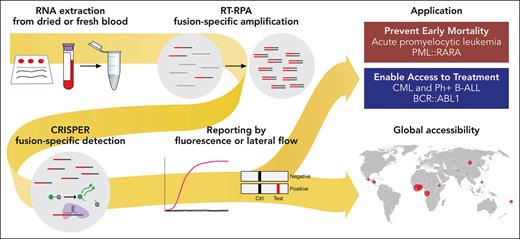
In this issue of Blood, Vedula et al report on a novel CRISPR-based RNA-fusion transcript detection assay, called SHERLOCK (specific high-sensitivity enzymatic reporter unlocking), for the rapid and reliable diagnosis of fusion-driven leukemias.1 SHERLOCK is a point-of-care (POC) test with 100% specificity and sensitivity, characterized by a readout time of about 1 to 2 hours. This novel test combines isothermal and target RNA reverse transcription and recombinase polymerase amplification with a customized Cas13-mediated detection that is coupled to cleavage of a collateral reporter. The diagnostic test is applicable in 2 assay readout methods: one uses fluorescence in a plate format, amenable to higher throughput, and the other lateral flow strips, also applicable as a POC test in lower-resource environments. SHERLOCK can be performed outside of molecular laboratory infrastructures because resource- and time-intensive processes such as thermocycling PCR and next-generation sequencing are eliminated (see figure).
SHERLOCK is a novel, highly sensitive and specific test for the detection of fusion-driven leukemia transcripts. It can be performed outside of molecular laboratories as a POC test and promises to be useful for the fast and reliable diagnosis of fusion-driven leukemias in low-resource environments. RT-RPA, reverse transcription and recombinase polymerase amplification. Professional illustration by Patrick Lane, ScEYEnce Studios.
SHERLOCK is a novel, highly sensitive and specific test for the detection of fusion-driven leukemia transcripts. It can be performed outside of molecular laboratories as a POC test and promises to be useful for the fast and reliable diagnosis of fusion-driven leukemias in low-resource environments. RT-RPA, reverse transcription and recombinase polymerase amplification. Professional illustration by Patrick Lane, ScEYEnce Studios.
Fusion oncogene-driven leukemias such as acute promyelocytic leukemia (APL), Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL), and chronic myeloid leukemia (CML) are rare hematological neoplasms.2 These types of leukemias are characterized by the cancer-defining fusion oncogene PML::RARA or BCR::ABL1 and respond exceptionally well to targeted treatments including arsenic trioxid combined with all-trans retinoic acid (ATRA) for APL3 or ABL tyrosine kinase inhibitors for Ph+ ALL4 and CML.5
Rapid and reliable detection is essential to confirm the diagnosis and select the appropriate targeted therapy. Metaphase cytogenetics, fluorescence in situ hybridization, or reverse transcription-polymerase chain reaction (RT-PCR) is currently used as the standard test for the diagnosis and to inform therapy. However, these tests are not available in most clinical laboratories and health care settings, and, consequently, require samples to be sent off to central testing laboratories or patients transferred to tertiary care centers.
Especially in APL, the delay of diagnosis as well as access to targeted treatment places patients at an unacceptable risk of increased mortality and morbidity.6 Once a highly fatal disease, APL is now a disease with long-term survival exceeding 90%. However, early death still reaches 30% in some real-world studies, which is mainly cause by generally preventable fatal hemorrhages affecting the cranial sites. High index of suspicion and recognition of APL with early administration of ATRA can be lifesaving. This can be fostered by rapid, reliable, and easily accessible diagnostic tests. In CML and Ph+ ALL, the urgency is less prominent, due to other effective cytoreductive agents that can be used as bridging therapy until diagnostic confirmation. However, these leukemias should not be missed because in Ph+ ALL prognosis can be substantially improved,7 and in CML half of patients with deep molecular responses maintain long-lasting treatment-free remission and the remaining reach a close to normal life expectancy with BCR-ABL1-targeted therapies.8
In high-income countries and tertiary care hospitals, the necessary molecular tests for the detection of fusion oncogenes are readily accessible. Specialized molecular laboratories provide these results within a turnaround time of 1 to 3 days. However, this is possible only if samples are prioritized, which may be increasingly problematic during times of shortage of staff also in higher-income countries. In contrast, community hospitals in lower-resource environments generally lack timely access to such costly infrastructures. Therefore, late detection or even missed diagnoses are relevant issues for patient care in lower-income environments.
Vedula et al further optimized SHERLOCK to enable the use outside of tertiary cancer centers and clinical laboratories. They validated their test using diagnostic samples from patients with APL and CML from academic centers as well as dried blood spots from 11 low-resource countries across 4 continents (Africa, Asia, Oceania, and North America). Their test demonstrated in both test environments 100% sensitivity and specificity as compared with RT-PCR standard-of-care tests. Although in the current study the test was applied in a tertiary care context and sample selection does not cover the full range of possible transcript variants, SHERLOCK can be adapted for the detection of other fusion oncogenes. In particular, the readout by a lateral flow strip is a simple and fast POC test with the potential to foster the rapid and reliable detection of disease-defining fusion-oncogenes outside of tertiary cancer centers or high-income countries.
Still, the utility and performance need to be confirmed in appropriately designed prospective studies within the targeted test environment. Moreover, the accessibility to the test should be ensured by adequate storage and shipment conditions as well as acceptable costs for the economically less privileged countries. Nonetheless, SHERLOCK represents a promising technological advance for the fast and reliable diagnosis of patients suffering from fusion-driven leukemias, especially in lower-resource environments.
Conflict-of-interest disclosure: N.B. declares no competing financial interests.