In this issue of Blood, Yang et al1 describe how ammonium produced by nitrogen-recycling gut microbial taxa propagates multiple myeloma–related bone disease.
Multiple myeloma is caused by proliferation of malignant plasma cells in the bone marrow niche. Skeletal manifestations of myeloma, namely focal lytic bone lesions, are common and morbid sequelae, affecting ≥80% of patients at diagnosis. Identification and treatment of myeloma-related bone disease is an important component of patient care.2 Several mechanisms underpin disruption of homeostatic bone remodeling in patients with myeloma.3 Although many basic and translational investigations of myeloma focus on tumor cells per se, untangling tumor-extrinsic drivers of bone disease is also relevant to identify pathways that could be therapeutically exploited in patients.
In their study, Yang and colleagues observe that patients with a higher burden of lytic bone lesions had a higher relative abundance of nitrogen-recycling taxa, such as Enterobacter cloacae and Klebsiella pneumoniae, in fecal specimens. Using elegant microbiota transplantation experiments, including with genetically modified microbial strains, the authors demonstrate that production of ammonium via microbial deaminase enzymes augmented osteoclast functions, resulting in bone degradation in a syngeneic murine myeloma model. They also show how supplementation of a probiotic cocktail of microbes blunted colonization by E cloacae and reduced myeloma-driven bone destruction.
The study builds on the group’s prior work demonstrating that gut flora capture and recycle nitrogen into amino acids, such as glutamine, that support the protein turnover required by proliferating myeloma cells.4 Metabolism of amino acids by myeloma cells provides nitrogen substrates that feed bacterial ammonium production—an elegant example of symbiosis, which leads to the downstream deleterious effects on bone described in the article. The findings are novel and shed light on an interesting pathological mechanism linking gut microbiome to myeloma biology. Although preliminary, it is compelling to hypothesize that reducing ammonium production by therapeutically targeting the gut microbiome could serve as a paradigm for secondary prevention that reduces the risk of evolution of plasma cell dyscrasias into symptomatic multiple myeloma.
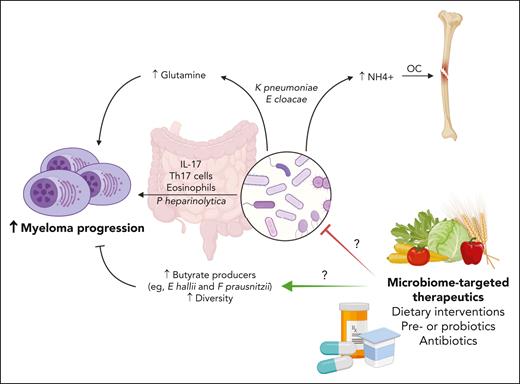
The study is among a growing body of work demonstrating a correlation between gut microbiome features, myeloma pathogenesis, and treatment responses (see figure). In addition to altered nitrogen recirculation, bacterial species that lead to an increase in interleukin-17 production (Prevotella heparinolytica) promote a microenvironment supportive of myelomagenesis (enriched with T-helper 17 cells and eosinophils).5 Increased microbial alpha diversity, generally considered a marker of overall microbiome health, and an increased relative abundance of butyrate-producing bacteria (eg, Eubacterium hallii and Faecalibacterium prausnitzii) seem to be features of individuals who attain deeper and more durable treatment responses after induction or in the context of maintenance treatment with the immunomodulatory drug lenalidomide.6,7 Gut microbial diversity has also been correlated with clinical outcomes in other contexts in myeloma, including after autologous hematopoietic cell transplantation.8 Those studies support the relevance of sampling gut flora in different disease and treatment contexts to discern relationships between bacterial metabolic pathways, myeloma proliferation, and host immune biology, with the goal of developing rationale for interventions that could synergize with evolving prevention and treatment approaches.
Role of gut microbiome in multiple myeloma. Targeting the microbiome may be a useful method to disrupt pathogenic processes mediated by gut microbes, such as the production of glutamine and ammonium (which activates osteoclasts) by the nitrogen-recycling bacteria E cloacae and K pneumoniae, or interleukin-17–mediated potentiation of tumor microenvironment via Th17 cells and eosinophils by P heparinolytica. Retrospective studies suggest strategies to increase the relative abundance of butyrate producers (eg, E hallii or F prausnitzii) or diversity may augment treatment responses. OC, osteoclasts; Th17, T-helper 17. Figure created with BioRender.com.
Role of gut microbiome in multiple myeloma. Targeting the microbiome may be a useful method to disrupt pathogenic processes mediated by gut microbes, such as the production of glutamine and ammonium (which activates osteoclasts) by the nitrogen-recycling bacteria E cloacae and K pneumoniae, or interleukin-17–mediated potentiation of tumor microenvironment via Th17 cells and eosinophils by P heparinolytica. Retrospective studies suggest strategies to increase the relative abundance of butyrate producers (eg, E hallii or F prausnitzii) or diversity may augment treatment responses. OC, osteoclasts; Th17, T-helper 17. Figure created with BioRender.com.
Manipulating the microbiome is a compelling approach to modify the natural history of myeloma, but prospective trials are needed.9 Ongoing clinical trials are testing whether empowering patients to improve their own health via dietary modifications can significantly alter the gut microbiome and affect the natural history of precursor plasma cell dyscrasias (refer to NCT05640843, NCT04920084, and NCT04497961 at https://clinicaltrials.gov/). Additional evaluations of targeted pre- or probiotics to promote or displace specific bacterial taxa, as well as targeted antibiotics, are warranted to determine whether these approaches can lead to a meaningful improvement in disease biomarkers or treatment responses. For example, a clinical trial testing the use of rifaximin to alter gut flora in patients with monoclonal gammopathy may find this intervention lowers the abundance of nitrogen recycling bacteria, reduces ammonium production, and disrupts the symbiotic pathway described by Yang et al (see NCT03820817). Thus, this work not only defines a potential therapeutic target for preventing myeloma-related bone disease but also provides confirmation that exploration of the complexities of the microbiome and its metabolic effects on health and disease is a rich area for continued study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.