In this issue of Blood, Hofer et al report 6-sulfo LacNAc (slan) as a marker for transcriptionally distinct subsets of CD16-positive (CD16+) monocytes that are expanded in male subjects with sarcoidosis (slan-negative CD16+) or depleted during hereditary diffuse leukodystrophy with axonal spheroids (HDLS) (slan-positive CD16+).1
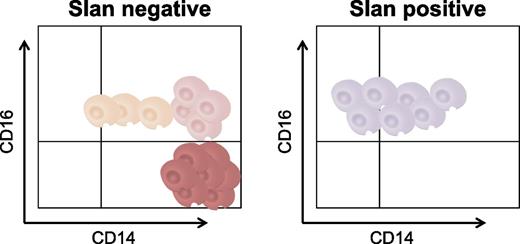
A new slan-based nomenclature for circulating CD16+ monocytes. slan identifies subsets of peripheral blood CD16+ monocytes that are differentially altered in frequency during sarcoidosis (increased frequency of slan-negative CD16+ monocytes in males) vs HDLS (reduced frequency of slan-positive CD16+ monocytes). This new slan-based classification offers a better understanding of monocyte development during homeostasis and disease pathogenesis.
A new slan-based nomenclature for circulating CD16+ monocytes. slan identifies subsets of peripheral blood CD16+ monocytes that are differentially altered in frequency during sarcoidosis (increased frequency of slan-negative CD16+ monocytes in males) vs HDLS (reduced frequency of slan-positive CD16+ monocytes). This new slan-based classification offers a better understanding of monocyte development during homeostasis and disease pathogenesis.
Monocytes originate from bone marrow precursors, circulate through the blood for a few days, and further differentiate into macrophages or dendritic cells upon recruitment into tissues under homeostatic or inflammatory conditions. Similar to other leukocyte types, peripheral blood monocytes are highly heterogeneous in morphology, phenotype, and immune functions. Monocyte differentiation may occur within the blood stream as a consequence of their abortive interactions with endothelial beds, a process known as marginalization2 or patrolling.3 In addition, certain studies in mice provide evidence that monocytes are recruited into nonlymphoid tissues, where they can take up antigens and then recirculate into lymph nodes with no major changes in their phenotype.4 Therefore, the pool of peripheral blood monocytes includes cells freshly derived from the bone marrow, as well as cells more advanced in their developmental program as a consequence of their marginalization/patrolling and likely recirculation.
In 1989, the development of flow cytometry and monoclonal antibodies facilitated the discovery of a subset of small monocytes expressing Fcγ receptor III/CD16.5 This discovery opened a new chapter in our understanding of monocyte heterogeneity during homeostasis and disease pathogenesis. In 2010, a panel of experts proposed a novel nomenclature for monocytes based on the differential expression of CD14 and CD16: classical (CD14++CD16−), intermediate (CD14++CD16+), and nonclassical (CD14+CD16++) monocytes.6 Indeed, these 3 monocyte subsets represent distinct stages of monocyte differentiation with distinct functional features.3 They are known to be recruited into different tissues via chemokine C-C motif receptor 2 for classical and intermediate monocytes, or via chemokine CX3C motif receptor 1 for nonclassical monocytes.6
slan is a carbohydrate modification of P-selectin glycoprotein ligand 1 (PSGL-1) recognized by the monoclonal antibody M-DC8.7 Although initially considered to be dendritic cells, the current consensus is that slan-positive CD16+ cells represent a fraction of nonclassical monocytes.1,3 In their article, Hofer et al revisit the CD14/CD16–based nomenclature6 and propose that a slan-based definition of CD16+ monocytes allows a better identification of monocyte subsets with unique transcriptional signatures and frequency variation during disease pathogenesis.1 Briefly, the authors identified and isolated monocyte subsets using 2 flow-cytometry gating approaches, CD14/CD16 and CD14/slan/CD16. In healthy subjects, the number of nonclassical CD14+CD16++ monocytes (48.1 ± 27.5 cells per microliter) was higher compared to that of slan-positive CD16++ monocytes (36.8 ± 23 cells per microliter; n = 10); this was because nonclassical monocytes include a fraction of CD14+ slan-negative CD16++ cells. However, the number of intermediate CD14++CD16+ monocytes (24.0 ± 11.2 cells per microliter) was lower compared to that of slan-negative CD16+ monocytes (41.7 ± 24.1 cells per microliter; n = 10) because the slan-negative fraction included both CD14++ and CD14+ monocytes. Intermediate vs nonclassical monocytes, together with slan-positive CD16++ vs slan-negative CD16+ monocytes, were sorted using magnetic beads, and their transcriptional profiling was characterized via massive analysis of complementary DNA ends and RNA sequencing with an Illumina HiSeq2000.1 Unique transcriptional signatures were identified using the CD14/CD16 and the slan/CD16 approach, with 676 and 385 differentially expressed genes, respectively, being identified (fold change cutoff, 1.2). Consistent with the role of carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (CHST2) in adding the slan residue on PSGL-1, CHST2 transcripts were upregulated in nonclassical vs intermediate monocytes.1 A particularity of slan-positive CD16++ vs slan-negative CD16+ monocytes was a ubiquitin C interactome1 and the superior expression of purinergic receptor P2Y, G-protein coupled, 10 (P2RY10), a G protein-coupled receptor demonstrated to bind sphingosine-1-phosphate and lysophosphatidic acid.8
All lines of evidence provided by Hofer et al1 clearly support the idea that slan-positive CD16++ monocytes represent a unique stage of monocyte differentiation. There remain many questions regarding the developmental origin of slan-positive CD16++ monocytes and their functional role.
Classical CD14++CD16− monocytes lack slan expression.1,3,7,9 However, a fraction of classical monocytes express a different epitope on PSGL-1: the cutaneous lymphocyte antigen (CLA),9 a motif involved in cell recruitment into the skin via binding to P- and E-selectin.7 If we consider that monocyte heterogeneity into the blood stream is the consequence of their differentiation from classical into intermediate and then into nonclassical monocytes, it is reasonable to assume that monocyte differentiation is associated with a switch in their trafficking potential from skin homing (via CLA) toward the ability to home into other lymphoid and/or nonlymphoid tissues (via slan). It was demonstrated that slan-positive cells fail to bind to P- and E-selectin,7 but the ligand for slan remains unknown. Whether slan acts as a new “zip code” for monocyte migration into specific tissues remains to be clarified. In addition to slan, defining the role of P2YR10 and other specific markers in regulating slan-positive CD16++ monocyte trafficking and function remains an important line of research with potential clinical applications for the treatment of pathological conditions associated with the expansion of these cells, such as HIV infection.10
Monocyte differentiation depends on macrophage colony-stimulating factor (M-CSF) signals via the M-CSF receptor (CD115). The expression of CD115 is indeed upregulated on CD16+ vs CD16− monocytes.9 Consistent with the fact that M-CSF controls slan expression, Hofer et al1 report that slan-positive CD16+ monocytes are almost absent in the peripheral blood of subjects with HDLS. The authors consistently identified several signaling molecules downstream from CD115 being upregulated in nonclassical monocytes of healthy individuals.1 In contrast to HDLS subsets, slan-negative, but not slan-positive, CD16+ monocytes were expanded in subjects with sarcoidosis.1
In conclusion, Hofer et al1 position slan on the expanding list of markers to be considered for the characterization of monocyte heterogeneity for immune monitoring studies. Of note, slan-positive and slan-negative CD16+ subsets are still heterogeneous in terms of intensity of CD14 expression, with the functional relevance of such differences remaining to be explored. This new insight will stimulate further investigations into the mechanisms regulating slan-positive monocyte development, the role of slan-positive monocytes in disease pathogenesis, and the possibility of using slan as a therapeutic target. On the basis of these studies, the recommendation that CD14 and CD16, together with slan, should be considered for the identification of functionally distinct monocyte subsets is now emerging (see figure).
Conflict-of-interest disclosure: The author declares no competing financial interests.