Key Points
Expansion of circulating monocytic myeloid-derived suppressor cells (MDSCs) correlates with clinical outcomes in patients with DLBCL.
Mechanisms of MDSC-dependent T-cell inhibition in DLBCL are related to IL-10, PD-L1, and S100A12.
Abstract
In diffuse large B-cell lymphoma (DLBCL), the number of circulating monocytes and neutrophils represents an independent prognostic factor. These cell subsets include monocytic and granulocytic myeloid-derived suppressor cells (M- and G-MDSCs) defined by their ability to suppress T-cell responses. MDSCs are a heterogeneous population described in inflammatory and infectious diseases and in numerous tumors including multiple myeloma, chronic lymphocytic leukemia, and DLBCL. However, their mechanisms of action remain unclear. We broadly assessed the presence and mechanisms of suppression of MDSC subsets in DLBCL. First, a myeloid suppressive signature was identified by gene expression profiling in DLBCL peripheral blood. Accordingly, we identified, in a cohort of 66 DLBCL patients, an increase in circulating G-MDSC (LinnegHLA-DRnegCD33posCD11bpos) and M-MDSC (CD14posHLA-DRlow) counts. Interestingly, only M-MDSC number was correlated with the International Prognostic Index, event-free survival, and number of circulating Tregs. Furthermore, T-cell proliferation was restored after monocyte depletion. Myeloid-dependent T-cell suppression was attributed to a release of interleukin-10 and S100A12 and increased PD-L1 expression. In summary, we identified expanded MDSC subsets in DLBCL, as well as new mechanisms of immunosuppression in DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin lymphoma, accounting for ∼40% of new cases. Although DLBCL is recognized as a single entity by the World Health Organization, several subgroups with different outcomes have been described.1 Despite introduction of immunotherapy, treatment failure is observed in ∼40% of DLBCL, emphasizing the need for prognostic biomarkers.2 The International Prognostic Index (IPI), with variants, still considered the standard prognostic factor for DLBCL, stratifies patients at diagnosis. However, this score does not take into account biological differences among subgroups.3-5 Additional prognostic factors, evaluated at the time of diagnosis, include various cytokines (soluble PD-L1,6 CxCL10,7 and interleukin-10 [IL-10]),7 tumor characteristics (cell of origin,8 LMO2,9 and MYC, BCL2, and BCL6 rearrangements),10 and components from the tumor microenvironment (CD3pos, CD4pos, and Foxp3 tumor-infiltrating T cells,11,12 CD68pos myeloid cells).2,13,14 Recently, an increase in circulating neutrophil15 or monocyte16,17 counts, combined with a decrease in lymphocyte count (so-called lymphocyte to monocyte ratio [LMR] and neutrophil to lymphocyte ratio [NLR]) have been proposed as independent prognostic markers in the rituximab era.
Specific granulocyte and monocyte subsets have demonstrated their immunosuppressive functions in cancers and in inflammatory diseases and are recognized as myeloid-derived suppressor cells (MDSCs).18,19 These cells are immature, arise from the myeloid lineage, and are defined by their immunosuppressive function. Human MDSCs, a heterogeneous population with a lack of specific markers, belong to 2 major types: granulocytic (G-MDSC; LinnegHLA-DRnegCD33posCD11bpos) and monocytic (M-MDSC; CD14posHLA-DRlow). Various additional markers (including CD116, CD124, vascular endothelial growth factor receptor [VEGFR], CD11c, CD11b, and CD274) have been associated with MDSCs.19 Interestingly, these markers are likely regulated through various stimuli, depending on the tumor type.20 MDSC induction and expansion are mediated by a combination of soluble factors (including VEGF, granulocyte-macrophage colony-stimulating factor, macrophage colony-stimulating factor, S100A8/A9, IL-4, IL-6, and IL-10) produced by tumor and/or surrounding cells such as stromal cells, T cells, or macrophages.19 These factors essentially trigger activation of the STAT3, STAT6, and STAT1 family, leading to the expression of genes involved in the blockade of the myeloid differentiation or in immune regulation. These multiple suppressive mechanisms converge to impair effector T cell and natural killer cell functions and also contribute to macrophage polarization toward a M2 anti-inflammatory phenotype. In humans, various mechanisms have been described including (1) regulatory T-cell (Treg) expansion, (2) depletion in amino acids essential for T-cell metabolism by expression of arginase 1 (ARG1) or indoleamine 2,3-dioxygenase (IDO), (3) production of reactive oxygen species (ROS) by expression of NADPH oxidase (NOX2), and (4) IL-10, transforming growth factor β (TGFβ) release, and PD-L1 expression.20,21
In most solid cancers studied (melanoma, renal, lung, or prostate cancer, hepatocellular carcinoma), although both G- and M-MDSCs were increased, a predominance of G-MDSCs was mostly detected.22,23 However, by using a model that allowed separate study of G- and M-MDSCs in tumor-bearing mice, Haverkamp et al demonstrated that M-MDSC was the most suppressive subset and that loss of G-MDSCs did not alter tumor incidence.24 In lymphoid malignancies, an enrichment in circulating MDSCs has been described in myeloma,25 T-cell lymphoma,26 chronic lymphocytic leukemia (CLL),27 and DLBCL.28,29 In DLBCL, G-MDSCs have never been studied, and only arginase 1 expression has been evaluated as a mechanism of M-MDSC–mediated immunosuppression.28,29 Here, we hypothesized that the increase in neutrophils and monocytes in peripheral blood of DLBCL patients might be related to an increase of G- and M- MDSCs. We found, using gene expression profiling (GEP), a myeloid suppressive signature in DLBCL peripheral blood. Accordingly, we identified an expansion in circulating G-MDSC and M-MDSC counts. Interestingly, M-MDSCs were immunosuppressive and correlated with the IPI and with event-free survival (EFS). Finally, we pointed out IL-10, PD-L1, and S100A12 as MDSC-related mechanisms of suppression in DLBCL.
Methods
Samples
Two cohorts of patients were used in this study. For GEP, 76 DLBCL patients at diagnosis from the Groupe Ouest-Est d'Etude des Leucémies et Autres Maladies du Sang (GOELAMS) 075 clinical trial (clinicaltrials.gov: NCT00561379) and 87 matched healthy donors (HDs) were enrolled. This cohort was already published by our group.6 Then, a prospective cohort was set with 66 DLBCL patients at diagnosis (BMS-LyTRANS study; clinicaltrials.gov: NCT01287923). Patients with previous corticosteroid treatment were excluded from this study. Forty-five age-matched HDs were also enrolled. Clinical characteristics of DLBCL patients enrolled in this second cohort are listed in Table 1. The research protocol was conducted under French legal guidelines and fulfilled the requirements of the local institutional ethics committee.
Whole blood GEP
Whole blood was collected into PAXgene Blood RNA tubes (Becton Dickinson BD Biosciences, San Jose, CA), ensuring blood stabilization and stored at −80°C before RNA extraction. High-quality total RNA was hybridized onto Affymetrix GeneChip Human Exon 1.0 ST oligonucleotide arrays (Affymetrix, Santa Clara, CA) according to the manufacturer’s instructions. Microarray quality control was assessed using Affymetrix Expression Control v1.1. Raw data were prefiltered by selection of core-annotated probe sets. Then, a Detection Above BackGround summary file was generated for the Robust Multiarray Average normalized data using Affymetrix Power Tools. DABG calculates the P value that the intensities in a probe set could have been observed by chance in a background distribution. A probe set was defined as present if its DABG P value was <5% in ≥50% of the samples in ≥1 condition. A transcript was preserved if ≥50% of the core probe sets of the transcript were present. Prefiltered data were RMA normalized with adjustment for GC content, and probe sets were mean-summarized to transcripts (Partek Genomics Suite version 6.5; Partek, St Louis, MO). Unsupervised analysis using hierarchical clustering analysis (HCA) was performed on the normalized data. Differentially expressed genes were identified by the Mann-Whitney nonparametric test, with adjustment for multiple testing using the Benjamini-Hochberg false discovery rate set at 5%. Array data are available in the Gene Expression Omnibus, accession number GSE83632. Interpretation of the functional roles was established using Ingenuity Pathway Analysis (IPA) (Ingenuity Systems) based on the 2 gene lists identified on the HCA as over- or underexpressed in DLBCL patients compared with HD. Canonical pathways analysis identified the pathways from the IPA knowledge database that were most significant to the data set (false discovery rate, P < 5%). Gene set enrichment assay (GSEA) was performed using the BROAD Institute GSEA software (http://www.broad.mit.edu/gsea/) on the DLBCL dataset using a panel of genes that we defined as related to myeloid suppressive cells based on literature knowledge (supplemental Table 1, available on the Blood Web site).30
Quantitative real-time polymerase chain reaction
Total RNA was extracted using PAXgene blood RNA kit (Qiagen, Valencia, CA). cDNA was then generated using Multiscribe Reverse Transcriptase and High-Capacity cDNA reverse Transcription kit (Invitrogen, Carlsbad, CA). For quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR), we used custom designed Taqman array microfluidic cards and the Taqman Universal Master Mix from Applied Biosystems (Invitrogen). Gene expression was measured by the ABI Prism 7900HT Sequence Detection System. 18S, CDKN1B, and ELF1 were determined as appropriate internal standard genes using TaqMan endogenous control assays and geNorm algorithm analysis. For each sample, the cycle threshold (CT) value for the gene of interest was determined, normalized to the geometric mean value of the 3 housekeeping genes, and compared with the median value obtained from healthy donors using the 2-ddCT method. Results were expressed as the ratio of DLBCL mean to HD mean for each gene. Experiments were also performed on sorted CD14posHLA-DRlow or CD14posHLA-DRhigh cells from 3 DLBCL peripheral blood samples. cDNA was prepared using Fluidigm Reverse Transcription Master Mix (Fluidigm, Sunnyvale, CA). The qPCR was performed in triplicate using 96.96 Dynamic Array integrated fluidics circuit and the BioMark HD System from Fluidigm. For each CD14posHLA-DRlow sample, the mean CT value for the gene of interest was calculated, normalized to the geometric mean value of the 2 housekeeping genes, and compared with the median value obtained from CD14posHLA-DRhigh samples using the 2-ddCT method. Results were expressed as the ratio of CD14posHLA-DRlow mean to CD14posHLA-DRhigh mean for each gene.
Fluorescent flow cytometry analysis
Blood samples were collected on heparin tubes. Plasma specimens were stored at −80°C until use. White blood cell (WBC) differential was performed using the Cytodiff panel (CD36, CD2, CD294, CD19, CD16, CD45; Beckman Coulter, Brea, CA) on 50 µL blood as previously described.31 Flow cytometry analysis of M-MDSCs, G-MDSCs, and dendritic cells were performed on whole blood (300 µL/tube) with the antibody panels described in supplemental Table 2. Absolute counts were obtained using Flow-Count beads (Beckman Coulter). An erythrocytes lysis (Uti-Lyse Dako, Carpinteria, CA) was performed before analysis by flow cytometry (Navios; Beckman Coulter). G-MDSCs and M-MDSCs were identified as shown in supplemental Figure 2 using Kaluza software (Beckman Coulter). Tregs and tumor circulating B cells were defined on blood mononuclear cells (PBMCs) after density gradient centrifugation (lymphocyte separation medium; Eurobio, CourtaBoeuf, France). For Treg determination, cells were fixed and permeabilized using the FOXP3 staining kit (BD Biosciences). Results were expressed as absolute values (106 or 109 cells/L), percentages, or mean florescence intensity.
Cytokines and enzymes assessment
Concentrations of IL-10 (eBioscience, San Diego, CA), TGF-β1 (R&D Systems, Minneapolis, MN), S100A8/9, S100A12, and Arginase I (Hycult Biotech, Ude, The Netherlands) in the plasma or culture supernatants were measured by enzyme-linked immunosorbent assay (ELISA), according to the manufacturer’s recommendations. Finally, arginase I and IDO activities were evaluated by high-performance liquid chromatography analysis32 as the ornithine to arginine and tryptophan to kynurenine ratio, respectively.
T-cell proliferation assay
Fresh PBMCs from DLBCL or HD samples were separated into 2 fractions. The first fraction was positively depleted for residual erythrocytes using glycophorin A beads (Miltenyi Biotech, Bergisch Gladbach, Germany), whereas the second fraction was depleted for erythrocytes and monocytes (CD14 microbeads; Miltenyi Biotech). The purity was >97%. The 2 negative fractions (PBMC and PBMC-CD14pos) were labeled with 0.2 μM carboxyfluorescein succinimidyl ester (CFSE; Interchim, Montlucon, France). PBMCs and PBMC-CD14poss were plated in triplicate in 96-well round-bottom plates (Corning Costar, Corning, NY) with RPMI 1640 medium enriched with 10% of human serum AB (Biowest, Courtaboeuf, France) and 1% penicillin-streptomycin (Gibco, Saint Aubin, France). Then, cells were stimulated with soluble anti-CD3 (0.6 μg/mL; Sanquin, Amsterdam, The Netherlands) and anti-CD28 (0.6 μg/mL; Sanquin) and cultured at 37°C in 5% CO2 and 100% humidity for 4 days. For some experiments, CD3pos T cells were purified using magnetic beads (CD3 microbeads, Miltenyi Biotec), labeled with CFSE, and cocultured with autologous CD14posHLA-DRlow– or CD14posHLA-DRhigh–sorted cells (FACS ARIA III; BD Biosciences) with a ratio of 2:1 in the presence of anti-CD3/anti-CD28 stimulation. CFSE dilution was assessed by flow cytometry and analyzed with ModFit LT software (Verity Software House, Topsham, ME). Results were expressed by the proliferation index.
T-cell proliferation assay with inhibitors
Cryopreserved PBMCs from DLBCL patients depleted for residual erythrocytes were CFSE labeled and then platted in triplicate at 106 cells/mL in RPMI 1640 medium enriched with 10% human serum AB and 1% penicillin-streptomycin in 96-well round-bottom plates and incubated at 37°C for 30 minutes with or without different neutralizing antibodies: anti–IL-10 (1-methyl-L-tryptophan; 10 μg/mL; R&D systems); anti-S100A12 (10 μg/mL; Acris, San Diego, CA), and anti-PD1 (20 μg/mL; clone 3.1 given by Daniel Olive)33 or with chemical analogs: L-1MT (1 mM; Sigma-Aldrich, Diegem, Belgium)34 and nor-NOHA (Nω-hydroxy-nor-L-Arginine; 20 mM; Enzo Life Sciences, Antwerpen, Belgium). At the concentration used, nor-NOHA inhibited >80% of the arginase 1 activity on a positive control (data not shown). Thereafter, cells were stimulated with soluble anti-CD3 and anti-CD28 and incubated at 37°C in 5% CO2 and 100% humidity for 4 days. CFSE dilution was assessed by flow cytometry and analyzed with ModFit LT software. Results were expressed as the percentage of proliferation of inhibitor-treated cells to control (anti-IgG1 isotype, 10 μg/mL, R&D for neutralizing antibodies, or medium plus vehicle for chemical analogs). DLBCL patients were classified into groups: without (Gr.1) or with (Gr.2) suppressive monocytes. The classification was based on suppressive assay or a number of M-MDSCs at more than threefold the median count.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA) using Spearman correlation, Wilcoxon, or Mann-Whitney tests as appropriate.
Results
Myeloid regulatory GEP is detected in DLBCL peripheral blood
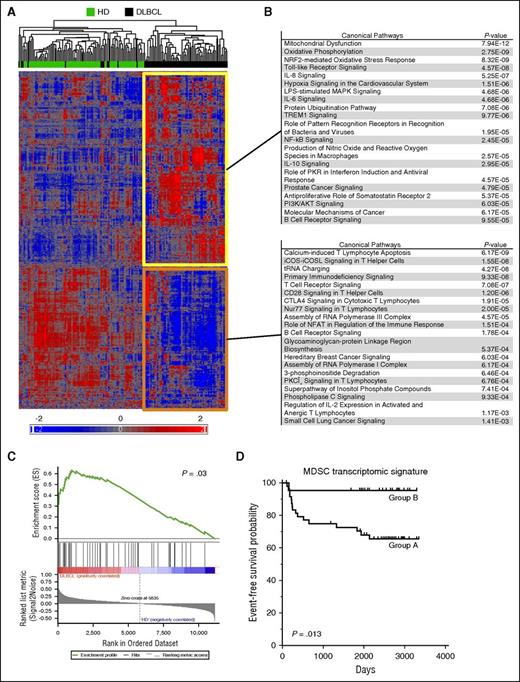
GEP performed on whole blood was evaluated using the Affymetrix Human Exon 1.0 ST Array on 76 DLBCL patients and 87 HDs. Following prefiltering, the 11 302 remaining probe sets (PSs) were classified by an unsupervised hierarchical clustering. Sixty-five of 76 DLBCL samples were clustered together with a set of 6792 upregulated PSs and a set of 4510 downregulated PSs compared with HDs (Figure 1A). To further identify the immune response pathways affected in DLBCL, these PSs were filtered (P < .05 and |fold change| > 1.5), highlighting 564 up- and 495 downregulated PSs in DLBCL compared with HD. These PSs were further separately analyzed with IPA (Figure 1B). Within the 20 most represented pathways in DLBCL, we identified an innate immune response signature (TLR, IL8, IL6, LPS, TREM1, NO/ROS signaling), as well as an anti-inflammatory response (IL-10 signaling). On the other hand, a significant underrepresentation of pathways involved in T-cell activation (ICOS, ICOSL, T cell receptor, CD28, CTLA4 signaling, regulation of IL-2 expression) was also revealed (Figure 1B). Then, by GSEA, we found in our DLBCL cohort a significant enrichment for a list of myeloid genes known to be upregulated in MDSCs (P = .03) (supplemental Table 1; Figure 1C). Finally, using the same list of myeloid genes (supplemental Table 1), we performed a hierarchical clustering analysis on the 76 DLBCL samples (supplemental Figure 1). A group of patients (group B) with low expressions of MDSC-related genes (including ARG1, S100A12, S100A8, MMP8, SLPI, MPO, CD274, TNFAIP6, CxCL5, and AIM2) showed a better EFS probability than the other group of patients (group A; P = .013; Figure 1D). Altogether, these results were in favor of a myeloid regulatory signature in DLBCL with a clinical impact on patients' prognosis.
A myeloid transcriptomic signature is detected in blood of DLBCL patients. (A) Hierarchical clustering of 76 DLBCL and 87 HD. After raw data normalization and prefiltering, 11 302 PSs were classified by an unsupervised hierarchical clustering. Up- (yellow) and downregulated (orange) gene lists were separately analyzed by IPA for canonical pathways. (B) Genes were filtered (P < .05 and |fold change| > 1.5) before IPA analysis and the top 20 significant pathways are shown for both gene lists. (C) GSEA plot in the studied cohort for a list of gene involved in MDSCs (supplemental Table 1). Genes were ranked by signal to noise ratio. (D) With the same list of genes, the 76 DLBCLs were classified by an unsupervised hierarchical clustering in group B with low expression of MDSC-related genes (supplemental Figure 1) or in group A. The EFS probability was calculated for both groups with a log-rank test.
A myeloid transcriptomic signature is detected in blood of DLBCL patients. (A) Hierarchical clustering of 76 DLBCL and 87 HD. After raw data normalization and prefiltering, 11 302 PSs were classified by an unsupervised hierarchical clustering. Up- (yellow) and downregulated (orange) gene lists were separately analyzed by IPA for canonical pathways. (B) Genes were filtered (P < .05 and |fold change| > 1.5) before IPA analysis and the top 20 significant pathways are shown for both gene lists. (C) GSEA plot in the studied cohort for a list of gene involved in MDSCs (supplemental Table 1). Genes were ranked by signal to noise ratio. (D) With the same list of genes, the 76 DLBCLs were classified by an unsupervised hierarchical clustering in group B with low expression of MDSC-related genes (supplemental Figure 1) or in group A. The EFS probability was calculated for both groups with a log-rank test.
MDSCs are expanded in peripheral blood of DLBCL patients
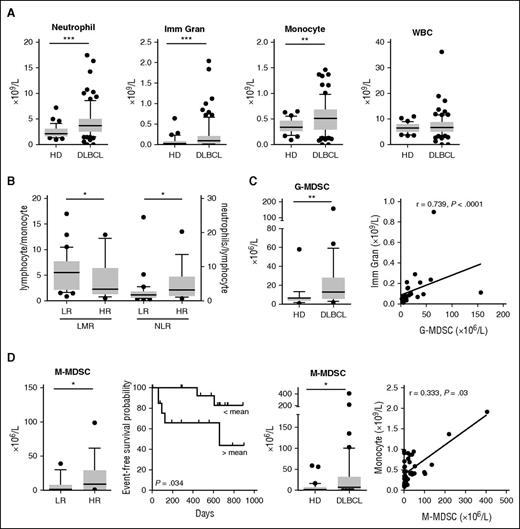
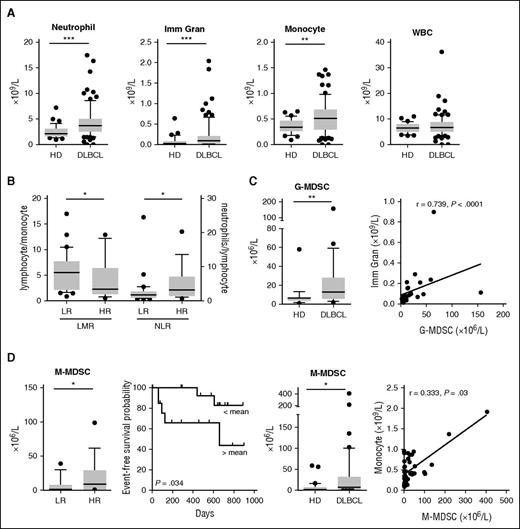
We next focused our attention on the quantitative modifications of myeloid cells in the peripheral blood of DLBCL patients. A new prospective cohort was set up with 66 DLBCL patients at diagnosis (BMS-LyTRANS study), as well as 45 HD samples. We noticed in DLBCL a significant increase in the monocyte (median: 0.51 vs 0.34 × 109/L, P = .002), neutrophil (median: 3.69 vs 2.14 × 109/L, P < .0001), and immature granulocyte counts (median: 0.09 vs 0.03 × 109/L, P = .0003) obtained on a regular complete blood count, whereas the WBC counts were not different (Figure 2A). Tumor circulating B cells were defined as CD19posCD10posCD38low and κpos or Λpos and were detected in 9 of 40 studied cases (22.5%), with a median count at 0.48 × 109/L representing 0.16% to 25.3% of the WBCs (data not shown). No correlation was found between tumor circulating cells and monocytes or granulocytes (data not shown). We found in patients with high-risk (HR; age-adapted [aa]IPI ≥3) disease a decrease of the LMR and an increase of the NLR compared with low-risk (LR) patients (P = .044 and 0.033, respectively; Figure 2B).
MDSC-like subsets accumulate in peripheral blood of DLBCL patients but only M-MDSCs correlate with the clinical status of DLBCL patients. (A) Neutrophils, immature granulocytes (Imm Gran), monocytes as determined on a complete blood count, and WBC counts are shown for 45 HD and 66 DLBCL samples. Box and whisker plots with the 10 to 90 percentiles and the outliers are shown. (B) DLBCL patients were separated in LR (n = 16) or HR (n = 34) groups based on the aaIPI score (<3 and ≥3, respectively). LMR and NLR ratio are shown. (C) (Left) G-MDSC and M-MDSC counts were performed in 23 HD and 31 DLBCL samples. (Right) Correlation (Spearman) between G-MDSC and Imm Gran counts and between M-MDSC and monocytes counts. (D) For 29 DLBCL samples, clinical information was available. (Top) Patients were separated in a LR (n = 12) or HR (n = 17) group based on the aaIPI score (<3 and ≥3, respectively). M-MDSCs are shown for these groups. (Bottom) Patients were split up between high (n = 14) and low (n = 15) count of M-MDSC (with a threshold at the median count), and the EFS probability was calculated for both groups with a log-rank test. *P < .05, **P < .01, ***P < .001.
MDSC-like subsets accumulate in peripheral blood of DLBCL patients but only M-MDSCs correlate with the clinical status of DLBCL patients. (A) Neutrophils, immature granulocytes (Imm Gran), monocytes as determined on a complete blood count, and WBC counts are shown for 45 HD and 66 DLBCL samples. Box and whisker plots with the 10 to 90 percentiles and the outliers are shown. (B) DLBCL patients were separated in LR (n = 16) or HR (n = 34) groups based on the aaIPI score (<3 and ≥3, respectively). LMR and NLR ratio are shown. (C) (Left) G-MDSC and M-MDSC counts were performed in 23 HD and 31 DLBCL samples. (Right) Correlation (Spearman) between G-MDSC and Imm Gran counts and between M-MDSC and monocytes counts. (D) For 29 DLBCL samples, clinical information was available. (Top) Patients were separated in a LR (n = 12) or HR (n = 17) group based on the aaIPI score (<3 and ≥3, respectively). M-MDSCs are shown for these groups. (Bottom) Patients were split up between high (n = 14) and low (n = 15) count of M-MDSC (with a threshold at the median count), and the EFS probability was calculated for both groups with a log-rank test. *P < .05, **P < .01, ***P < .001.
As described in solid tumors22 and multiple myeloma,25 both granulocyte and monocyte compartments contain MDSC subsets. In CLL27 and DLBCL,28,29 only M-MDSCs were already detected in patients. Thus, we next evaluated the absolute count of circulating MDSCs and identified, in whole blood, G-MDSCs (LinnegCD123lowHLA-DRnegCD33posCD11bpos) and M-MDSCs (CD14posHLA-DRlow). MDSC counts were significantly increased in DLBCL compared with HD samples with a median count at 13 vs 6 × 106/L (P = .001) for G-MDSCs and 7.42 vs 2 × 106/L (P = .01) for M-MDSCs (Figure 2C). Interestingly, G-MDSCs and M-MDSCs correlate with immature granulocyte and monocyte counts (P < .0001 and P = .03), respectively (Figure 2C). No correlation was found between G-MDSC and M-MDSC counts (data not shown). Finally, M-MDSCs were increased in HR compared with LR DLBCL (P = .049) and were associated with a worse EFS (P = .034, hazard ratio = 0.19; Figure 2D). G-MDSCs did not correlate with risk factors or EFS (data not shown). As the myeloid regulatory compartment in peripheral blood also includes dendritic cells, we assessed the 3 subsets of dendritic cells as follows: LinnegHLA-DRposCD123posCD141negCD1cneg pDC, LinnegHLA-DRposCD123negCD141posCD1cneg type 1 cDCs and LinnegHLA-DRposCD123negCD141negCD1cpos type 2 cDCs.35 These cell types were significantly decreased in DLBCL compared with HDs, without correlation with any type of MDSCs (supplemental Figure 3).
M-MDSCs from DLBCL patients suppress T-cell response
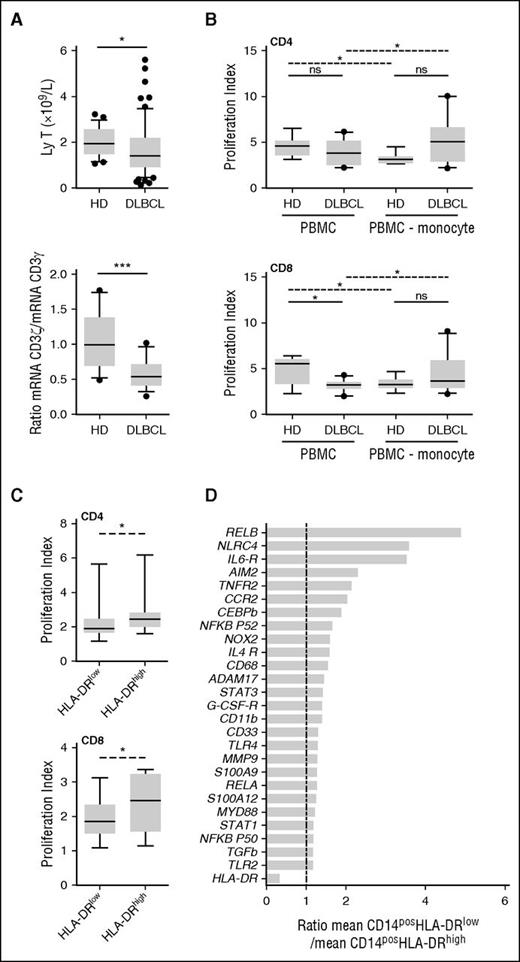
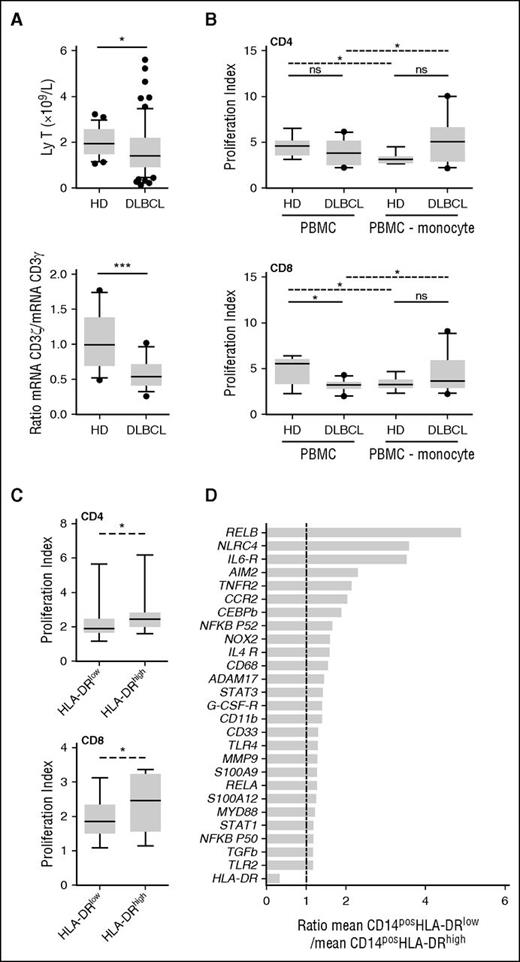
MDSC immunosuppressive activities lead to a T-cell proliferation blockade or apoptosis. Noteworthy T-cell count and the ratio of CD3ζ (CD247) to CD3γ mRNA were decreased in DLBCL (P = .01 and P = .0009, respectively; Figure 3A). Given the fact that (1) in DLBCL, only M-MDSCs were correlated with prognostic, and (2) this subset has been described being the dominant suppressive population of MDSCs,24 we evaluated the suppressive capability of the monocyte fraction. We noticed that CD8pos T lymphocytes issued from DLBCL patients proliferated less than those from HD in response to polyclonal activation (P = .01; Figure 3B). Interestingly, when CD14pos monocytes were depleted from PBMCs, the proliferation index ratio (monocyte-depleted PBMCs to PBMCs) was increased for DLBCL patients, whereas it was decreased for HDs, demonstrating a specific suppressive activity for monocytes in lymphoma patients (Figure 3B). To show the specific in vitro suppressive capacity of CD14posHLA-DRlow cells, autologous CD3pos T cells were stimulated in their presence or with CD14posHLA-DRhigh cells. CD4pos and CD8pos T-cell proliferation index was lower in the presence of CD14posHLA-DRlow cells compared with DRhigh monocytes (Figure 3C). Then, by qPCR analysis performed on 3 DLBCL patients, we found a panel of genes involved in MDSC functions to be slightly increased in CD14posHLA-DRlow compared with CD14posHLA-DRhigh (Figure 3D).
M-MDSCs are immunosuppressive in DLBCL. (A) (Left) T-cell count. (Right) Ratio of CD3ζ to CD3γ mRNA evaluated by qRT-PCR (TLDA). Box and whisker plots with the 10 to 90 percentiles and the outliers are shown. *P < .05, ***P < .001. (B) Activated CFSE-labeled T lymphocytes were cultured in the presence (PBMC) or absence (PBMC-monocyte) of autologous monocytes isolated from DLBCL patients and HD. Proliferation index for CD4pos (top) and CD8pos (bottom) cells in presence of autologous monocyte (PBMC) or after monocyte depletion (PBMC-monocyte) are represented on box and whisker plots with the 10 to 90 percentiles and the outliers (n = 10 DLBCL and n = 8 HD). Wilcoxon (dashed line) or Mann-Whitney (solid line) tests were used for paired nonparametric and nonpaired nonparametric analyses, respectively. (C) Activated CFSE-labeled T lymphocytes were cultured in the presence of autologous sorted M-MDSCs (HLA-DRlow) or monocytes (HLA-DRhigh) isolated from DLBCL patients (n = 7). Proliferation index for CD4pos (top) and CD8pos (bottom) cells are represented on box and whisker plots with the 10 to 90 percentiles. (D) Gene expression evaluated by qPCR on CD14posHLA-DRlow and CD14posHLA-DRhigh cells sorted from 3 DLBCL. For each gene, the relative expression (mRNA) of CD14posHLA-DRlow and CD14posHLA-DRhigh PBMC was compared (ratio of the mean expression on CD14posHLA-DRlow to the mean expression on CD14posHLA-DRhigh). ns, nonsignificant; *P < .05.
M-MDSCs are immunosuppressive in DLBCL. (A) (Left) T-cell count. (Right) Ratio of CD3ζ to CD3γ mRNA evaluated by qRT-PCR (TLDA). Box and whisker plots with the 10 to 90 percentiles and the outliers are shown. *P < .05, ***P < .001. (B) Activated CFSE-labeled T lymphocytes were cultured in the presence (PBMC) or absence (PBMC-monocyte) of autologous monocytes isolated from DLBCL patients and HD. Proliferation index for CD4pos (top) and CD8pos (bottom) cells in presence of autologous monocyte (PBMC) or after monocyte depletion (PBMC-monocyte) are represented on box and whisker plots with the 10 to 90 percentiles and the outliers (n = 10 DLBCL and n = 8 HD). Wilcoxon (dashed line) or Mann-Whitney (solid line) tests were used for paired nonparametric and nonpaired nonparametric analyses, respectively. (C) Activated CFSE-labeled T lymphocytes were cultured in the presence of autologous sorted M-MDSCs (HLA-DRlow) or monocytes (HLA-DRhigh) isolated from DLBCL patients (n = 7). Proliferation index for CD4pos (top) and CD8pos (bottom) cells are represented on box and whisker plots with the 10 to 90 percentiles. (D) Gene expression evaluated by qPCR on CD14posHLA-DRlow and CD14posHLA-DRhigh cells sorted from 3 DLBCL. For each gene, the relative expression (mRNA) of CD14posHLA-DRlow and CD14posHLA-DRhigh PBMC was compared (ratio of the mean expression on CD14posHLA-DRlow to the mean expression on CD14posHLA-DRhigh). ns, nonsignificant; *P < .05.
In DLBCL, M-MDSC suppressive activity is independent from arginase I and IDO activities
In B-cell malignancies, arginase 1 and IDO activities were reported in DLBCL29 and CLL,27 respectively. MDSCs mediate T-cell suppression through various mechanisms.21 Therefore, we compared in DLBCL vs HD peripheral blood the gene expression of 5 key enzymes (ARG1, IDO1, NOS2, HO-1, and PTGS2) involved in MDSC biology. Among those, only ARG1 was significantly increased in DLBCL (P = .003). IDO1, although not significant, showed a trend of upregulation (1.8-fold) in DLBCL (Figure 4A). To evaluate arginase 1 and IDO activities, we measured the concentration of arginine and tryptophan, as well as their degradation products, ornithine and kynurenine, respectively. Arginase 1 and IDO activities were increased in DLBCL (P = .02 and P = .04, respectively; Figure 4B). Finally, we measured by ELISA the arginase 1 enzyme in plasma and found increased levels in DLBCL samples compared with HD (median at 157.4 vs 16.66 ng/mL; P < .0001; Figure 4C). Then, based on the presence or absence of suppressive monocytes, 2 groups of DLBCL were constituted, and we showed that in DLBCL the proliferation of CD4pos and CD8pos cells issued from PBMCs was not increased in the presence of arginase or IDO inhibitors regardless of the presence of M-MDSCs (Figure 4D). Interestingly, arginase 1 in plasma did not correlate with M-MDSCs, but instead was correlated with the number of immature granulocytes, which contains G-MDSCs36 (r = 0.33, P = .049; Figure 4E).
Arginase 1 and IDO are not M-MDSC mechanisms of immunosuppression in DLBCL. (A) Expression of ARG1, IDO1, NOS2, HO-1, and PTGS2 was evaluated by TLDA on 17 DLBCL and 15 HD samples. The relative gene expression (mRNA) of PBMCs from DLBCL and HD was compared (ratio of the mean expression on DLBCL to the mean expression on HD). (B) Arginase 1 and IDO activities were evaluated by high-performance liquid chromatography with the ratio ornithine to arginine and kynurenine to tryptophan, respectively (n = 31 DLBCL and 21 HD). (C) Arginase 1 in plasma was evaluated by ELISA (n = 43 DLBCL and 33 HD). *P < .05, **P < .01, ***P < .001. (D) Percentage of increase or decrease in T-cell proliferation (proliferation index) between treated (L-1MT or nor-NOHA) and untreated cells, evaluated for CD4pos and CD8pos cells by CFSE labeling (n = 8 DLBCL for L-1MT and n = 7 for nor-NOHA). DLBCL patients were classified into groups: without (Gr.1, n = 5) or with (Gr.2, n = 2 or 3) suppressive monocytes. The classification was based on suppressive assay or a number of M-MDSCs at more than threefold the median count. (E) Correlation of the arginase 1 in plasma and M-MDSC count (left) or immature granulocytes count (Imm gran) (right) count in DLBCL peripheral blood (n = 36 DLBCL). ns, not significant.
Arginase 1 and IDO are not M-MDSC mechanisms of immunosuppression in DLBCL. (A) Expression of ARG1, IDO1, NOS2, HO-1, and PTGS2 was evaluated by TLDA on 17 DLBCL and 15 HD samples. The relative gene expression (mRNA) of PBMCs from DLBCL and HD was compared (ratio of the mean expression on DLBCL to the mean expression on HD). (B) Arginase 1 and IDO activities were evaluated by high-performance liquid chromatography with the ratio ornithine to arginine and kynurenine to tryptophan, respectively (n = 31 DLBCL and 21 HD). (C) Arginase 1 in plasma was evaluated by ELISA (n = 43 DLBCL and 33 HD). *P < .05, **P < .01, ***P < .001. (D) Percentage of increase or decrease in T-cell proliferation (proliferation index) between treated (L-1MT or nor-NOHA) and untreated cells, evaluated for CD4pos and CD8pos cells by CFSE labeling (n = 8 DLBCL for L-1MT and n = 7 for nor-NOHA). DLBCL patients were classified into groups: without (Gr.1, n = 5) or with (Gr.2, n = 2 or 3) suppressive monocytes. The classification was based on suppressive assay or a number of M-MDSCs at more than threefold the median count. (E) Correlation of the arginase 1 in plasma and M-MDSC count (left) or immature granulocytes count (Imm gran) (right) count in DLBCL peripheral blood (n = 36 DLBCL). ns, not significant.
IL-10, PDL-1, and S100A12 are involved in M-MDSC suppressive activity
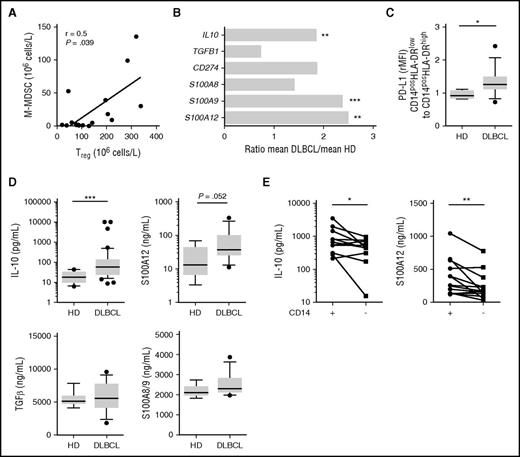
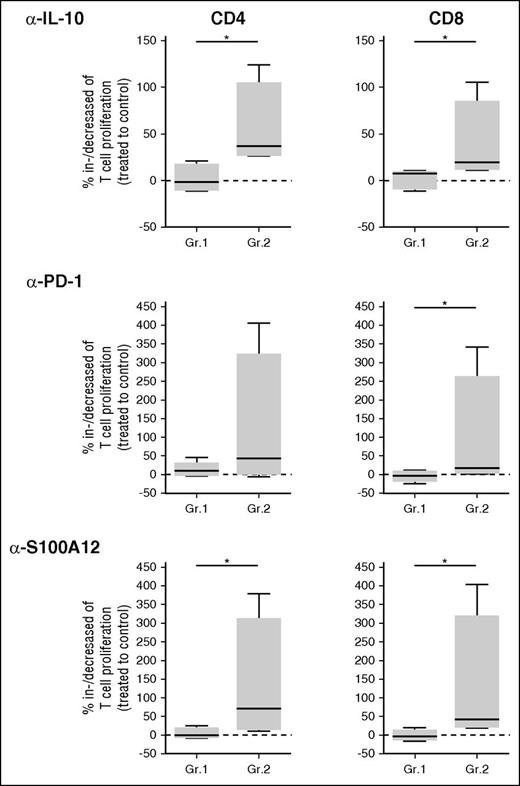
Because of the lack of correlation between M-MDSCs and arginase 1 or IDO in DLBCL, we sought for additional mechanisms of immunosuppression already described for MDSCs, including Treg induction, IL-10 and TGFβ release, and PD-L1 expression.21 First, we found that Treg and M-MDSC counts were correlated in DLBCL patients (r = 0.5, P = .039; Figure 5A). We then compared in DLBCL and HD peripheral blood, the gene expression of immunosuppressive molecules. IL10, S100A9, and S100A12 were significantly overexpressed in DLBCL unlike TGFB1 and S100A8. In addition, although not significant, CD274/PD-L1 was also increased in DLBCL by 1.85-fold (Figure 5B). Interestingly, although the expression of PD-L1 on M-MDSCs was increased in DLBCL (P = .015; Figure 5C), CD274 expression did not correlate with the circulating malignant B-cell count (data not shown). Similarly, IL-10 was increased at the protein level in DLBCL peripheral blood (P < .001), and a trend was also noticed for S100A12 (P = .052) unlike TGF-β1 and S100A8/A9 (Figure 5D). We thus evaluated the levels of IL-10 and S100A12 in culture supernatants from stimulated PBMCs, depleted or not from CD14pos cells. These molecules were decreased after CD14pos depletion (P < .05; Figure 5E). Finally, on the 2 groups of DLBCL classified depending on M-MDSC content, we analyzed the percentage of proliferating T cells after CD3/CD28 stimulation in the presence of various blocking antibodies (IL-10, PD-1, and S100A12; Figure 6). In the group of patients with circulating MDSCs, neutralizing IL-10, PDL-1, and S100A12 resulted in an increase of CD4 and/or CD8 T-cell proliferation.
In DLBCL, IL-10, PD-L1, and S100A9/12 are expressed by monocytes. (A) Correlation of the Treg and M-MDSC counts in DLBCL peripheral blood (n = 17). (B) Expression of the immunomodulatory genes IL10, TGFβ1, CD274, S100A8, S100A9, and S100A12 evaluated by TLDA. The relative gene expression (mRNA) of PBMC from DLBCL and HD was compared (ratio of the mean expression on DLBCL to the mean expression on HD; n = 17 DLBCL and 15 HD). (C) Surface PD-L1 by flow cytometry expressed as ratio of mean fluorescence intensity for CD14posHLA-DRlow to CD14posHLA-DRhigh (n = 13 DLBCL and 8 HD). (D) Cytokines detection by ELISA in HD (n = 8) and DLBCL plasma: IL-10 (n = 45), TGF-β (n = 14), S100A8/9 (n = 14), and S100A12 (n = 14). (E) Cytokine (IL-10 and S100A12) detection by ELISA in supernatants of PBMCs not depleted (+) or depleted (−) in CD14pos cells (n = 13 DLBCL). *P < .05, **P < .01, ***P < .001.
In DLBCL, IL-10, PD-L1, and S100A9/12 are expressed by monocytes. (A) Correlation of the Treg and M-MDSC counts in DLBCL peripheral blood (n = 17). (B) Expression of the immunomodulatory genes IL10, TGFβ1, CD274, S100A8, S100A9, and S100A12 evaluated by TLDA. The relative gene expression (mRNA) of PBMC from DLBCL and HD was compared (ratio of the mean expression on DLBCL to the mean expression on HD; n = 17 DLBCL and 15 HD). (C) Surface PD-L1 by flow cytometry expressed as ratio of mean fluorescence intensity for CD14posHLA-DRlow to CD14posHLA-DRhigh (n = 13 DLBCL and 8 HD). (D) Cytokines detection by ELISA in HD (n = 8) and DLBCL plasma: IL-10 (n = 45), TGF-β (n = 14), S100A8/9 (n = 14), and S100A12 (n = 14). (E) Cytokine (IL-10 and S100A12) detection by ELISA in supernatants of PBMCs not depleted (+) or depleted (−) in CD14pos cells (n = 13 DLBCL). *P < .05, **P < .01, ***P < .001.
In DLBCL, IL-10, PDL-1, and S100A12 are involved in M-MDSC suppressive activity. DLBCL patients were classified into groups: without (Gr.1, n = 5) or with (Gr.2, n = 4) suppressive monocytes based on suppressive assay or a number of M-MDSCs at more than threefold the median count. The percentage of increase or decrease in T-cell proliferation (proliferating index) between cells treated by blocking antibodies (anti–IL-10, –PD-1, and -S100A12) and an irrelevant antibody is shown for CD4pos and CD8pos cells. *P < .05.
In DLBCL, IL-10, PDL-1, and S100A12 are involved in M-MDSC suppressive activity. DLBCL patients were classified into groups: without (Gr.1, n = 5) or with (Gr.2, n = 4) suppressive monocytes based on suppressive assay or a number of M-MDSCs at more than threefold the median count. The percentage of increase or decrease in T-cell proliferation (proliferating index) between cells treated by blocking antibodies (anti–IL-10, –PD-1, and -S100A12) and an irrelevant antibody is shown for CD4pos and CD8pos cells. *P < .05.
Discussion
Circulating MDSCs have been described in various solid tumors with, in most cases, a predominance of G-MDSCs.22,23 Regarding lymphoid neoplasias, both G- and M- MDSCs are increased in Hodgkin lymphoma and in multiple myeloma.25,37,38 Few studies were conducted on B-cell lymphomas or CLL, which focused exclusively on the M-MDSC subtype.27-29,39 Because the majority of blood biomarkers in DLBCL are linked to tumor microenvironment and in particular to myeloid cell biology,6,7,14-17 further characterization of the MDSC compartment is highly relevant in this disease. Our aim, in this study, was to characterize the various MDSC subsets in peripheral blood from DLBCL patients.
As previously described we found an increase in neutrophil and monocyte counts, whereas the number of lymphocytes was reduced. This was also reflected in the whole blood GEP from DLBCL by an overexpression of myeloid-related genes and a downregulation of lymphoid-related genes. Additionally, we found enrichment in pathways involved in the innate immunity (TLR, IL8, IL6, LPS, TREM1, NO/ROS signaling) or in the anti-inflammatory response (IL-10 signaling).
Because there is no specific lineage marker, MDSC definitions are divergent, as such we explored by flow cytometry various panels on the first 15 patients: G-MDSC1 (LinnegHLA-DRnegCD33posCD11bpos), G-MDSC2 (LinnegHLA-DRnegCD33posCD11bposCD34pos), G-MDSC3 (LinnegCD123lowHLA-DRnegCD33posCD11bpos), G-MDSC4 (SSChighCD66bpos) and also: M-MDSC1 (CD14posHLA-DRlow), M-MDSC2 (CD14posCCR2high), and M-MDSC3 (CD14posCD163pos).40,41 We retained the phenotype LinnegCD123lowHLA-DRnegCD33posCD11bpos and CD14posHLA-DRlow as the most informative (data not shown), in particular we found CD123 helpful in discarding polynuclear basophils during the gating strategy as their phenotype overlaps G-MDSCs. We then found higher counts of G-MDSCs (LinnegCD123lowHLA-DRnegCD33posCD11bpos). During the preparation of this manuscript and in agreement with our results, an expansion of myeloid cells with a G-MDSC phenotype has been reported in a small cohort of DLBCL patients (n = 5).42 In our study, G-MDSCs did not correlate with clinical outcomes or Treg count. Additionally, the suppressive fraction of granulocyte was recently identified as remaining granulocytes in the PBMC fraction after density cell separation and called low density neutrophils.43 In DLBCL, G-MDSCs were detected only in whole blood but were absent from PBMCs (data not shown). Thus, we focused our attention to M-MDSCs.
M-MDSCs (CD14posHLA-DRlow) were expanded in DLBCL patients, in concordance with 2 other studies with altogether 41 DLBCL patients.28,29 Finally, an additional monocyte subset (CD14posCD16pos) was found increased in DLBCL, although partially overlapping with the M-MDSCs (data not shown). Whether, this latter subset displays suppressive function is controversial. For some authors, these cells do produce the antiproliferative cytokine IL-10 or induce Tregs,44-46 whereas for others, they highly express HLA-DR and release the proinflammatory cytokine tumor necrosis factor α.47 One explanation might be their heterogeneity and gating challenge by flow cytometry; thus, the CD14posCD16pos compartment likely contains different subsets including M-MDSCs. Interestingly, in our DLBCL cases, CD14posCD16pos cells downregulated HLA-DR, a hallmark of MDSCs (data not shown). Altogether, our results were in favor of an increase in various MDSC subsets in DLBCL, with a clinical relevance for the M-MDSC population. In contrast to most solid tumors, where G-MDSCs are the predominant population of MDSCs,23 in lymphoid disease the M-MDSCs are preeminent.27-29,39 M-MDSCs were recently defined, in tumor-bearing mice, as the most immunosuppressive subset.24 In DLBCL, we have shown that CD14pos cells exerted a suppressive activity on T cells because their proliferation is restored when CD14pos cells are depleted.
In lymphoid malignancies, arginase 1 and IDO are 2 mechanisms of suppression already described.27,29 In our study, neither arginase 1 nor IDO activity was involved in the MDSC suppressive activity. The arginase 1 protein level was found highly expressed in the plasma without any relationship to M-MDSC subsets but rather correlated to the immature granulocyte fraction. Consequently, arginase 1 might likely be released by granulocytes but is not responsible for the M-MDSC suppressive activity. Two previous papers already reported an expansion of M-MDSCs in DLBCL. One paper did not demonstrate functionally the suppressive activity of these cells, which is essential for the MDSC characterization.28 The second one did not focus on the DLBCL but rather involved a large variety of non-Hodgkin’s lymphomas. Moreover, the functional assays were limited, and with the exception of arginase 1, no other mechanisms of T-cell suppression were explored.29
These results prompted us to seek for additional suppressive mechanisms in MDSCs from DLBCL. Release of IL-10, TGF-β, and activation of the PD-L1/PD-1 axis are known MDSC mechanisms of suppression.48-51 Interestingly, in DLBCL, we and others have shown that IL-10 and sPD-L1 can be elevated compared with HD.6,52 Herein, we demonstrated that monocyte suppressive activity was mediated by a release of IL-10 by CD14pos cells and an expression of PD-L1 on monocytes. These suppressive activities were impaired by neutralizing antibodies.53 Nevertheless, in most DLBCL we found beside G- and M-MDSCs, an increase of neutrophils, immature granulocytes, and monocytes, potentially including suppressive subsets; thus, we cannot rule out that the mechanisms described in our study could be shared between various subsets. We also demonstrated that S100A12 is responsible for T-cell suppression in DLBCL. In accordance with this finding, S100A8, S100A9, and S100A12 are expressed in MDSCs from hepatocellular and colorectal carcinomas,54,55 and S100 proteins, among other abilities, enhance the suppressive activity of MDSCs via an increase in ROS production.56-58 The number of Tregs, another mediator of T-cell proliferation arrest, was correlated with M-MDSC count but not with G-MDSC count. Taken altogether, our results indicate that several regulatory mechanisms might be involved by MDSCs to block the immune response in DLBCL patients.
Here, we demonstrated that in DLBCL at diagnosis, M-MDSCs are predominant and are correlated with a worse prognosis. It would be interesting to monitor these populations during the course of the treatment; indeed, accumulation or depletion of MDSCs is influenced by chemotherapy and/or immunotherapy.59 Our study also points out the potential interest of MDSCs as targets in future therapeutic trials on DLBCL patients. Controlling the expansion and accumulation of MDSCs or blocking their suppressive functions, for instance, by CSF-R160 or an S100 family member61 targeting, represents promising novel approaches in cancer therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are indebted to the clinicians of the Bretagne Reseau Expertise en Hématologie (BREHAT) network and to the French Blood Bank of Rennes for providing samples. The authors acknowledge the Centre de Ressources Biologiques of Rennes (BB-0033-00056) for managing samples. Affymetrix microarrays were processed in the Microarray Core Facility of the Institute of Research on Biotherapy, Centre Hospitalier Universitaire, INSERM, Université de Montpellier 1, Montpellier, France. The authors thank Daniel Olive (INSERM 1068, Marseille, France) for the gift of the neutralizing anti–PD-1.
This work was supported by a research grant from the National Institute of Cancer (INCa Recherche Translationnelle 2010), the Groupe Ouest-Est des Leucémies et des Autres Maladies du Sang, the Lymphoma Study Association (LYSA) group, and the Ligue Nationale Contre le Cancer (équipe labellisée). M.R. is a recipient of a fellowship from the Nuovo-Soldati Fundation.
Authorship
Contribution: I.A. designed and performed experiments, analyzed data, and contributed to the writing; F.U. designed and performed experiments and analyzed data; D.R., C.P., J.D., and J.L.P. analyzed data; T.L., R.H., S.L.G., G.C., P.G., K.B., N.M., and G.D. provided samples; K.T. and T.F. raised the funds, designed and supervised research, analyzed data, and contributed to the writing; M.R. designed and supervised research, analyzed data, and wrote the paper; and all authors revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karin Tarte, INSERM U917, Faculté de Médecine, 2 Avenue du Pr Léon Bernard, F-35043 Rennes, France; e-mail: karin.tarte@univ-rennes1.fr; Thierry Fest, INSERM U917, Faculté de Médecine, 2 Avenue du Pr Léon Bernard, F-35043 Rennes, France; e-mail: thierry.fest@univ-rennes1.fr; or Mikael Roussel, Laboratoire Hématologie, CHU Pontchaillou, 2 rue Henri Le Guilloux, F-35033 Rennes, France; e-mail: mikael.roussel@chu-rennes.fr.