Abstract
Hypodiploidy <40 chromosomes is an uncommon genetic feature of acute lymphoblastic leukemia (ALL) in both children and adults. It has long been clear by cytogenetic analyses, and recently confirmed by mutational profiling, that these cases may be further subdivided into 2 subtypes: near-haploid ALL with 24 to 30 chromosomes and low-hypodiploid ALL with 31 to 39 chromosomes. Both groups are associated with a very poor prognosis, and these patients are among those who could benefit most from novel treatments.
Introduction
Near haploid (24-30 chromosomes) and low hypodiploid (31-39 chromosomes) are rare subtypes of B-cell precursor childhood and adult acute lymphoblastic leukemia (ALL). Initially recognized as harboring massive losses of chromosomes, with specific patterns of retained disomies,1-3 these 2 groups have recently been shown to also have distinct mutational profiles, with TP53 mutations turning out to be a hallmark of low-hypodiploid cases.4 In spite of their rarity, near haploidy and low hypodiploidy are important in clinical risk classification. As early as 1981, Brodeur et al5 concluded that ALL with low chromosome numbers had a particularly poor prognosis. Whereas the survival rates of ALL in general since then have risen (for pediatric cases, dramatically), hypodiploidy <40 chromosomes remains a poor prognostic marker across all age groups, although a recent report showed that minimal residual disease–negative pediatric cases may be curable with chemotherapy.6 In this review, we summarize the current knowledge of hypodiploid ALL.
Characteristic chromosomal patterns in near-haploid and low-hypodiploid ALL
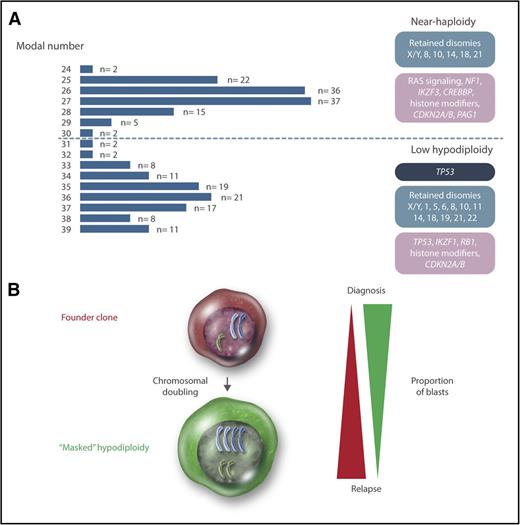
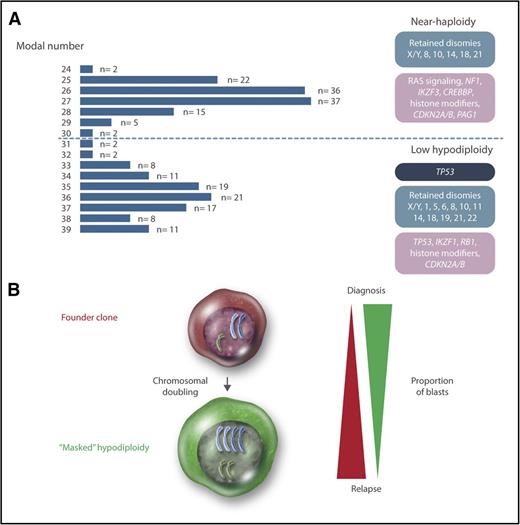
Cytogenetically, hypodiploid <46 chromosomes ALL may be divided into high-hypodiploid (40-45 chromosomes), low-hypodiploid, and near-haploid ALL. High-hypodiploid cases are heterogeneous cytogenetically and not a clear separate group and therefore not further discussed in this review. Chromosome numbers below 40 display a clear bimodal distribution, with peaks at 27 and 36 chromosomes, corresponding to the near-haploid and low-hypodiploid subtypes, respectively (Figure 1). In near-haploid ALL, retained disomies primarily comprise chromosomes X/Y, 8, 10, 14, 18, and 21, whereas retention of disomies X/Y, 1, 5, 6, 8, 10, 11, 14, 18, 19, 21, and 22 is seen in low hypodiploid ALL (Figure 1). Notably, chromosome 21 is always retained in both groups.
Genetics of hypodiploid less than 40 chromosomes ALL. (A) Modal numbers of all 218 hypodiploid ALL cases with <40 chromosomes reported in the literature, ascertained from the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer.7 Cases where the modal number was given as a range, with a known T-cell immunophenotype, or with t(1;19)(q23;p13), t(9;22)(q34;q22), or t(14;18)(q32;q21) were excluded from the analysis. There are 2 clear peaks centered on 27 and 36 chromosomes, corresponding to near-haploid and low-hypodiploid ALL, respectively. Chromosomal patterns are shown in blue, constitutional mutations in dark blue, and somatic mutations in purple. (B) Chromosomal doubling is frequent in both near-haploid and low-hypodiploid ALL, sometimes leading to masked hypodiploidy (ie, only the clone with doubled chromosomes are detected). The relative proportions of the founder and doubled clone often differ between diagnosis and relapse, with the doubled clone generally dominating at diagnosis and the founder clone at relapse. Professional illustration by Somersault18:24.
Genetics of hypodiploid less than 40 chromosomes ALL. (A) Modal numbers of all 218 hypodiploid ALL cases with <40 chromosomes reported in the literature, ascertained from the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer.7 Cases where the modal number was given as a range, with a known T-cell immunophenotype, or with t(1;19)(q23;p13), t(9;22)(q34;q22), or t(14;18)(q32;q21) were excluded from the analysis. There are 2 clear peaks centered on 27 and 36 chromosomes, corresponding to near-haploid and low-hypodiploid ALL, respectively. Chromosomal patterns are shown in blue, constitutional mutations in dark blue, and somatic mutations in purple. (B) Chromosomal doubling is frequent in both near-haploid and low-hypodiploid ALL, sometimes leading to masked hypodiploidy (ie, only the clone with doubled chromosomes are detected). The relative proportions of the founder and doubled clone often differ between diagnosis and relapse, with the doubled clone generally dominating at diagnosis and the founder clone at relapse. Professional illustration by Somersault18:24.
The diagnostic challenge of masked hypodiploidy
A frequent phenomenon in hypodiploid ALL is doubling of the chromosomal content, resulting in clones with 50 to 78 chromosomes (ie, in the hyperdiploid and near-triploid range) (Figure 1). Such chromosomal doubling has been seen in 76 out of 119 (64%) near-haploid and 44 out of 99 (44%) low-hypodiploid reported cases.7 The doubled clone contains tetrasomies for chromosomes that were disomic in the hypodiploid clone and uniparental disomies with complete loss of heterozygosity (LOH) for chromosomes that were monosomic. Hypodiploid ALL sometimes harbors blast cells of 2 different sizes,3,8-11 and based on one such case, where the smaller blasts harbored near haploidy and the larger ones hyperdiploidy, the chromosomal doubling is believed to occur via endoreduplication (ie, replication of the genome without subsequent cytokinesis).10 Notably, the doubled clone may be the only one detected at diagnosis (sometimes denoted “masked hypodiploidy”). In fact, low-hypodiploid and near-triploid adult ALL are often considered to be one genetic subgroup.12-14
The presence of a doubled clone does not affect the prognosis of hypodiploid ALL.12,15,16 However, doubling of a near-haploid chromosomal set leads to a clone with 50 to 60 chromosomes, overlapping with the high-hyperdiploid (51-67 chromosomes) ALL subtype, which is associated with a favorable prognosis.17 Thus, mistaking masked near haploidy for high hyperdiploidy could lead to erroneous risk classification and hence risk of treatment failure.8,10,15,18-20 Cytogenetically, masked near-haploid ALL may be distinguished by having mainly tetrasomies, in contrast to “true” high hyperdiploidy, which mainly will have trisomies along with tetrasomy 21 (Table 1). Alternatively, interphase fluorescence in situ hybridization or DNA indexing by flow cytometry may be used (Table 1).21 Notably, masked near-haploid cases are readily detectable by SNP array analysis as having LOH involving all chromosomes that are not gained, whereas high-hyperdiploid cases mostly will display retained heterozygosity (Table 1).19,22 Thus, as SNP array analysis and, in the future, next-generation sequencing are increasingly being used for genetic testing of ALL, the risk of misclassifying masked near-haploid cases will decrease.
Near-haploid and low-hypodiploid ALL harbor distinct mutational profiles
Hypodiploid ALL generally has simple karyotypes with few structural aberrations, although chromosomal rearrangements seem to be more common in low hypodiploidy than in near haploidy.15,16,23 No recurrent structural abnormality and no common fusion gene have been identified.4,7,15,16,18,23 Relatively few microdeletions are seen, possibly because so much genetic material has already been lost through the hypodiploidy.4,19,24
Recently, Holmfeldt et al4 showed that near-haploid and low-hypodiploid ALL also harbor distinct mutational profiles (Figure 1). Characteristic of near-haploid ALL were somatic alterations targeting receptor tyrosine kinase and RAS signaling (71% of cases), in particular involving NF1 (44%), histone modifiers (64%), mainly CREBBP (32%), CDKN2A/B (20%), the 6p22 histone gene cluster (19%), IKZF3 (13%), and PAG1 (10%). In low-hypodiploid ALL, on the other hand, 91% of both childhood and adult cases harbored loss-of-function mutations in TP53; subsequent studies have confirmed that such mutations are a hallmark of this subtype.24,25 Notably, these mutations were also detected in nonmalignant cells in almost half of the pediatric cases, suggesting that childhood low-hypodiploid ALL may often be associated with Li-Fraumeni syndrome. Thus, patients presenting with low-hypodiploid ALL should be investigated for constitutional TP53 mutations and, if found, offered genetic counseling. In contrast, TP53 mutations in low-hypodiploid adult ALL always appear to be somatic.24,25 Other targeted genes comprised IKZF2 (53% of pediatric cases and 36% of adult cases), RB1 (41% of pediatric cases and 19% of adult cases), histone modifiers (60%), and CDKN2A/B (20%).4,19
Possible leukemogenic impact of hypodiploidy
The reason for the selective advantage that massive loss of chromosomes apparently provides in leukemogenesis is not known. It is possible that the hypodiploidy is a passenger event, but the fact that severe hypodiploidy is seen across a wide spectrum of neoplasms and involves different chromosomes in different diagnoses indicates that it does affect tumorigenesis.26 A common assumption has been that the widespread LOH resulting from hypodiploidy leads to the unmasking of recessive alleles.5,23,27,28 A candidate could be TP53, since the loss of chromosome 17, and thereby 1 TP53 allele, together with a mutation in the remaining copy in low-hypodiploid ALL, could result in complete absence of a functioning protein. However, this would only explain the monosomy for this particular chromosome.
A perhaps more likely outcome is global gene dosage effects resulting from the hypodiploidy. In line with this, differential expression for genes on retained chromosomes has been reported.4,25 The leukemogenic effect of the endoreduplication may be to restore the gene copy numbers to more normal levels, increasing the fitness of the cell, and to decrease the risk of nondisjunction events resulting in nullisomy. It is notable that the doubled clones frequently are no longer seen at relapse,28,29 which may indicate that they are more sensitive to treatment than the founder hypodiploid clones (Figure 1), although it cannot be excluded that cases where the doubled clone still dominates at relapse may be misclassified as high hyperdiploid and thus remain undetected, introducing a bias.
Clinical features
Near haploidy and low hypodiploidy are each seen in ∼0.5% of childhood ALL.15,23,29-32 In adult ALL, near haploidy has never been reported, whereas 3% to 4% of cases harbor low hypodiploidy or near triploidy.12-14,33 Although low-hypodiploid ALL was initially reported to be more common in males, subsequent studies have shown roughly equal incidence in males and females, similar to near-haploid ALL.3,4,13,15 All near-haploid ALL cases have been 1 to 19 years old at diagnosis; the median age is 5 years.7 Low-hypodiploid ALL, on the other hand, occurs at all ages but with a higher median age than pediatric ALL in general; in cases 1 to 15 years old, it was 11.5 years.7 In adults, the incidence of low hypodiploidy increases with age.12,33 Thus, the age profiles of patients with near-haploid and low-hypodiploid ALL differ, with the former being restricted to childhood and the latter becoming more frequent with increasing age. Both groups display relatively low white blood cell counts of <50 × 109/L at diagnosis.8,12,14-16,23,30,31,34
Outcome and future treatment options
Generally, complete remission (CR) is achieved with induction therapy for childhood hypodiploid ALL,6,8,30 whereas CR rates are lower in low hypodiploid adult ALL.12,13 The event-free survival (EFS) rates are very low, with 5- to 8-year EFS rates of 25% to 40% for near-haploid ALL, 30% to 50% for childhood low hypodiploid ALL, and 0% to 20% for adult low-hypodiploid ALL,13,15,16,29-31,33 despite stratification to high-risk protocols. Overall survival rates are not much higher: 35% to 50% for childhood hypodiploid ALL and 0% to 20% for adult low-hypodiploid ALL.12,13,16,30,31,33,35 It is not clear whether hematopoietic stem cell transplantation in CR1 is beneficial in near-haploid and low hypodiploid ALL.16,35,36 In contrast, negative minimal residual disease status at the end of induction therapy was recently reported to be associated with improved EFS in hypodiploid childhood ALL, and it was suggested that such patients may be curable with intensive chemotherapy.6
Considering the consistently poor outcome of hypodiploid ALL, patients in this group are prime candidates for novel treatment regimens. Noting that both near-haploid and low-hypodiploid ALL have activated RAS and phosphatidylinositol 3-kinase (PI3K) signaling, Holmfeldt et al4 investigated the sensitivity of cell lines and xenografts to MEK (functioning downstream of RAS), PI3K, and PI3K/mammalian target of rapamycin (mTOR) inhibitors. Whereas MEK inhibition did not have an effect, PI3K and PI3K/mTOR inhibitors reduced proliferation in all investigated tumors. Thus, inhibition of the PI3K pathway may be a novel treatment option for hypodiploid ALL.
Concluding remarks
There are still many questions remaining regarding the role of hypodiploidy in leukemogenesis. Clearly, the massive loss of chromosomes must promote leukemia development, but the exact mechanism remains elusive. The recent mapping of additional genetic changes showed that near-haploid and low-hypodiploid ALL indeed are different subtypes with unique mutational patterns. Notably, the high frequency of constitutional TP53 mutations in childhood patients with low-hypodiploid ALL prompts further genetic testing of family members and counseling. The possibly increased risk of future malignancies in survivors of low-hypodiploid ALL must also be considered. Finally, the still very poor outcome of hypodiploid ALL with current treatment protocols must be addressed, and it will be interesting to see whether novel targeted treatments may increase survival in these patients.
Acknowledgments
This work was supported by the Danish Council for Independent Research, the Swedish Cancer Fund, the Swedish Childhood Cancer Foundation, and the Swedish Research Council.
Authorship
Contribution: S.S. and K.P. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kajsa Paulsson, Division of Clinical Genetics, BMC C13, SE-221 84, Lund, Sweden; e-mail: kajsa.paulsson@med.lu.se.