Key Points
Ezh2 is dispensable for fetal HSCs.
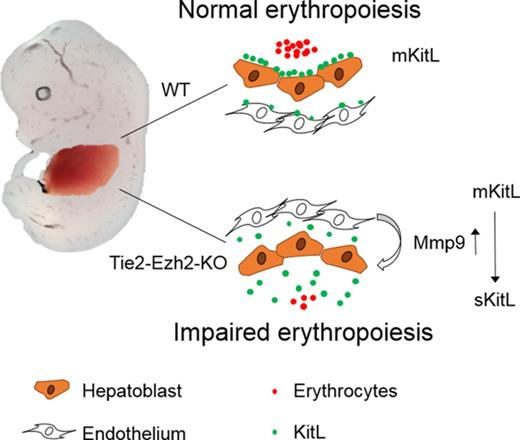
Inactivation of Ezh2 in FL endothelium leads to cell-extrinsically mediated anemia.
Abstract
Despite the well-established cell-intrinsic role of epigenetic factors in normal and malignant hematopoiesis, their cell-extrinsic role remains largely unexplored. Herein we investigated the hematopoietic impact of inactivating Ezh2, a key component of polycomb repressive complex 2 (PRC2), in the fetal liver (FL) vascular niche. Hematopoietic specific (Vav-iCre) Ezh2 inactivation enhanced FL hematopoietic stem cell (HSC) expansion with normal FL erythropoiesis. In contrast, endothelium (Tie2-Cre) targeted Ezh2 inactivation resulted in embryonic lethality with severe anemia at embryonic day 13.5 despite normal emergence of functional HSCs. Ezh2-deficient FL endothelium overexpressed Mmp9, which cell-extrinsically depleted the membrane-bound form of Kit ligand (mKitL), an essential hematopoietic cytokine, in FL. Furthermore, Mmp9 inhibition in vitro restored mKitL expression along with the erythropoiesis supporting capacity of FL endothelial cells. These data establish that Ezh2 is intrinsically dispensable for FL HSCs and provides proof of principle that modulation of epigenetic regulators in niche components can exert a marked cell-extrinsic impact.
Introduction
Ezh2 is a histone transferase and core component of the polycomb repressive complex 2 (PRC2) which mediates trimethylation of H3K27, a mark of transcriptionally silent chromatin.1 PRC2 is essential for early mouse embryonic development, as embryos that carry a null mutation of any of its core components die prior to gastrulation.2-4 Studies of conditional knockout (KO) models have provided fundamental insights into the role of Ezh2 in the regulation of hematopoiesis, revealing an essential role for PRC2 in lymphocyte differentiation.5,6 Subsequently, several groups have described the role of PRC2 in hematopoietic stem cell (HSC) function.7-11 Loss of Ezh2 has minimal impact on adult HSCs, likely because of redundancy with its homolog, Ezh1, which is crucial for preventing the senescence of adult HSCs.8-10 In contrast, the role of Ezh2 and PRC2 in fetal hematopoiesis remains less clear,8-12 with studies using different targeting approaches to inactivate Ezh2 implicating either no role8 or a major cell-intrinsic role10 of Ezh2 in the regulation of fetal HSCs and erythropoiesis. Importantly, deletion of PCR2 core components Suz1211 or Eed8 with a hematopoietic-specific Cre line (Vav-Cre) led to minor or no impact on fetal hematopoiesis, respectively.
As HSCs emerge from hemogenic endothelium,13 endothelium-specific Cre lines such as Tie2-Cre10,14 induce deletion in both endothelial and hematopoietic cells. Furthermore, endothelial and other vascular cells have a well-characterized role in the regulation of the adult bone marrow (BM) HSC niche,15 and a vascular HSC niche has also recently been identified in fetal liver (FL).16 We therefore reasoned that loss of Ezh2 function in FL vascular cells might impact FL hematopoiesis, either through disruption of HSC emergence from the hemogenic endothelium or by a cell-extrinsic microenvironmental impact of Ezh2-inactivated vascular cells on hematopoietic cells by regulating the expression of important hematopoietic niche factors. To explore this possibility, we inactivated Ezh2 in hematopoietic cells with (Tie2-Cre; Tie2-Ezh2-KO) or without (Vav-iCre; Vav-Ezh2-KO) simultaneous deletion in endothelial cells, allowing us to distinguish the hematopoietic impact of Ezh2 deletion in the FL vascular HSC niche from deletion in HSCs alone.
Methods
Mice
Ezh2fl/fl,5 mKitLfl/fl,17 Tie2-Cre,14 Vav-iCre,18 Gata1-EGFP,19 Rosa26-LSL-tdTomato,20 and Mmp9−/−21 mice have been described and had all been backcrossed to a C57BL/6 background. All mouse experiments were performed in accordance with UK Home Office project license 30/3103. Embryonic development was estimated considering the day of vaginal plug formation as 0.5 days postcoitum and somite pairs. Embryonic day 10.5 (E10.5) embryos have somite pairs that range from 31 to 36.
In vivo experiments
For transplantation experiments, C57BL/6 (CD45.1) recipient mice were lethally irradiated (10 Gy, split dose) before IV transplantation of 500 000 E12.5 total FL cells of indicated genotypes, together with 1 000 000 CD45.1 unfractionated BM competitor cells. CD45.2 and CD45.1 contributions to blood cell lineages were monitored every 4 weeks posttransplantation for 16 weeks. Analysis of hematological parameters was performed using a Sysmex KX-21N analyzer.
FACS analysis and sorting
Fluorescence-activated cell sorting (FACS) experiments were performed using LSRII, LSRFortessa, LSRFortessa X20, AriaII, AriaIII, and Aria Fusion cytometers (BD Biosciences). Data were analyzed using FlowJo software (TreeStar). Cells were incubated in Fc-block before being stained with antibodies. Gates were set using a combination of fluorescence-minus-one controls and also populations that are known to be negative for the antigen. Antibodies used are detailed in supplemental Table 1 (available on the Blood Web site).
FL tissue preparation
Dissected FLs at indicated fetal stage were digested in a mix of Collagenases IV (2 mg/mL; Worthington) and DNase I (200 U/mL; Calbiochem) at 37°C for 15 minutes. The dissociated cells were centrifuged at 300g for 6 minutes and resuspended in 10% fetal bovine serum/phosphate-buffered saline.
Whole-mount immunofluorescence staining and analysis
The yolk sac, head, limb buds, and lateral body wall were removed from the E10.5 embryos and fixed for 45 minutes in 4% paraformaldehyde/phosphate-buffered saline on ice. Whole-mount immunofluorescence staining was performed as described previously.22 Images were acquired with a Zeiss 780 confocal upright microscope and analyzed using Fiji and Imaris software. Antibodies used are detailed in supplemental Table 1.
Immunofluorescence staining and imaging
Freshly isolated E12.5 FLs were included in Tissue-Tek optimum cutting temperature compound for snap freezing, sliced at 7-µm thickness, and processed as described previously.17 Cytospin preparations were performed using Cytospin 4 (Thermo Shandon) by centrifuging at 350 rpm for 10 minutes. Slides were air-dried, fixed with acetone for 5 minutes at 4°C, and processed as described previously.17 Images were acquired with a Zeiss 780 confocal inverted microscope and analyzed using Fiji software. Antibodies used are detailed in supplemental Table 1.
Hematoxylin and eosin staining
Embryos were dissected and fixed in 4% paraformaldehyde for 2 hours at 4°C and processed as described previously.23
Chromatin immunoprecipitation (ChIP)
C166 (ATCC) and MS-5 (DSMZ) were cultured in Dulbecco’s modified Eagle medium (Gibco) and Iscove modified Dulbecco medium (Gibco), respectively, and supplemented with 10% fetal calf serum (Hyclone), 1% penicillin/streptomycin (PAA Laboratories), and 1% l-glutamine (Gibco). For H3K27ac and H3K27me3 ChIP, cells were fixed with 1% formaldehyde for 10 minutes. For Ezh2 ChIP, cells were fixed with 2 mM disuccinimidyl glutarate (Sigma) and 1% formaldehyde for 1 hour each. Fixed chromatin samples were fragmented using an S220 Focused-ultrasonicator (Covaris) to an average size of 200 to 500 bp. Immunoprecipitation was then performed using Protein A/G Dynabeads (Life Technologies). Antibodies and primers used are detailed in supplemental Table 1.
Quantitative real-time PCR analysis
Total FL RNA was purified using TRIzol (Invitrogen) as recommended. Reverse transcription was performed using SuperScript III (Invitrogen) with random hexamers. Complementary DNA was amplified and analyzed with specific Taqman probes (listed in supplemental Table 2) on an ABI 7500 Fast real-time polymerase chain reaction (PCR) instrument. Data were normalized to Hprt.
In vitro coculture and methylcellulose colony-forming assay
Six thousand to 10 000 E12.5 FL endothelial or stromal cells were sorted by FACS and cocultured with 150 FACS-sorted E12.5 FL Lin−Sca1+c-Kit+ (LSK) cells for 5 days at 37°C in 200 µL of StemSpan Serum-Free Expansion Medium (StemCell Technologies) supplemented with 1% penicillin/streptomycin, 1% l-glutamine, and cytokines mIL3 (5 ng/mL; PeproTech) and hTHPO (10 ng/mL; PeproTech). Media were half-changed at day 3. Nonadherent hematopoietic cells were harvested at day 5 and cultured with Methocult M3434 (Stem Cell Technologies) for 7 days at 37°C. Burst forming unit–erythroid colony formation was determined by 2,7-diaminofluorene (Sigma) staining. For Mmp9 inhibitor24 (10 nM, CAS 1177749-58-4; Millipore) treatment, the inhibitor or dimethyl sulfoxide (DMSO; control) were added to the coculture from day 1 and replenished at day 3 while changing media.
Five thousand MS-5 cells were cultured in 10% fetal calf serum/Iscove modified Dulbecco medium for 24 hours at 37°C prior to treatment with 10 mM GSK126, 10 μM Mmp9 inhibitor, or DMSO for 24 hours. Two hundred FACS-sorted BM LSK cells were then added to the coculture with or without drug treatment of 72 hours at 37°C in 200 µL of StemSpan Serum-Free Expansion Medium complete medium as indicated previously. Nonadherent hematopoietic cells were harvested at day 3 and cultured with Methocult M3436 (Stem Cell Technologies) for 7 days at 37°C. Burst forming unit–erythroid colony formation was determined by 2,7-diaminofluorene staining.
Mmp9 ELISA
Supernatant of coculture was collected by centrifuging for 5 minutes at 500 g to remove any nonadherent cells. Samples were assessed by murine Mmp9 enzyme-linked immunosorbent assay (ELISA) kits (R&D systems) according to the manufacturer's instructions. Optical density measurements were determined using a microplate reader (Molecular Devices-SpectraMaxM2e) set to 450 nm with wavelength correction set to 570 nm.
Statistical evaluation
Statistical analyses was performed using GraphPad Prism 7.0. All results are presented in graphs are the mean ± standard error of the mean (SEM) of at least 3 independent experiments, unless otherwise noted in the figure legends. Each exact n value is indicated in the corresponding figure legend. A 2-tailed unpaired Student t test was used for all analysis. FACS and immunofluorescence data presented are representative of at least 3 independent experiments that yielded similar results, unless otherwise noted in the figure legends.
Results
Loss of Ezh2 cell-intrinsically promotes fetal HSC expansion
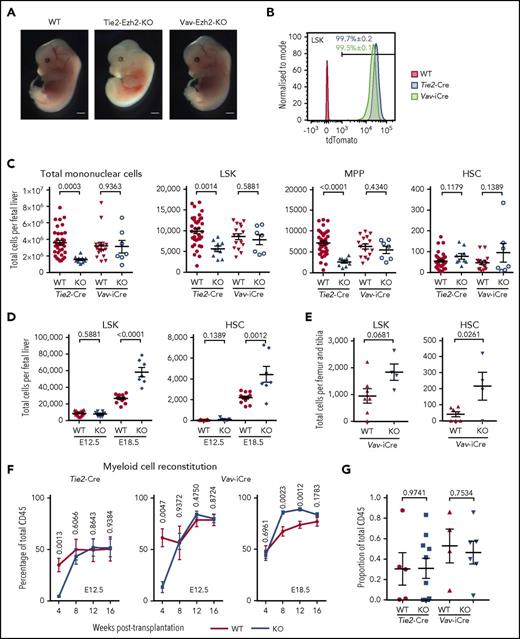
Comparison of Tie2-Ezh2-KO and Vav-Ezh2-KO embryos revealed a striking difference in survival, with embryonic lethality at E13.5 in Tie2-Ezh2-KO embryos in contrast to Vav-Ezh2-KO embryos, which survived at expected Mendelian ratios to E18.5 (P < .0001; Table 1). Tie2-Ezh2-KO embryos showed marked anemia in comparison with Vav-Ezh2-KO despite close to 100% deletion efficiency in all hematopoietic cells using both Cre lines at E12.5 (Figure 1A-B). As a previous study had implicated loss of HSCs as the cause of the anemia in Tie2-Ezh2-KO FL hematopoiesis,10 we first set out to compare numbers of hematopoietic stem/progenitor cells in Tie2-Ezh2-KO and Vav-Ezh2-KO embryos (supplemental Figure 1). E12.5 FL cellularity was reduced in Tie2-Ezh2-KO embryos (2.3-fold reduction; P = .0003), in contrast to normal cellularity in Vav-Ezh2-KO embryos (Figure 1C). Numbers of LSK cells, which encompass a heterogeneous mix of progenitor cells and infrequent HSCs,25,26 were reduced in Tie2-Ezh2-KO FL (1.8-fold reduction; P = .0014) but were not significantly affected in Vav-Ezh2-KO FL (Figure 1C). The reduction of LSK cells in Tie2-Ezh2-KO embryos was primarily accounted for by a reduction in LSKCD150−CD48+ multipotent progenitor cells (Figure 1C). In contrast, using a more stringent definition of HSCs,26 the number of LSKCD150+CD48− phenotypic HSCs was similar to wild type (WT) (Cre-negative Ezh2fl/fl or Ezh2fl/+) embryos in both Tie2-Ezh2-KO and Vav-Ezh2-KO FL (Figure 1C). Importantly, numbers of phenotypically defined HSCs were much closer to the number of repopulating units per E12.5 FL as compared with LSK cell numbers.27 HSCs emerge from the aorta-gonad-mesonephros region, and Tie2-Cre but not Vav-iCre will delete in the hemogenic endothelium.28 However, numbers of intra-aortic c-Kit+ cells and clusters were similar to WT embryos for both Tie2-Ezh2-KO and Vav-Ezh2-KO (supplemental Figure 2), supporting that Tie2-Cre mediated Ezh2 inactivation does not affect the emergence of HSCs.
Ezh2 is dispensable for fetal HSC. (A) Representative image of wild-type (WT; n = 21; 5 independent experiments), Tie2-Ezh2-KO (n = 4; 2 independent experiments), and Vav-Ezh2-KO (n = 7; 3 independent experiments) embryos at E13.5. Scale bars, 1 mm. (B) Representative FACS histogram plots showing the recombination efficiency of Tie2-Cre (n = 6; 1 experiment) and Vav-iCre (n = 3; 1 experiment) in E12.5 FL LSK cells using Rosa26-LSL-tdTomato reporter. (C) Absolute numbers of Tie2-Ezh2-WT (n = 37), Tie2-Ezh2-KO (n = 9), Vav-Ezh2-WT (n = 16), and Vav-Ezh2-KO (n = 7) total mononuclear cells, LSK, multipotent progenitor (MPP) (LSKCD150−CD48+), and HSC (LSKCD150+CD48−) per FL at E12.5. Data pooled from 5 (Tie2-Cre) and 4 (Vav-iCre) independent experiments. (D) Absolute numbers of Vav-Ezh2-WT (E12.5, n = 16; E18.5, n = 13) and Vav-Ezh2-KO (E12.5 and E18.5, n = 7) LSK cells and HSCs per FL at E12.5 (4 independent experiments) and E18.5 (3 independent experiments). (E) Absolute numbers of Vav-Ezh2-WT (n = 7) and Vav-Ezh2-KO (n = 4) LSK cells and HSC per femur and tibia at E18.5 (2 independent experiments). (F) Peripheral myeloid cell (Mac1+) reconstitution kinetics in CD45.1 mice transplanted with 500 000 CD45.2 Tie2-Ezh2-WT (n = 5), Vav-Ezh2-WT (n = 4), Tie2-Ezh2-KO (n = 10) or Vav-Ezh2-KO (n = 6) total E12.5 FL cells; Vav-Ezh2-WT (n = 11) or Vav-Ezh2-KO (n = 13) E18.5 FL cells, in all cases together with 1 000 000 CD45.1 competitor BM cells. Data are pooled from 2 (E12.5) or 3 (E18.5) independent experiments. (G) Contribution of CD45.2 E12.5 FL cells to HSC compartment at 16 weeks following transplantations shown in panel F. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
Ezh2 is dispensable for fetal HSC. (A) Representative image of wild-type (WT; n = 21; 5 independent experiments), Tie2-Ezh2-KO (n = 4; 2 independent experiments), and Vav-Ezh2-KO (n = 7; 3 independent experiments) embryos at E13.5. Scale bars, 1 mm. (B) Representative FACS histogram plots showing the recombination efficiency of Tie2-Cre (n = 6; 1 experiment) and Vav-iCre (n = 3; 1 experiment) in E12.5 FL LSK cells using Rosa26-LSL-tdTomato reporter. (C) Absolute numbers of Tie2-Ezh2-WT (n = 37), Tie2-Ezh2-KO (n = 9), Vav-Ezh2-WT (n = 16), and Vav-Ezh2-KO (n = 7) total mononuclear cells, LSK, multipotent progenitor (MPP) (LSKCD150−CD48+), and HSC (LSKCD150+CD48−) per FL at E12.5. Data pooled from 5 (Tie2-Cre) and 4 (Vav-iCre) independent experiments. (D) Absolute numbers of Vav-Ezh2-WT (E12.5, n = 16; E18.5, n = 13) and Vav-Ezh2-KO (E12.5 and E18.5, n = 7) LSK cells and HSCs per FL at E12.5 (4 independent experiments) and E18.5 (3 independent experiments). (E) Absolute numbers of Vav-Ezh2-WT (n = 7) and Vav-Ezh2-KO (n = 4) LSK cells and HSC per femur and tibia at E18.5 (2 independent experiments). (F) Peripheral myeloid cell (Mac1+) reconstitution kinetics in CD45.1 mice transplanted with 500 000 CD45.2 Tie2-Ezh2-WT (n = 5), Vav-Ezh2-WT (n = 4), Tie2-Ezh2-KO (n = 10) or Vav-Ezh2-KO (n = 6) total E12.5 FL cells; Vav-Ezh2-WT (n = 11) or Vav-Ezh2-KO (n = 13) E18.5 FL cells, in all cases together with 1 000 000 CD45.1 competitor BM cells. Data are pooled from 2 (E12.5) or 3 (E18.5) independent experiments. (G) Contribution of CD45.2 E12.5 FL cells to HSC compartment at 16 weeks following transplantations shown in panel F. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
To determine whether Ezh2-inactivated HSCs expand normally during fetal development, we next assessed Vav-Ezh2-KO embryos at E18.5, at which stage LSK cell and HSC numbers were increased relative to WT embryos (2.2-fold increase for LSK cells, P < .0001, and 2.0-fold increase for HSCs, P = .0012; Figure 1D), supporting that Ezh2 KO FL HSCs expand more rapidly than WT counterparts. Analysis of Vav-Ezh2-KO fetal BM at E18.5 revealed a similar increase in number of LSK cells (1.9-fold; P = .0681) and HSCs (5.3-fold; P = .0261), indicating that the migration from FL and colonization of HSCs in fetal BM was not compromised by the loss of Ezh2 (Figure 1E). To further examine any potential cell-intrinsic defect of HSC function caused by Ezh2 inactivation, we performed in vivo competitive repopulation assay using Tie2-Ezh2-KO and Vav-Ezh2-KO FL donor cells at E12.5 and Vav-Ezh2-KO FL donor cells at E18.5. We found that Ezh2 KO FL was functionally comparable to WT FL (Figure 1F-G) for long-term myeloid cell production. In line with the previously reported role of Ezh2 in early lymphoid development,5,6 where loss of Ezh2 blocks the maturation of lymphocytes and leads to accumulation of immature pro-B and DN3 T cells, the engraftment of the mature lymphoid lineage was severely compromised in recipients of Tie2-Ezh2-KO and Vav-Ezh2-KO FL cells (supplemental Figure 3A). However, both Tie2-Ezh2-KO and Vav-Ezh2-KO FL cells were capable of sustaining long-term engraftment of the myeloid lineage (Figure 1F), including presence of phenotypic HSCs at week 16 following transplantation (Figure 1G; supplemental Figure 3B), indicating that Ezh2 KO FL HSCs are capable of long-term self-renewal. Furthermore, in line with the increased numbers of phenotypic HSCs in E18.5 Vav-Ezh2-KO FL (Figure 1D), myeloid engraftment was higher in recipients of equal numbers of FL cells from Vav-Ezh2-KO (Figure 1F). Taken together, these data provide definitive evidence that Ezh2 is dispensable for the emergence and expansion of phenotypic and functional HSCs during fetal hematopoiesis.
Ezh2 inactivation in endothelial cells leads to cell-extrinsically mediated anemia
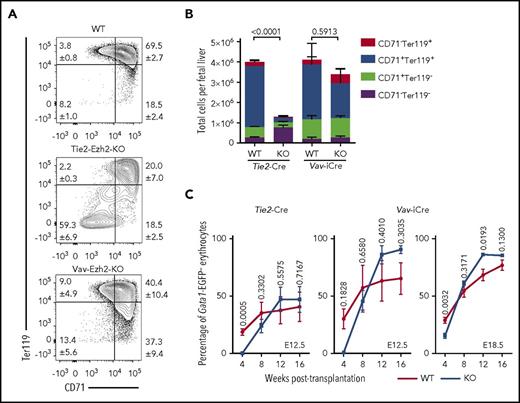
As Tie2-Ezh2-KO but not Vav-Ezh2-KO E12.5 embryos were severely anemic, we next focused our analysis on erythroid development. Erythroid staging revealed a marked loss (>24-fold) of CD71+Ter119+ erythroid precursors in Tie2-Ezh2-KO but not Vav-Ezh2-KO embryos (Figure 2A-B), indicating a Tie2-Ezh2-KO specific failure of late stages of fetal erythropoiesis. To determine whether this disruption in erythropoiesis was cell-intrinsically mediated, we traced the erythroid-lineage chimerism posttransplantation with a recently generated Gata1-EGFP reporter mouse.19 This analysis demonstrated that Ezh2 inactivation did not cell-intrinsically affect the erythroid reconstitution following transplantation of FL (Figure 2C). Collectively, these data do not support that the anemia phenotype of Tie2-Ezh2-KO embryos is because of a cell-intrinsic impact of Ezh2 deletion on FL HSCs or progenitor cells. Therefore, we hypothesized that depletion of Ezh2 in FL endothelium was causing a cell-extrinsic suppression of FL progenitors, particularly those of erythropoietic lineage.
Absence of Ezh2 leads to cell-extrinsically mediated anemia. (A) Representative FACS plots showing distinct block in E12.5 FL erythropoiesis in Tie2-Ezh2-KO but not Vav-Ezh2-KO embryos. Shown are the mean values with SEM for the frequencies of the indicated population across all experiments. (B) Absolute numbers of E12.5 FL erythroid progenitor populations (Tie2-Ezh2-WT, n = 26, 5 independent experiments; Tie2-Ezh2-KO, n = 7, 4 independent experiments; Vav-Ezh2-WT, n = 6, 2 independent experiments; Vav-Ezh2-KO, n = 5, 2 independent experiments). Two-way analysis of variance was used to assess statistical significance. Error bars represent SEM. (C) Peripheral Gata1-EGFP− erythroid (Ter119+) reconstitution kinetics in CD45.1 Gata1-EGFP recipient mice transplanted with 500 000 CD45.2 Tie2-Ezh2-WT (n = 7), Vav-Ezh2-WT (n = 2), Tie2-Ezh2-KO (n = 12) or Vav-Ezh2-KO (n = 3) total E12.5 FL cells; Vav-Ezh2-WT (n = 5) or Vav-Ezh2-KO (n = 7) E18.5 FL cells, together with 1 000 000 CD45.1 Gata1-EGFP competitor BM cells. Data are pooled from 1 (Vav-iCre; E12.5) or 2 (Tie2-Cre; E12.5 and Vav-iCre; E18.5) independent experiments. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
Absence of Ezh2 leads to cell-extrinsically mediated anemia. (A) Representative FACS plots showing distinct block in E12.5 FL erythropoiesis in Tie2-Ezh2-KO but not Vav-Ezh2-KO embryos. Shown are the mean values with SEM for the frequencies of the indicated population across all experiments. (B) Absolute numbers of E12.5 FL erythroid progenitor populations (Tie2-Ezh2-WT, n = 26, 5 independent experiments; Tie2-Ezh2-KO, n = 7, 4 independent experiments; Vav-Ezh2-WT, n = 6, 2 independent experiments; Vav-Ezh2-KO, n = 5, 2 independent experiments). Two-way analysis of variance was used to assess statistical significance. Error bars represent SEM. (C) Peripheral Gata1-EGFP− erythroid (Ter119+) reconstitution kinetics in CD45.1 Gata1-EGFP recipient mice transplanted with 500 000 CD45.2 Tie2-Ezh2-WT (n = 7), Vav-Ezh2-WT (n = 2), Tie2-Ezh2-KO (n = 12) or Vav-Ezh2-KO (n = 3) total E12.5 FL cells; Vav-Ezh2-WT (n = 5) or Vav-Ezh2-KO (n = 7) E18.5 FL cells, together with 1 000 000 CD45.1 Gata1-EGFP competitor BM cells. Data are pooled from 1 (Vav-iCre; E12.5) or 2 (Tie2-Cre; E12.5 and Vav-iCre; E18.5) independent experiments. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
Endothelial Ezh2 inactivation causes global loss of FL mKitL
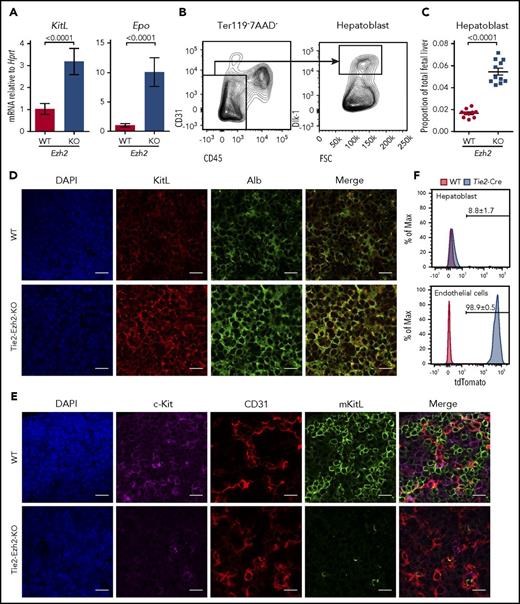
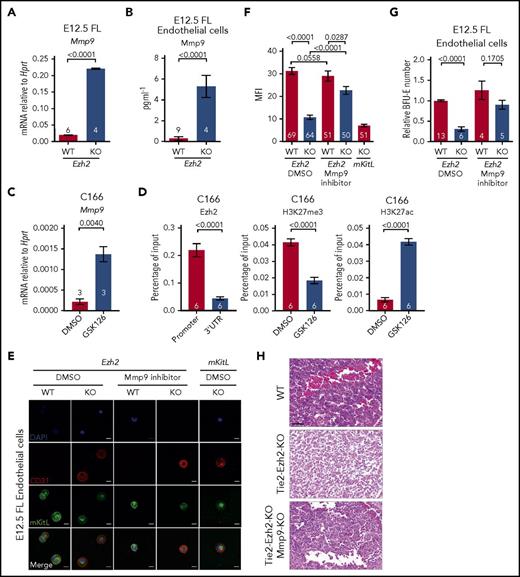
We next assessed levels of erythropoietin (Epo) and KitL expression in Tie2-Ezh2-KO as these are 2 crucial cytokines for cell-extrinsic regulation of normal fetal erythropoiesis.29,30 However, both Epo (9.8-fold; P < .0001) and KitL (3.6-fold; P < .0001) messenger RNA (mRNA) levels were upregulated in Tie2-Ezh2-KO E12.5 FL (Figure 3A), likely because of the increased proportion of hepatoblasts (threefold; P < .0001; Figure 3B-D), which are the major producer of these cytokines in the FL,31 as a consequence of the marked reduction of erythroid precursors in Tie2-Ezh2-KO FL (Figure 2B). KitL is expressed as 2 major isoforms, the membrane-bound (mKitL) and the soluble (sKitL) isoforms.32 The mKitL isoform is important for supporting erythropoiesis.33 Therefore, we further examined the expression of mKitL by immunofluorescence staining using an intracellular KitL-specific antibody that is specific for mKitL.34 Strikingly, despite the increased total KitL mRNA and protein expression (Figure 3A,D), there was an almost complete loss of mKitL in Tie2-Ezh2-KO E12.5 FL (Figure 3E). Importantly, the loss of mKitL was not endothelium-specific, but also affected hepatoblasts (Figure 3D-E; supplemental Figure S4), which express KitL but are not targeted by Tie2-Cre (Figure 3F), suggesting the presence of Ezh2 KO endothelium-derived cell-extrinsic factor(s) causing a loss of mKitL expression.
Tie2-Cre mediated deletion of Ezh2 leads to a specific loss of mKitL expression. (A) KitL and Epo mRNA expression levels in E12.5 FL cells (WT, n = 6; KO, n = 4; 2 independent experiments). (B) Gating strategy used to define hepatoblast (Ter119−CD45−CD31−Dlk-1+) population within the total FL live (7AAD−) singlet population. (C) Proportion of hepatoblasts in E12.5 FL (WT, n = 10; KO, n = 11; 4 independent experiments). Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM. (D-E) Immunofluorescence staining of E12.5 FL (n = 3 each) using antibodies against KitL (D, red), Alb (D, green, hepatoblast marker), c-Kit (E, magenta, hematopoietic cell marker), CD31 (E, red, endothelial cell marker), mKitL (E, green). Nuclei was stained with 4′,6-diamidino-2-phenylindole (DAPI; D-E, blue). Scale bars, 20 µm. (F) Representative FACS histogram plots showing the recombination efficiency of Tie2-Cre in E12.5 FL hepatoblasts (n = 5, 2 independent experiments) and endothelial cells (n = 6, 1 experiment; Ter119−CD45−CD31+Lyve1+) using Rosa26-LSL-tdTomato reporter. Shown are the mean values with SEM for the frequencies of the indicated population across all experiments.
Tie2-Cre mediated deletion of Ezh2 leads to a specific loss of mKitL expression. (A) KitL and Epo mRNA expression levels in E12.5 FL cells (WT, n = 6; KO, n = 4; 2 independent experiments). (B) Gating strategy used to define hepatoblast (Ter119−CD45−CD31−Dlk-1+) population within the total FL live (7AAD−) singlet population. (C) Proportion of hepatoblasts in E12.5 FL (WT, n = 10; KO, n = 11; 4 independent experiments). Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM. (D-E) Immunofluorescence staining of E12.5 FL (n = 3 each) using antibodies against KitL (D, red), Alb (D, green, hepatoblast marker), c-Kit (E, magenta, hematopoietic cell marker), CD31 (E, red, endothelial cell marker), mKitL (E, green). Nuclei was stained with 4′,6-diamidino-2-phenylindole (DAPI; D-E, blue). Scale bars, 20 µm. (F) Representative FACS histogram plots showing the recombination efficiency of Tie2-Cre in E12.5 FL hepatoblasts (n = 5, 2 independent experiments) and endothelial cells (n = 6, 1 experiment; Ter119−CD45−CD31+Lyve1+) using Rosa26-LSL-tdTomato reporter. Shown are the mean values with SEM for the frequencies of the indicated population across all experiments.
Loss of Ezh2 impairs endothelium erythropoietic supporting ability via overexpression of Mmp9
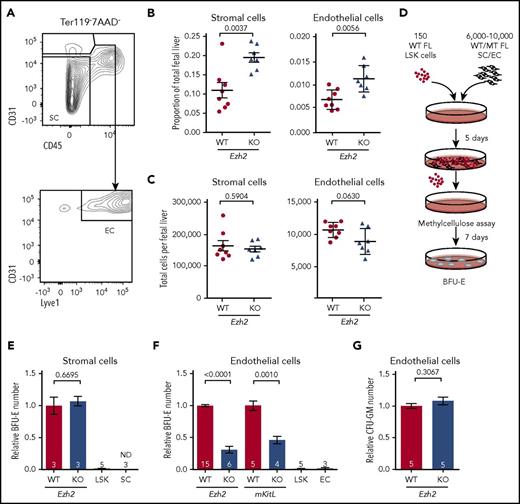
As various FL HSC niche populations play an essential role in supporting fetal hematopoiesis, we further examined the Tie2-Ezh2-KO E12.5 FL HSC niche components and found that in addition to hepatoblasts, the proportions of both stromal (1.8-fold; P = .0037) and endothelial cells (1.5-fold; P = .0056) were also increased in the FL (Figures 3E and 4A-B), likely because of relative loss of the erythroid lineage cells (Figure 2A-B) with overall normal numbers of stromal and endothelial cells (Figure 4C). Therefore, the impaired fetal erythropoiesis was because of defective niche populations rather than a reduction of their cell numbers.
Reduced erythropoietic supporting ability of Ezh2-deficient endothelium. (A) Gating strategy used to define stromal cell (SC; Ter119−CD45−CD31−) and endothelial cell (EC; Ter119−CD45−CD31+Lyve1+) populations within the total FL live (7AAD−) singlet population. Proportion (B) and absolute numbers (C) of SCs and ECs (WT, n = 8; KO, n = 7; 2 independent experiments) per FL at E12.5. (D) Experimental design of the coculture assay. (E) Measurement of WT or Tie2-Ezh2-KO E12.5 FL SC erythropoietic supporting ability with coculture assay (2 independent experiments). LSK indicates LSK cells only. SC indicates SCs only. (F) Measurement of WT, Tie2-Ezh2-KO, or mKitL-KO E12.5 FL EC (Ter119−CD45−CD31+) erythropoietic supporting ability with coculture assay (7 independent experiments). LSK indicates LSK cells only. EC indicates ECs only. (G) Measurement of WT or Tie2-Ezh2-KO E12.5 FL EC myeloid supporting ability with coculture assay (2 independent experiments). Numbers of replicates are indicated at the bottom of each column. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
Reduced erythropoietic supporting ability of Ezh2-deficient endothelium. (A) Gating strategy used to define stromal cell (SC; Ter119−CD45−CD31−) and endothelial cell (EC; Ter119−CD45−CD31+Lyve1+) populations within the total FL live (7AAD−) singlet population. Proportion (B) and absolute numbers (C) of SCs and ECs (WT, n = 8; KO, n = 7; 2 independent experiments) per FL at E12.5. (D) Experimental design of the coculture assay. (E) Measurement of WT or Tie2-Ezh2-KO E12.5 FL SC erythropoietic supporting ability with coculture assay (2 independent experiments). LSK indicates LSK cells only. SC indicates SCs only. (F) Measurement of WT, Tie2-Ezh2-KO, or mKitL-KO E12.5 FL EC (Ter119−CD45−CD31+) erythropoietic supporting ability with coculture assay (7 independent experiments). LSK indicates LSK cells only. EC indicates ECs only. (G) Measurement of WT or Tie2-Ezh2-KO E12.5 FL EC myeloid supporting ability with coculture assay (2 independent experiments). Numbers of replicates are indicated at the bottom of each column. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
To gain a deeper mechanistic understanding of Ezh2 KO stromal and endothelial cells’ erythropoietic supporting ability, we established an in vitro coculture assay with WT E12.5 FL LSK cells and Tie2-Ezh2-KO E12.5 FL-derived stromal or endothelial cells (Figure 4D). In agreement with previous studies,35 the presence of stromal or endothelial cells was crucial for maintaining progenitors in vitro as LSK cells that were cultured alone lost their erythroid colony forming potential (Figure 4E-F). Stromal cells (in the absence of endothelial cells) from Tie2-Ezh2-KO FL showed a normal erythropoietic supporting ability in vitro (Figure 4E). In contrast, erythropoietic supporting ability of Tie2-Ezh2-KO FL-derived endothelial cells was markedly reduced (3.2-fold reduction; P < .0001; Figure 4F). Moreover, E12.5 FL endothelium derived from a mKitL-specific KO mouse line17 also showed a similar defect as for Tie2-Ezh2-KO endothelium (2.2-fold reduction; P = .001; Figure 4F). Interestingly, the myeloid supporting ability of Tie2-Ezh2-KO endothelium remained normal, in keeping with a key role of Kit signaling in erythropoiesis29,36 (Figure 4G). Together, these data support that a major cause of the defective FL erythropoiesis associated with Tie2-Ezh2-KO was the loss of mKitL expression in FL niche populations.
As mKitL is a known target of Mmp9,37 a metallopeptidase that is capable of cleaving the transmembrane domain of mKitL protein, causing release of sKitL and loss of mKitL expression, we next explored the possibility that Ezh2 inactivation might lead to the overexpression of Mmp9 in FL endothelial cells. We found that Mmp9 gene expression was markedly upregulated in Tie2-Ezh2-KO FLs (11.3-fold; P < .0001; Figure 5A). This result was further supported by the significantly higher Mmp9 protein level (18.4-fold; P < .0001; Figure 5B) that was present in the supernatant of cocultured Tie2-Ezh2-KO endothelium. To determine whether Ezh2 directly regulated Mmp9 expression, an embryonic-derived endothelial cell line (C166) was used. Similarly to the impact of Ezh2-KO in FL endothelial cells, treatment of C166 cells with an Ezh2 specific inhibitor (GSK126) led to a marked increase in Mmp9 expression (6.3-fold; P = .004; Figure 5C). ChIP–quantitative PCR (qPCR) revealed an enriched binding of Ezh2 at the Mmp9 promoter (5-fold; P < .0001), associated with reduced H3K27me3 (2.3-fold reduction; P < .0001) and increased H3K27ac levels (6.3-fold; P < .0001) upon GSK126-mediated Ezh2 inhibition (Figure 5D). Taken together, these data support a direct role of Ezh2 on the regulation of Mmp9 in FL endothelial cells.
EZH2 directly regulates Mmp9 expression. (A) Mmp9 mRNA expression level in E12.5 FL cells (2 independent experiments). (B) Mmp9 protein level in coculture supernatant in panel G quantified by ELISA. (C) Mmp9 mRNA expression level in DMSO and 10 μM GSK126 (Ezh2 inhibitor) treated C166 cells (1 experiment). (D) ChIP-qPCR for Ezh2, H3K27me3, and H3K27ac at Mmp9 locus in DMSO or GSK126 treated C166 cells (3 independent experiments). (E) Representative immunofluorescence staining (2 independent experiments) of E12.5 FL endothelial cells collected post in vitro treatment with DMSO or 10 nM Mmp9 inhibitor using antibodies against CD31 (red, endothelial marker) and mKitL (green). Nuclei were stained with DAPI (blue). Scale bars, 20 µm. (F) Quantification of mKitL mean fluorescence intensity (MFI) in panel E. (G) In vitro coculture assay with DMSO or Mmp9 inhibitor treatment (5 independent experiments). (H) Hematoxylin and eosin–stained E13.5 FL sections from indicated genotypes, representative of 3 embryos per genotype. Scale bars, 50 µm. Numbers of replicates are indicated at the bottom of each column. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
EZH2 directly regulates Mmp9 expression. (A) Mmp9 mRNA expression level in E12.5 FL cells (2 independent experiments). (B) Mmp9 protein level in coculture supernatant in panel G quantified by ELISA. (C) Mmp9 mRNA expression level in DMSO and 10 μM GSK126 (Ezh2 inhibitor) treated C166 cells (1 experiment). (D) ChIP-qPCR for Ezh2, H3K27me3, and H3K27ac at Mmp9 locus in DMSO or GSK126 treated C166 cells (3 independent experiments). (E) Representative immunofluorescence staining (2 independent experiments) of E12.5 FL endothelial cells collected post in vitro treatment with DMSO or 10 nM Mmp9 inhibitor using antibodies against CD31 (red, endothelial marker) and mKitL (green). Nuclei were stained with DAPI (blue). Scale bars, 20 µm. (F) Quantification of mKitL mean fluorescence intensity (MFI) in panel E. (G) In vitro coculture assay with DMSO or Mmp9 inhibitor treatment (5 independent experiments). (H) Hematoxylin and eosin–stained E13.5 FL sections from indicated genotypes, representative of 3 embryos per genotype. Scale bars, 50 µm. Numbers of replicates are indicated at the bottom of each column. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM.
We therefore assessed whether inhibition of Mmp9 could restore expression of mKitL and the erythropoiesis supporting capacity of FL endothelial cells. Indeed, Mmp9-specific inhibitor (CAS 1177749-58-4) treatment of Tie2-Ezh2-KO endothelial cells in vitro rescued the cell-surface expression of mKitL (Figure 5E-F) and almost completely restored their erythropoietic supporting capability (Figure 5G). We next sought to determine whether KO of Mmp9 in the Tie2-Ezh2-KO embryo also rescue the FL anemia phenotype. In agreement with our in vitro findings, genetic KO of Mmp9 led to a restoration of erythropoiesis in Tie2-Ezh2-KO embryos as demonstrated by histology (Figure 5H) and also in line with previously reported rescue of anemia associated with Tie2-Ezh2-KO by Mmp9 genetic KO as observed macroscopically.23 Collectively, our data support that Ezh2 inactivation in FL endothelial cells leads to a cell-extrinsic disruption of erythropoiesis, mediated by overexpression of Mmp9 with consequent loss of mKitL expression.
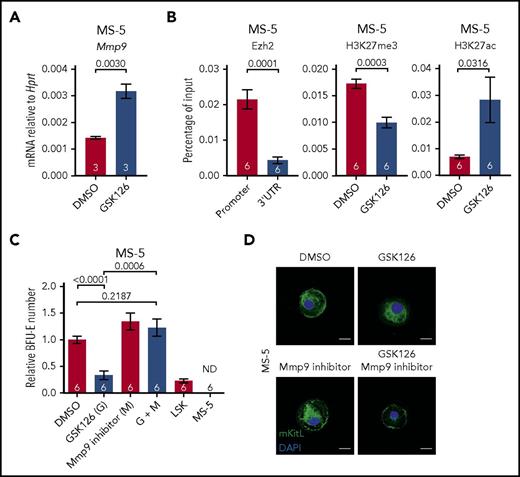
Finally, to determine whether inactivation of Ezh2 in niche cells might also cause a cell-extrinsic disruption of adult hematopoiesis, we examined the impact of Ezh2 inactivation in MS-5 cells, an adult murine BM derived stromal cell line. In keeping with our findings in fetal hematopoiesis, GSK126-mediated Ezh2 inhibition led to an upregulation of Mmp9 expression in MS-5 cells (2.2-fold; P = .003; Figure 6A). ChIP-qPCR analysis also showed enriched binding of Ezh2 at the Mmp9 promoter (4.9-fold; P = .0001; Figure 6B). Furthermore, Ezh2 inhibition with GSK126 was accompanied by reduced H3K27me3 (1.7-fold reduction; P = .0003) and increased H3K27ac (4.1-fold; P = .0316) levels at the Mmp9 promoter (Figure 6B), once again supporting a direct role of Ezh2 in the regulation of Mmp9 expression. Using an in vitro coculture assay, we determined that the erythropoietic supporting ability of MS-5 was severely impaired by GSK126-mediated Ezh2 inhibition, which could be rescued by Mmp9 inhibitor treatment (Figure 6C). Moreover, cell-surface expression mKitL in MS-5 cells was lost upon Ezh2 inhibition and could be restored by concurrent inhibition of Mmp9 (Figure 6D). These results support that Ezh2 also has an important role in the regulation of Mmp9 in adult BM derived stromal cells.
Reduced erythropoietic supporting ability of Ezh2-inactivated adult BM stromal cells. (A) Mmp9 mRNA expression level in DMSO and 10 μM GSK126 (Ezh2 inhibitor) treated MS-5 cells (1 experiment). (B) ChIP-qPCR for Ezh2, H3K27me3, and H3K27ac at Mmp9 locus in DMSO or GSK126 treated MS-5 cells (3 independent experiments). (C) In vitro coculture assay of MS-5 cells treated with DMSO, GSK126, and/or Mmp9 inhibitor (2 independent experiments). Numbers of replicates are indicated at the bottom of each column. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM. (D) Representative immunofluorescence staining (n ≥ 30 cells, 2 independent experiments) of MS-5 cells collected post in vitro treatment with DMSO, GSK126, and/or 10 nM Mmp9 inhibitor using antibodies against mKitL (green). Nuclei were stained with DAPI (blue). Scale bars, 20 µm.
Reduced erythropoietic supporting ability of Ezh2-inactivated adult BM stromal cells. (A) Mmp9 mRNA expression level in DMSO and 10 μM GSK126 (Ezh2 inhibitor) treated MS-5 cells (1 experiment). (B) ChIP-qPCR for Ezh2, H3K27me3, and H3K27ac at Mmp9 locus in DMSO or GSK126 treated MS-5 cells (3 independent experiments). (C) In vitro coculture assay of MS-5 cells treated with DMSO, GSK126, and/or Mmp9 inhibitor (2 independent experiments). Numbers of replicates are indicated at the bottom of each column. Two-tailed Student t tests were used to assess statistical significance. Error bars represent SEM. (D) Representative immunofluorescence staining (n ≥ 30 cells, 2 independent experiments) of MS-5 cells collected post in vitro treatment with DMSO, GSK126, and/or 10 nM Mmp9 inhibitor using antibodies against mKitL (green). Nuclei were stained with DAPI (blue). Scale bars, 20 µm.
Discussion
Epigenetic factors have been shown to be important cell-intrinsic regulators of normal and malignant hematopoiesis.38 For example, Ezh2 loss-of-function mouse models have demonstrated a key role for Ezh2 during early lymphoid development.5,6 Furthermore, somatic inactivating mutations of Ezh2 occur not only in patients with chronic myeloid malignancies39 but also fetal origin juvenile myelomonocytic leukemia,40 supporting the notion that hematopoietic cell specific Ezh2 inactivation might confer a clonal advantage to fetal HSCs. Moreover, loss of function of Ezh2 is associated with reactivation of a fetal-like gene expression program in adult HSCs.41 However, a previous study using Tie2-Cre to induce Ezh2 inactivation suggested an essential role of Ezh2 in fetal but not adult HSCs.10 This finding has not been supported by other studies using different approaches to inactivate PRC2.8,9 Therefore, we directly compared the phenotype of Tie2-Ezh2-KO and Vav-Ezh2-KO embryos. In keeping with previous work,10 we observed a reduced number of FL LSK cells, disrupted erythropoiesis, and severe anemia, specifically in Tie2-Ezh2-KO embryos. However, it is well established that LSK is a highly heterogeneous population containing a small minority of bona fide HSCs.42 By incorporating signaling lymphocytic activation molecule markers,26 we established that numbers of phenotypic FL HSCs were not affected by the loss of Ezh2. Moreover, in functional (transplantation) assays, Tie2-Ezh2-KO FL could sustain long-term myeloid and erythroid cell reconstitution, with a defect in lymphoid reconstitution as expected from previous studies.5,6 We also observed altered early myeloid reconstitution kinetics following transplantation of both Tie2-Ezh2-KO and Vav-Ezh2-KO FL, with reduced myeloid cell engraftment at 4 weeks following transplantation that was fully recovered by 8 weeks. As this defect was present following transplantation of both Tie2-Ezh2-KO and Vav-Ezh2-KO FL (Figure 1F), this would support a cell-intrinsic cause for this observation, which does not help to explain the anemia phenotype specifically associated with Tie2-Ezh2-KO, and may relate to numbers, or differentiation kinetics, of Ezh2-deficient FL progenitor cells. Furthermore, Vav-Ezh2-KO FLs showed increased numbers of HSCs at later stages of gestation, suggesting that Ezh2 inactivation cell-intrinsically promotes the expansion of FL HSCs, a finding in line with the presence of Ezh2 inactivating mutations in hematological malignancies of fetal origin, or those showing fetal-like gene expression programs.40
In contrast to the severe anemia phenotype of Tie2-Ezh2-KO embryos, hematopoietic specific inactivation of Ezh2 with Vav-iCre did not result in anemia at E12.5. As Tie2-Cre also targets endothelial cells,14 our data strongly suggested an endothelial cell mediated mechanism of defective fetal hematopoiesis in the Tie2-Ezh2-KO model. One possibility we considered is that Ezh2 function is critical for emergence of HSCs from the hemogenic endothelium but is dispensable for HSC function following emergence, similar to findings with the transcription factor Runx1.28 However, we showed that Ezh2 is dispensable for HSC emergence through demonstration of morphologically normal numbers of hematopoietic clusters in the aorta-gonad-mesonephros region at E10.5 and as Tie2-Ezh2-KO FL contained normal numbers of phenotypic and functional HSCs at E12.5. We therefore concluded that the impact of Ezh2 inactivation in endothelial cells was likely to be cell-extrinsically mediated.
As the major hematopoietic defect in Tie2-Ezh2-KO embryos was anemia, we focused our analysis on known cell-extrinsic growth factors that are crucial to support FL erythropoiesis and identified a dramatic and almost complete loss of mKitL, a crucial cytokine for supporting erythropoiesis.29,33 Crucially, this loss of mKitL was not restricted to the endothelial cells but was also present on hepatoblasts which were not targeted by Tie2-Cre. This global loss of mKitL in Tie2-Ezh2-KO FL was caused by overexpression of Mmp9 by Ezh2-deleted FL endothelium, which exerts a hematopoietic impact by cleaving mKitL.23,37 We also demonstrate that Ezh2 directly regulates the expression of Mmp9 in embryonic endothelial cells and adult BM stromal cells. Notably, mKitL has been suggested to be more efficient at supporting erythropoiesis than sKitL,33 potentially because of a more persistent activation of c-Kit signaling,43 helping to explain the failure of fetal erythropoiesis upon the loss of mKitL in the Tie2-Ezh2-KO model. We also used an mKitL-specific KO mouse17 to demonstrate that mKitL loss recapitulated the loss of erythroid supporting capability of endothelial cells in a novel coculture assay. Furthermore, global loss of mKitL in utero has been reported to be associated with severe developmental and hematopoietic abnormalities normally associated with Steel alleles (with complete loss of Kit signaling)17,36 ; Steel null mice die in utero at E14 to E16 with severe anemia.29 Moreover, Mmp9-specific inhibitor treatment was able to fully rescue the erythropoietic supporting capability of Tie2-Ezh2-KO endothelial cells in vitro by restoring the mKitL cell-surface expression, thus providing conclusive evidence that cell-extrinsically mediated loss of mKitL is a key mechanism of anemia in Tie2-Ezh2-KO embryos, although other mechanisms may also be involved.
To our knowledge, this is the first description of a cell-extrinsic role of an epigenetic factor in the regulation of hematopoiesis. This is of relevance for a number of reasons. First, this highlights the importance of considering possible cell-extrinsic mechanism when studying models of epigenetic regulators to avoid making false assumptions. Second, because of the key role of Ezh2 in the development of hematological cancers, 3 Ezh2-specific inhibitors (GSK2816126,44 EPZ-6438,45 and CPI-120546 ) are in clinical trials. However, despite the encouraging outcome from both GSK2816126 and EPZ-6438 treatment on patients with advanced non-Hodgkin lymphoma or solid tumors, they have some undesired hematopoietic side effects including anemia.47,48 Our findings provide a mechanistic explanation for some of these unfavorable adverse effects where global suppression of Ezh2 expression will increase MMP9 production, which subsequently depletes mKitL in the HSC niche. This “off-target” toxicity could be particularly important when combining such agents with cytotoxic chemotherapy.
Collectively, our study has definitively established that Ezh2 is dispensable for HSC function in fetal hematopoiesis and has also identified a previously undescribed potential for disruption of epigenetic regulators to exert a major cell-extrinsic impact on hematopoiesis mediated through nonhematopoietic cell types. This finding has potentially important implications for analysis of mouse models and understanding of hematological toxicity of epigenetic therapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Peter Besmer for the intracellular KitL antibody and Biomedical Services at University of Oxford for expert animal support.
The authors acknowledge the contributions of the Medical Research Council (MRC) Weatherall Institute of Molecular Medicine (WIMM) Flow Cytometry Facility, supported by the MRC Human Immunology Unit, MRC Molecular Haematology Unit (MC_UU_12009), National Institute for Health Research Oxford Biomedical Research Centre (131/030), John Fell Fund (101/517), EPA fund (CF182 and CF170), and WIMM Strategic Alliance awards G0902418 and MC_UU_12025. This work was also supported by the MRC and Wolfson Foundation (grant 18272) funded Wolfson Imaging Centre Oxford. M.F.T.R.d.B. and E.A. are funded through a program in the MRC Molecular Haematology Unit Core award (MC_UU_12009/2). S.E.W.J. is funded through an MRC Unit Grant MC_UU_12009/5 and an international recruitment grant from the Swedish Research Council (2013-8995). This study was funded by Kay Kendall Leukaemia Fund Project Grant (KKL811) and MRC Senior Clinical Fellowship (MR/L006340/1) (A.J.M.).
Authorship
Contribution: W.H.N. designed, performed, and analyzed mouse genetic experiments, in vitro coculture experiments, and imaging data; C.A.G.B. performed and analyzed mouse genetic experiments; E.A. conducted and interpreted whole-mount immunofluorescence staining data; L.C. and P.D.-O. performed hematoxylin and eosin staining; M.F.T.R.d.B. and S.E.W.J. provided scientific advice; and A.J.M. conceived and designed the research, interpreted the data, secured funding for this project, and wrote the manuscript with W.H.N.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adam J. Mead, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, Oxford OX3 9DS, United Kingdom; e-mail: adam.mead@imm.ox.ac.uk.