Abstract
Background
To analyze whether some distinct gene mutations could predict treatment response to Hypomethylation Agent---Decitabine.
Patients and Methods
Targeted next-generation sequencing was performed on reported 30 recurrently mutated genes for 106 MDS patients before they accepted Decitabine therapy. For those who presented satisfactory response (CR) or poor response (NR or PD), their gene sequencing were repeated after Decitabine therapy to analyze the alteration of the gene mutation.
Results
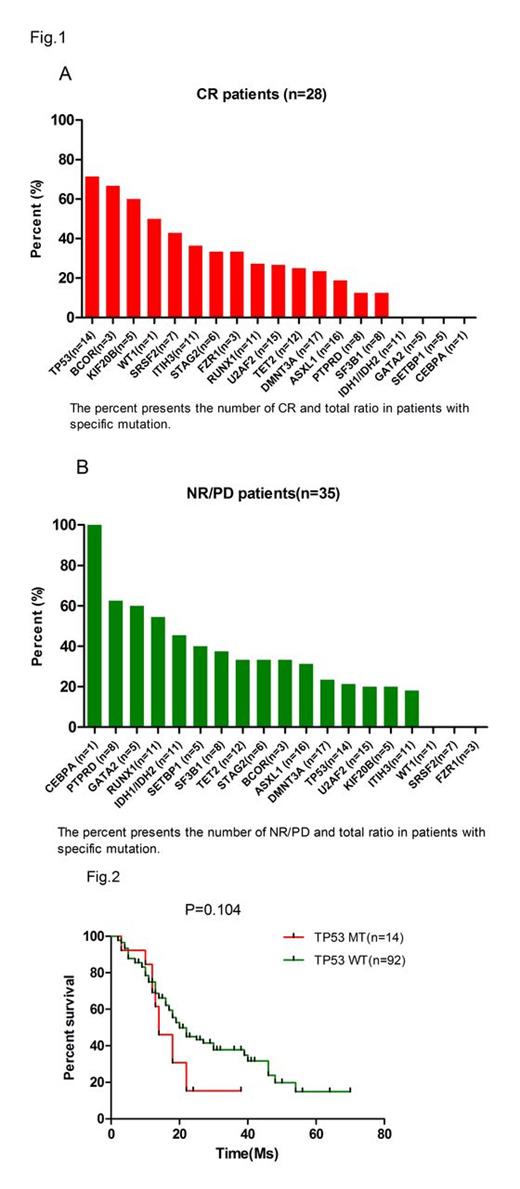
The overall response rate was 67.0%, with 26.4% patients (28 cases) achieving CR. The median survival of patients who achieved CR was longer than those who just achieved NR/PD (22 months VS 14 months, p =0.021). Among the 14 TP53 mutant patients, ten achieved CR (71.4%). Moreover, two of 3 BCOR mutant patients (66.7%) and three of 5 KIF20B mutant patient (60.0%) also achieved CR. Zero of the patients who were with IDH1/2(n=11), GATA2 (n=5), SETBP1 (n=5), EZH2 (n=3) or CEBPa (n=1) mutations could get CR response (Figure1). On the other hand, total five of the 8 PTPRD mutant patients (62.5%) and five of 11 ASXL1mutant patients did not responded to Decitabine. In addition, 16 among the 28 CR patients accepted the target sequencing after the stop of decitabine therapy. Ten of the 16 patients showed decreased number of mutant genes (Table1). Besides, the TP53 mutation was completely fade away in five of 7 patients after Decitabine treatment (Table 2). Finally, the TP53 mutant and wild type of MDS patients achieved comparable overall survival, suggesting decitabine treatment could significantly alter the prognostic impact of adverse genetic alterations.
Conclusion
Some gene mutation (specially TP53) seemed predictive for Decitabine response. Further studies with large samples are needed to identify it.
The number of mutant genes on CR patients before and after Decitabie therapy
| Sample . | Number of mutations . | response . | |
|---|---|---|---|
| Before | After | ||
| Sample 586 | 3 | 2 | CR |
| Sample 967 | 3 | 2 | CR |
| Sample 1761 | 4 | 2 | CR |
| Sample 1619 | 2 | 3 | CR |
| Sample 1864 | 7 | 4 | CR |
| Sample 1854 | 3 | 1 | CR |
| Sample 1818 | 3 | 2 | CR |
| Sample 1522 | 2 | 4 | CR |
| Sample 2038 | 2 | 2 | CR |
| Sample 2055 | 2 | 1 | CR |
| Sample 702 | 3 | 1 | CR |
| Sample 828 | 2 | 0 | CR |
| Sample 1361 | 0 | 0 | CR |
| Sample 1002 | 2 | 1 | CR |
| Sample 1917 | 1 | 2 | CR |
| Sample 1154 | 0 | 0 | CR |
| Sample . | Number of mutations . | response . | |
|---|---|---|---|
| Before | After | ||
| Sample 586 | 3 | 2 | CR |
| Sample 967 | 3 | 2 | CR |
| Sample 1761 | 4 | 2 | CR |
| Sample 1619 | 2 | 3 | CR |
| Sample 1864 | 7 | 4 | CR |
| Sample 1854 | 3 | 1 | CR |
| Sample 1818 | 3 | 2 | CR |
| Sample 1522 | 2 | 4 | CR |
| Sample 2038 | 2 | 2 | CR |
| Sample 2055 | 2 | 1 | CR |
| Sample 702 | 3 | 1 | CR |
| Sample 828 | 2 | 0 | CR |
| Sample 1361 | 0 | 0 | CR |
| Sample 1002 | 2 | 1 | CR |
| Sample 1917 | 1 | 2 | CR |
| Sample 1154 | 0 | 0 | CR |
Status of TP53 mutation after hypomethylating agents in CR patients.
| Sample . | Before . | After . | response . |
|---|---|---|---|
| Sample 586 | TP53/8/D242H | TP53-WT | CR |
| Sample 967 | TP53/6/R174L | TP53-WT | CR |
| Sample 1864 | TP53/8/R244H | TP53/8/R244H | CR |
| Sample 1854 | TP53/4/W91 | TP53-WT | CR |
| Sample 2038 | TP53/8/F231S | TP53/8/F231S | CR |
| Sample 2055 | TP53/6/R196* | TP53-WT | CR |
| Sample 1002 | TP53/8/R234H | TP53-WT | CR |
| Sample . | Before . | After . | response . |
|---|---|---|---|
| Sample 586 | TP53/8/D242H | TP53-WT | CR |
| Sample 967 | TP53/6/R174L | TP53-WT | CR |
| Sample 1864 | TP53/8/R244H | TP53/8/R244H | CR |
| Sample 1854 | TP53/4/W91 | TP53-WT | CR |
| Sample 2038 | TP53/8/F231S | TP53/8/F231S | CR |
| Sample 2055 | TP53/6/R196* | TP53-WT | CR |
| Sample 1002 | TP53/8/R234H | TP53-WT | CR |
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.