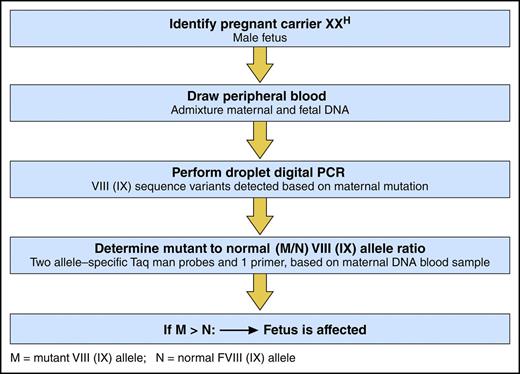
In this issue of Blood, Hudecova and colleagues describe a simple, noninvasive assay for prenatal detection of hemophilia by droplet digital polymerase chain reaction (ddPCR) on maternal peripheral blood.1
ddPCR to detect a fetus with hemophilia. Professional illustration by Patrick Lane, ScEYEnce Studios.
ddPCR to detect a fetus with hemophilia. Professional illustration by Patrick Lane, ScEYEnce Studios.
Prenatal diagnosis is a must!
In digital PCR we trust.
With blood from carrier moms,
Alleles are compared, mutant and non.
For a fetus of male sex
With mutant allele excess,
Diagnosis is elemental,
And compared with amnio, so gentle!
Hemophilia in the neonate
Known before the birth date,
Allows for factor injection
For circumcision or C-section.
Whether intron 22 mutation,
Missense, or other alteration,
Carrier blood collection
Is the key to prenatal detection.
Hemophilia is an X-linked disorder caused by deficient or defective factor VIII (hemophilia A) or factor IX (hemophilia B) and is characterized by spontaneous or traumatic bleeding into joints and muscles. Infants born with hemophilia may have circumcision bleeding or life-threatening intracranial hemorrhage, for which factor prophylaxis may be protective.2 Thus, early diagnosis, preferably prenatal diagnosis, is critical to prevent disabling complications. Currently, hemophilia is diagnosed by the factor VIII or IX activity on a cord blood sample obtained at birth. By this approach, however, a diagnosis may be missed in up to one-third of hemophilia cases, which arise as a spontaneous mutation, or may be unsuspected in up to two-thirds of cases with a family history, if the mother is unaware of her carrier status, even despite a bleeding tendency.3,4
The recently established nationwide program, My Life, Our Future (MLOF), to genotype individuals with hemophilia A and B and their female carrier relatives, has begun to alleviate the latter problem.5 Knowledge of genotypic carrier status is critical to prenatal diagnosis, as prenatal tests for hemophilia are increasingly being developed to test carrier mothers during pregnancy. In 2011, Tsui et al introduced a microfluidic assay for prenatal diagnosis that analyzes maternal plasma DNA for F8 or F9 sequence variants, such that an allelic imbalance with more mutant (M) than normal/wild-type (N) allele in maternal carrier plasma indicates an affected child.6 The latter technique, although highly accurate, is not widely used, as, due to molecular complexity, it cannot detect the most common mutation causing severe hemophilia A, the intron 22 inversion mutation.7
In this study, Hudecova and her colleagues employed powerful new genomic techniques, including ddPCR and massively parallel sequencing, to analyze maternal plasma DNA samples from 18 at-risk pregnancies (see figure). Sampling from 18 to 42 weeks’ gestation, they correctly diagnosed hemophilia in the fetus of 15 pregnancies, including hemophilia B in 8 and hemophilia A in 7, 3 of which were identified to have the intron 22 inversion mutation. In 3 at-risk pregnancies, no classification was possible. Findings in the 15 correctly identified fetuses were confirmed by blood from probands in 2 of the identified families and by placental tissue in 2 families with no available probands, and all were confirmed by cord blood sample for factor VIII and IX activity.
To assess the accuracy of their ddPCR approach, computer simulations were conducted that showed >99% accuracy if there were at least 5 test wells of maternal DNA and if at least 15% of the sample was fetal DNA. To assess haplotype dosage imbalance, relative haplotype dosage analysis and sequential probability analysis were performed, using an odds ratio of 100 to determine the presence of overrepresentation or underrepresentation of maternal or fetal haplotypes.
What are the implications of these findings? First, these assays provide a noninvasive approach to prenatal detection of hemophilia A or B on a peripheral blood sample from hemophilia carriers. The use of digital droplet PCR for prenatal diagnosis of hemophilia represents a major improvement over current invasive methods, eg, chorionic villus sampling, amniocentesis, and cordocentesis. Second, knowledge of a hemophilia diagnosis before birth provides for an opportunity for early hemostatic intervention to promote hemostasis before procedures, eg, circumcision, are performed, or to prevent central nervous system and related morbidity and mortality8 by cesarean delivery, which is recommended to reduce the risk of intracranial hemorrhage when the birth of a child with severe hemophilia is anticipated.9 Third, prenatal testing is also important to ensure hemostatic support for the mother for whom it may be necessary to prevent bleeding with perinatal anesthesia and/or postpartum bleeding.10 Fourth, these prenatal assays depend on knowledge of the mother’s carrier genotype, which is potentially more accurate than factor levels, which may increase with hormone use or the increasing hormone levels of pregnancy and mask carrier diagnosis. Finally, the development of these assays is timely in view of the ongoing MLOF genome project in hemophilia and underscores the need for carrier testing and genetic counseling of female members from hemophilia kindreds.
Conflict-of-interest disclosure: The author declares no competing financial interests.