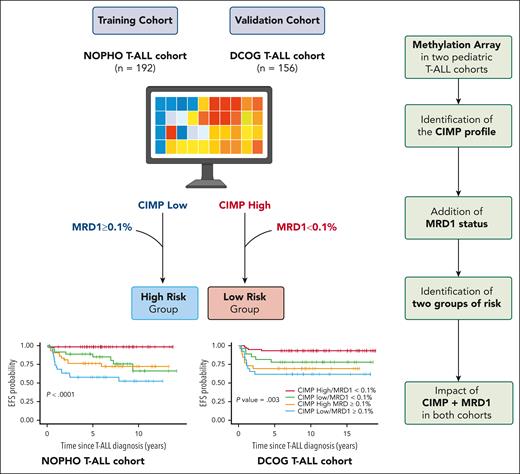
In this issue of Blood, Schäfer Hackenhaar et al1 propose a DNA methylation-based stratification for pediatric patients with T-cell acute lymphoblastic leukemia (T-ALL). The authors identified 2 distinct epigenetic phenotypes, namely CpG island methylator phenotype (CIMP) low and high, and demonstrated the prognostic relevance of this CIMP classification, identifying both high-risk and low-risk groups of patients when combined with minimal residual disease (MRD) status at the end of induction (EOI).
T-ALLs remain a significant challenge in both pediatric and adult hematology. Despite substantial improvements in overall survival (OS) over the past decade,2 relapses continue to pose a formidable obstacle due to the highly refractory nature of the disease at recurrence. Identifying patients at high risk of relapse is an urgent and unmet need. Although numerous studies have previously identified potential molecular markers to predict for a high risk of relapse and also to predict low-risk profiles,3 so far, only MRD levels are used for stratification in T-ALL. Unlike B-cell precursor ALL, T-ALL still lacks robust biomarkers that reliably predict for relapse risk but also are able to identify patients at low risk who could benefit from deescalated therapy.
Over the past decade, advancements in genomic research have demonstrated the critical role of gene alterations in T-ALL. More recently, next-generation sequencing, single-nucleotide polymorphism array, and RNA-sequencing analyses have uncovered novel gene alterations or deregulations with potential prognostic value.4-6 Pölönen et al conducted a comprehensive analysis of over 1300 pediatric T-ALL cases, leading to the proposal of an integrative but complex stratification framework based on nearly 15 distinct genetic subgroups of T-ALL.7
T-ALL is among a subset of cancers with the highest frequency of mutations in enzymes regulating epigenetic processes, suggesting that deregulation of the epigenetic landscape may play an essential role in T-ALL leukemogenesis.8 However, epigenetic features remained an underexplored area in pediatric T-ALL. Although an earlier study reported that an epigenetic approach could help to determine different clusters with prognostic impact in adult T-ALL,9 To our knowledge, Schäfer Hackenhaar et al were the first to report the impact of epigenetic profiles in limited cohorts of pediatric T-ALL.10
In this study the authors performed array-based methylome analysis in pediatric T-ALL based on 2 pediatric cohorts: the Nordic Society of Paediatric Haematology and Oncology (NOPHO) AALL2008 (n = 192) and the Dutch Childhood Oncology Group (DCOG) ALL10/11 (n = 156). CIMP classification was performed using a CpG panel consisting of nearly 1000 CpG sites previously identified as the most differentially methylated in pediatric T-ALL. The authors identified 2 groups of patients, a CIMP-low group (40% of the patients in both cohorts) and a CIMP-high group (60%). The main clinical and biological characteristics at diagnosis, including sex, age, central nervous system involvement, and MRD status, were comparable between the 2 groups. However, the CIMP-low group exhibited significantly higher white blood cell counts. In term of prognosis, the CIMP profile and the level of MRD at EOI were associated with prognosis in univariate Cox model. The most significant finding of this study is that combining CIMP classification at diagnosis with MRD status at EOI significantly improved outcome prediction in both cohorts. These findings were identified in the Nordic NOPHO ALL2008 cohort and were validated in the pediatric patients with T-ALL from the Dutch DCOG ALL-10/ALL-11.
The poor-prognosis subgroup, characterized by a CIMP-low profile and MRD at EOI ≥ 0.1% (22% of NOPHO patients and 19% of DCOG patients), showed a cumulative incidence of relapse (CIR) of 29% and 23% and a 5-year OS of 59.7% and 65.4%. The good-prognosis subgroup had a CIMP-high profile and MRD at EOI < 0.1% (33% of NOPHO patients and 42% of DCOG patients). This subgroup had an exceptionally low CIR of 0% and 3.4% and a 5-year OS of 98.2% and 94.8%, respectively (see figure).
Epigenetic-based stratification in pediatric T-ALL. In this study the authors performed array-based methylome analysis in pediatric T-ALL issued from 2 pediatric cohorts: the Nordic NOPHO AALL2008 (n = 192) and the Dutch DCOG ALL10/11 (n = 156). The authors identified 2 distinct epigenetic phenotypes, namely, CIMP low and high, and demonstrated the prognostic relevance of this CIMP classification, identifying both high-risk and low-risk groups of patients when combined with MRD status at the EOI. Created with BioRender.com.
Epigenetic-based stratification in pediatric T-ALL. In this study the authors performed array-based methylome analysis in pediatric T-ALL issued from 2 pediatric cohorts: the Nordic NOPHO AALL2008 (n = 192) and the Dutch DCOG ALL10/11 (n = 156). The authors identified 2 distinct epigenetic phenotypes, namely, CIMP low and high, and demonstrated the prognostic relevance of this CIMP classification, identifying both high-risk and low-risk groups of patients when combined with MRD status at the EOI. Created with BioRender.com.
Multivariate Cox model analysis revealed that both a CIMP-low profile and MRD at EOI ≥ 0.1% were independent prognostic markers of relapse in both cohorts. An additional important observation from the study was that integrating DCOG MRD measurements at day 15 with the CIMP profile provided a strong predictor of relapse, allowing for earlier risk stratification and the potential for timely, adapted treatment interventions.
The authors integrated their epigenetic analysis with a whole-genome transcriptome approach, uncovering notable associations between epigenetic profiles and gene expression patterns. Patients classified as CIMP-low were enriched for TAL1 and BEX1 expression, and CIMP-high patients exhibited increased deregulation of homeobox oncogenes such as TLX3, HOXA9, HOXA10, and NKX2.1. They also identified significant gene expression changes in CIMP-low patients, particularly involving genes related to cell proliferation and migration.
A key limitation of this study lies in the lack of integration with previously identified molecular alterations (eg, NOTCH1, FBXW7, RAS, PTEN)4,5 and transcriptomic predictive signatures.6 Future research is essential to determine the most relevant and predictive alterations by combining these molecular markers with the current findings.
Despite these limitations, Schäfer Hackenhaar and colleagues have introduced the first epigenetics-based classifier for pediatric T-ALL. This classifier, when combined with MRD at EOI, demonstrates a robust ability to identify high-risk and low-risk patient groups, paving the way for improved risk stratification and personalized treatment strategies. We now have comprehensive insights into the mutational, transcriptomic, and epigenetic landscapes of this disease. The current challenge is to prioritize and integrate these markers to enhance predictive accuracy, hopefully finally proving a comprehensive molecular-based classification of pediatric T-ALL.
Conflict-of-interest disclosure: The authors declare no competing financial interests.