KarMMa trial, BCMA-negative relapse was detected in 4% (3 of 71) of relapses after idecabtegene-vicleucel, with 1 confirmed case of biallelic TNFRSF17 deletion.9 In contrast, GPRC5D antigen-null or -low escape appears to occur in the majority of patients post–anti-GPRC5D CAR-T therapy.3,4,8 This raises the question: Is the GPRC5D gene and its regulatory regions more susceptible to genomic instability or epigenetic reprogramming? Notably, even among immunotherapy-naive patients, GPRC5D monoallelic deletions are observed in 15% of cases, whereas TNFRSF17 monoallelic deletions are seen in only 4%.10 Although cognizant of the differences in CAR T-cell function and persistence among variable CAR T-cell products, the stark contrast in BCMA vs GPRC5D antigen escape rates post–CAR-T therapy suggests inherent differences in the genomic and epigenetic landscape of these target genes, as well as their differential mutability. This disparity may, in part, reflect their essentiality for plasma cell survival, with BCMA being indispensable and thus subject to stricter regulatory controls or protective mechanisms that limit its loss, whereas GPRC5D, with no known function in clonal plasma cells, may be more prone to silencing or deletion.
Given the propensity for GPRC5D antigen escape, combining anti-GPRC5D therapies with other anti-MM agents, such as immunomodulatory drugs or Cereblon E3-ligase modulators, anti-CD38 antibodies, or proteasome inhibitors, as well as dual immune targeting, are being investigated in clinical trials. The rationale for these combination strategies is to promote T-cell expansion, enhance immunogenic cell death, modulate the immunosuppressive environment, as well as eliminate antigen-negative escape clones. Whether similar combinatorial approaches can extend the durability of anti-GPRC5D CAR-T therapies will need to be evaluated in future clinical trials.
The potential role of hypomethylating agents is particularly noteworthy in MM immunotherapy, as they may reverse epigenetic silencing and restore suppressed target antigen expression. Especially if the global hypermethylation signature observed in GPRC5D antigen–negative clones extends to other antigens, such as BCMA silencing as the authors suggest, hypomethylating agents could potentially enhance the efficacy and durability of BCMA-targeted therapies as well. Further studies, however, are necessary to determine whether BCMA is similarly prone to epigenetic silencing. Of note, other transcriptional regulatory agents, such as all-trans retinoic acid, have been studied in combination with anti-GPRC5D CAR-T therapy with limited clinical benefit despite preclinical evidence of GPRC5D upregulation.3
Future research will need to provide additional insight into the clonal dynamics and kinetics of these GPRC5D mutant and/or silent clones. In particular: (1) What are the mutational processes driving GPRC5D genomic alterations and the mediators of its epigenetic imprinting? (2) Are these events stochastically occurring and only emerging due to the high therapeutic pressure of immune therapy? and (3) Are these mutant clones detectable prior to or during immune therapy exposure before biochemical or clinical relapse? As with all scientific inquiry, our ability to detect these events is dependent on the limit of detection of the screening assays such as is the case with bulk whole genome sequencing and quantitative polymerase chain reaction. Longitudinal bulk or single cell genomic and epigenetic profiling of clonal plasma cells before, during, and after relapse will be required to provide deeper insights into the dynamics of antigen escape.
Conflict-of-interest disclosure: The authors declare no competing financial interests.