Abstract
Mutations in genes encoding RNA splicing factors constitute the most common class of alterations in patients with myelodysplasticsyndromes (MDS). These occur predominantly as heterozygous point mutations at restricted residues in SF3B1, SRSF2, and U2AF1 in a mutually exclusive manner, suggesting that spliceosomal gene mutations confer gain-of-function with converging biological effects. However, recent studies suggest that mutations in each splicing factor result in distinct alterations in pre-mRNA splicing. It is therefore unclear if such mutual exclusivity is due to overlapping biological effects and/or synthetic lethal interactions. Furthermore, although cells bearing mutant splicing factors have been shown to require the wildtype allele for survival, whether these mutations can exist in a homozygous state is unknown. Here we addressed these questions by analyzing the effects of expressing SF3B1 and SRSF2 mutations simultaneously or in a homozygous state in vivo.
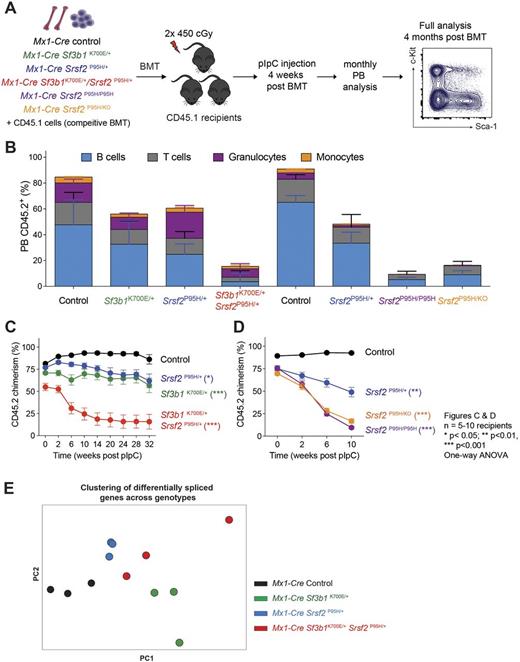
Re-analysis of published sequencing data revealed that only 2% of MDS patients (86/4,032) have mutations in >1 splicing factor simultaneously (whether these mutations are present in the same cell or not is unclear). Of these 86 patients, co-mutations in SRSF2 and SF3B1 represent the most prevalent combination (n=23/86) with all SRSF2 mutations affecting the P95 residue while all co-existing SF3B1 mutations occurred outside of the most commonly mutated K700 residue. To understand the basis for exclusivity of SRSF2P95 and SF3B1K700 mutations, we generated mice for inducible heterozygous expression of Sf3b1K700E/+ and Srsf2P95H/+ mutations simultaneously (Mx1-cre Sf3b1K700E/+/Srsf2P95H/+). We next performed competitive bone marrow transplantation (BMT) assays where each mutation was induced, alone or together, following stable engraftment (Figure 1A). Simultaneous expression of Sf3b1K700E and Srsf2P95H mutations resulted in severe defects on the self-renewal and differentiation of hematopoietic stem and progenitor cells (HSPC), which were outcompeted by wildtype and single-mutant HSPCs (Figure 1B). In noncompetitive BMT assays, HSPCs co-expressing Sf3b1K700E and Srsf2P95H mutations had severe defects in multi-lineage reconstitution (Figure 1C). Analyses of hematopoietic organs 6 months post-BMT revealed a near complete absence of Sf3b1K700E/+/Srsf2P95H/+ double-mutant cells, which was distinct from expression of Sf3b1K700E or Srsf2P95H mutationalone. In addition, mice with conditional homozygous expression of the SRSF2P95H mutation (Mx1-cre Srsf2P95H/P95H) had severe defects in HSPC self-renewal as well as multi-lineage reconstitution, analogous to those seen with hemizygous Srsf2P95H expression (Mx1-cre Srsf2P95H/KO) (Figure 1D).
As noted earlier, SF3B1 and SRSF2 mutations cause different effects on mRNA splicing. However, there has never been a direct comparison of the effects of each of these mutations in an isogenic context. To address this and to understand the mechanistic basis for exclusivity of these mutations, we performed RNA-seq on lineage- c-Kit+ cells from Mx1-cre Sf3b1K700E/+/Srsf2P95H/+ andcontrols 2 weeks after conditionally expressing each mutation alone or together. As evidence of the intolerability of combined SF3B1/SRSF2 mutations, mean allelic ratio of Sf3b1K700E and Srsf2P95H expressed in double-mutant mice was 20.7% and 33.5%, respectively, markedly lower than the near 50% expression seen in single-mutant controls. Despite this, principle component analysis of differentially spliced genes revealed distinct changes mediated by expression of Sf3b1K700E and Srsf2P95H mutations (Figure 1E). Moreover, previously described changes to alternative 3" splice site selection as well as cassette exon splicing were seen in cells bearing Sf3b1K700E and Srsf2P95H mutation, respectively, as well as in Sf3b1K700E/+/Srsf2P95H/+ double-mutantcells.
These findings indicate thatspliceosomalgene mutations, despite imparting distinct alterations on gene expression and splicing, are not tolerated when co-expressed in the same cell, thus providing a basis for their strong mutual exclusivity in MDS. These data, combined with the fact that neitherhemizygousnor homozygous expression of splicing factor mutations is tolerated, further establishes the unique requirement ofspliceosomalmutant cells on the remaining function ofwildtypespliceosomecomponents.
Palacino:H3 Biomedicine Inc.: Employment. Seiler:H3 Biomedicine: Employment. Buonamici:H3 Biomedicine: Employment. Smith:H3 Biomedicine Inc.: Employment.
Author notes
Asterisk with author names denotes non-ASH members.