In this issue of Blood, Chong et al report a comprehensive landscape of structural rearrangements (SRs) of the programmed death ligand (PDL) locus (9p24.1), which harbors PDL1 (CD274) and PDL2 (PDCD1LG2) providing novel information about the breakpoint anatomy of the SRs in a large cohort of B-cell lymphomas (eg, non-Hodgkin lymphoma [NHL]). Importantly, the studies were performed by using DNA obtained from formalin-fixed paraffin-embedded tissues.1
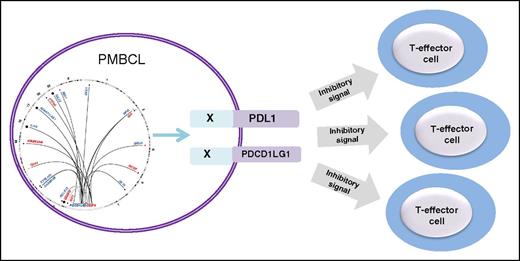
A complex landscape of structural rearrangements involving the PDL locus revealed by capture sequencing results in alteration of the tumor immune environment in PMBCL.
A complex landscape of structural rearrangements involving the PDL locus revealed by capture sequencing results in alteration of the tumor immune environment in PMBCL.
The exploitation of immune regulatory checkpoints that lead to evasion of anti-tumor responses2 is emerging as an important mechanism in tumor initiation, progression, and therapy resistance. Members of the B7 family, including PDL1 (CD274) and PDL2 (PDCD1LG2), have been implicated in the pathogenesis of lymphomas via several mechanisms,3 including transcriptional regulation, copy-number variations (CNVs),4 and chromosomal SRs.5 To date, fluorescence in situ hybridization (FISH) studies have revealed CNVs of PDL1/2 frequently in classical Hodgkin lymphoma and SRs of PDL1/2 in primary mediastinal B-cell lymphoma (PMBCL) and primary testicular lymphoma. However, the specific location of the breakpoints in either PDL1 or PDL2 remains elusive because of the technical challenges associated with evaluating these rare neoplasms, most of which are available for study as formalin-fixed paraffin-embedded tissues. The study by Chong et al demonstrates the feasibility of applying similar methodologies to characterize the landscape of PDL SRs in other lymphomas.
By using an oligonucleotide capture sequencing strategy targeting the 9p24.1 locus in 67 clinical specimens and 1 cell line, the study successfully identified 36 novel PDL SRs, 14 involving CD274, 19 involving PDCD1LG2, and 3 involving both PDLs concurrently. Importantly, 13 (36%) of 36 SRs are observed in cases in which FISH failed to identify an SR. In addition to the translocation partners previously identified, such as CIITA,6 IGHV7-81, IGHG2, and NRG1, the Chong et al study significantly expands the number of partner genes to PDL1/2.
Despite the highly promiscuous nature of the PDL locus, the exact translocation breakpoints are not conserved, but translocations involving PDL1/2 occur in well-defined cluster breakpoint regions 1 (CBR1) and 2 (CBR2) in addition to recurrent intrachromosomal PDL SRs that include duplications, deletions, and inversions. The figure shows the landscape of SRs of the PDL locus identified in PMBCL and their functional consequences. Adding to the clinical relevance of the findings is the observation that PDL SRs, including those with deletions of the 3′ untranslated region (3′UTR) of either CD274 or PDCD1LG2, demonstrated increased membrane expression of the proteins. Similarly, a microinversion displacing the CD274 3′UTR and disrupting the PDCD1LG2 coding sequence demonstrated CD274 protein expression. Functional relevance of the PDL SR is provided by the observation that expression of wild-type PDCD1LG2 and the PDCD1LG2-IGHV7-81 3′ fusion in a non–PDL-expressing B-cell lymphoma is associated with cell membrane expression of the protein, which impairs the level of activation of cocultured T cells implying suppression of the T-cell–mediated immune response.
Recent developments in checkpoint inhibitors and clinical trials that evaluate these agents in lymphoma heighten the importance of identifying genetic alterations that have an impact on clinical outcomes. The work by Chong et al, although it provides a previously unrecognized genetic landscape of a plethora of PDL SRs that involve numerous partner genes, deletions, and microinversions, also raises important questions. Future studies will need to address whether PDL SRs are associated with distinct clinical outcomes, as has been recently reported in classical Hodgkin lymphoma7 and, more intriguingly, the possibility that CBR1 and CBR2 may impart distinct clinical responses to standard therapies as well as checkpoint blockade.
The observation that PDL SRs are highly associated with protein expression in PMBCL suggests the possibility that standard laboratory techniques such as immunohistochemistry may prove to be sufficient in selecting patients for checkpoint blockade therapy. It is anticipated that future studies will investigate whether PDL SRs play a role in contributing to resistance to checkpoint inhibitors. The ability to carry out comprehensive base-pair resolution breakpoint mapping of PDL SRs from formalin-fixed paraffin-embedded tissues provides an opportunity for the identification of similar genetic alterations in other lymphomas. In addition, the robust nature of oligonucleotide capture sequencing demonstrated in the Chong et al study will likely lead to an investigation into the identification of SRs of other members of the novel B7 family molecules.8
Conflict-of-interest disclosure: The author declares no competing financial interests.