Key Points
CD166 identifies human and murine long-term repopulating stem cells.
CD166 is a functional marker of stem cells and the hematopoietic niche.
Abstract
We previously showed that immature CD166+ osteoblasts (OB) promote hematopoietic stem cell (HSC) function. Here, we demonstrate that CD166 is a functional HSC marker that identifies both murine and human long-term repopulating cells. Both murine LSKCD48−CD166+CD150+ and LSKCD48−CD166+CD150+CD9+ cells, as well as human Lin−CD34+CD38−CD49f+CD166+ cells sustained significantly higher levels of chimerism in primary and secondary recipients than CD166− cells. CD166−/− knockout (KO) LSK cells engrafted poorly in wild-type (WT) recipients and KO bone marrow cells failed to radioprotect lethally irradiated WT recipients. CD166−/− hosts supported short-term, but not long-term WT HSC engraftment, confirming that loss of CD166 is detrimental to the competence of the hematopoietic niche. CD166−/− mice were significantly more sensitive to hematopoietic stress. Marrow-homed transplanted WT hematopoietic cells lodged closer to the recipient endosteum than CD166−/− cells, suggesting that HSC-OB homophilic CD166 interactions are critical for HSC engraftment. STAT3 has 3 binding sites on the CD166 promoter and STAT3 inhibition reduced CD166 expression, suggesting that both CD166 and STAT3 may be functionally coupled and involved in HSC competence. These studies illustrate the significance of CD166 in the identification and engraftment of HSC and in HSC-niche interactions, and suggest that CD166 expression can be modulated to enhance HSC function.
Introduction
How hematopoietic stem cell (HSC)–hematopoietic niche (HN) interactions maintain HSC function remains unknown. Several markers on HSC have a ligand on cells of the HN. However, these markers are neither obligatory for HSC function nor are they universally expressed on HSC across species, or on cells of the HN. A role for osteoblasts (OB) in maintaining HSC is well-documented.1-3 We previously showed that more immature OB with high Runx2 expression maintain hematopoietic function.4 Recently, we found that anti-activated leukocyte cell adhesion molecule (or CD166) expression on OB directly correlates with Runx2 expression and high hematopoiesis–enhancing activity.5 CD166 expression decreased with OB maturation concomitant with a decline in Runx2 expression and OB-mediated ex vivo maintenance of HSC.5
Expression of CD166 on niche cells has been reported.6 CD166, which can mediate CD166-CD166 homophilic interactions, is a member of the immunoglobulin superfamily and can also bind the only other known ligand, CD6.7 CD166 was originally used to identify a subset of human adult bone marrow (BM) and mobilized peripheral blood (PB) CD34+ cells enriched for progenitor activity.8 However, functional studies with CD166 were not pursued. CD166 expression on Stro-1+ stromal cells9 and binding of hematopoietic cells via CD166 to a yolk sac-derived stromal cell line were also demonstrated.10 These, and our data2,4,5,11 confirmed that CD166 is expressed on hematopoietic progenitors and on OB, suggesting the unique possibility that these cells may interact with one another through CD166-CD166 interactions.
Recently, Jeannet et al12 reported that CD166 is differentially regulated in adult hematopoiesis and that CD166−/− HSC have an engraftment defect, although young CD166−/− mice displayed normal hematopoietic counts and numbers of phenotypically defined HSC. However, these studies12 did not examine the potential of CD166 to identify bona fide normal murine and human HSC, nor did they investigate the functional capacity of CD166 in the niche.
In this report, we demonstrate that CD166 is a universal functional marker of murine and human HSC and OB within the HN. We also demonstrate that it is involved in modulating HSC-niche interactions and HSC fate. The conserved homology between murine and human CD166 provides an excellent translational bridge between these systems to advance future interventions for enhancing HSC engraftment and clinical benefit.
Methods
Mice, human cord blood, and transplantation
Breeding pairs of CD166−/− mice (B6.129[FVB]-Alcamtm1Jawe/J mice; stock No. 010635) were obtained from The Jackson Laboratories. CD166−/−, B6.SJL-Pt-cqPep3b/BoyJ (BoyJ), C57BL/6, C57BL/6-BoyJ F1, and NOD scid γ (NSG) mice were bred and housed at Indiana University. Mice 8- to 12-weeks-old were used. Recipients other than NSG mice received 1100 cGy (700 and 400 cGy split-dose); NSG mice received 275 cGy. All procedures were approved by the Institutional Animal Care and Use Committee of the Indiana University School of Medicine and followed National Institutes of Health guidelines. Human umbilical cord blood (CB) was obtained from St. Vincent’s Hospital (Indianapolis, IN) with Institutional Review Board approval. This study was conducted in accordance with the Declaration of Helsinki. Test cells were injected IV in 200 µL phosphate-buffered saline and where needed, 10e5 competitor cells per recipient were used. In murine and human studies, chimerism was assessed monthly in the PB and at 4- or 6-months posttransplantation (PT) in the BM of primary recipients, as indicated. Secondary recipients received the equivalent of half a femur from primary recipients without competitor cells.
Homing and mobilization
Homing of transplanted cells to the BM of irradiated recipients was performed as previously described.13 Briefly, low-density BM cells were stained with CellTrace Violet (CTV) tracking dye (Invitrogen), washed, and injected IV into nonirradiated recipients to avoid radiation-induced injury to the marrow microenvironment. Recipient mice were sacrificed 16 hours PT, and BM cells were recovered and analyzed for CTV+ cells. Mobilization of hematopoietic stem and progenitor cells (HSPC) was performed as previously described.14
Cell staining, flow cytometry, and cell sorting
Cells washed with stain wash (phosphate-buffered saline, 1% bovine calf serum, and 1% penicillin-streptomycin) were stained for 15 minutes on ice. Low-density BM cells were stained with fluorescein isothiocyanate-conjugated CD3, CD4, CD45R, Ter119, and Gr1; allophycocyanin-conjugated c-Kit (CD117); phycoerythrin-Cy7-conjugated Sca-1; phycoerythrin-conjugated CD166 (clone-eBioALC48; eBiosciences, catalog No. 12-1661-82); AF700-conjugated CD48; and PerCpCy5.5-conjugated CD150. In some experiments, fluorescein isothiocyanate-conjugated CD48 was used to simultaneously select Lin−CD48− cells. Cell sorting was performed on a BD FACSAria and flow cytometric analysis was done on a BD LSRII.
ChIP
Chromatin immunoprecipitation (ChIP) was done using IL-7-stimulated TAIL7 cells with immunoprecipitation of protein-bound DNA sequences performed using antibodies for STAT3 (Cell Signaling), RNA polymerase-II (Millipore), or an irrelevant Ab (Jagged2; Santa Cruz Biotechnology). After purification of eluted DNA, ChIP-quantitative polymerase chain reaction was performed using sequence-specific probes for promoter regions of CD166, Survivin, glyceraldehyde-3-phosphate dehydrogenase, or the control open reading frame (ORF)-free intergenic region IGX1A (SABiosciences), in a 7900HT PCR system (Applied Biosystems).
Two-photon microscopy
Mice were injected with carboxyfluorescein succinimidyl ester (CFSE)-labeled cells as described in “Methods.” At 16-hours PT, mice were anesthetized and a jugular vein catheter was inserted to allow fluorescent tracer injections on the microscope stage. The dorsal skull surface was exposed, placed in a custom-designed atraumatic stereotaxic device, and submerged in microscope oil. Images were acquired with an Olympus FV1000 confocal system custom-modified for multiphoton imaging. Images were collected in a nondescanned mode using Olympus XLUMPLFL 20×W, NA 0.95 objective, and 830 nm excitation wavelength. Imaging was performed at a scan rate of 4 µs per pixel. Sections through the depth of tissue (Z-stacks) were collected from 6 regions of the calvarium. Z-stacks were collected at step-size settings of 1 µm and 512 × 512 pixels. Image analysis was performed using Amira 3D Visualization and Image Analysis software, version 4.1 (FEI, Burlington, MA).
Statistical analysis
Data are presented as mean ± standard deviation and where applicable, triplicate samples were measured. Two-tailed Student t tests were performed when only 2 groups were compared. One-way factorial analyses of variances were used for multiple group comparisons. Significance was set at 0.05.
Results
CD166 identifies murine and human long-term BM repopulating cells
We examined the repopulating potential of CD166+ and CD166− subsets of putative HSC rigorously identified by Lin−Sca1+c-Kit+ (LSK) and signaling lymphocyte activation molecule markers.15 As shown in Figure 1A, CD166 and CD150 fractionated LSK48− cells into 4 distinct groups (fluorescence minus one [FMO] controls are shown in supplemental Figure 1A, available on the Blood Web site). A total of 25, 50, and 100 cells from groups 1 to 4 were competitively transplanted. Chimerism at 4 months PT (data from 25 cells/mouse shown in Figure 1B) demonstrated that engraftment of CD150+ cells was predominantly restricted to cells coexpressing CD166, although some low-level engraftment was observed among CD166−CD150+ cells (group 4). The reconstitution kinetics of cells in groups 2, 3, and 4 were strikingly different (data from 100 cells/mouse shown in Figure 1C). CD166+CD150+ cells (group 3) displayed long-term HSC (LT-HSC) kinetics, whereas CD166+CD150− cells (group 2) provided declining levels of repopulating potential, reminiscent of short-term HSC (ST-HSC) activity. CD166−CD150+ cells (group 4) appear to be intermediate-term HSC16 or LT-HSC with a limited, but consistent, repopulating potential. Limiting dilution analysis (LDA) (Figure 1D) revealed that the frequency of LT-HSC was highest among CD166+CD150+ cells (1:17). Serial transplantation in which primary recipients received 10 cells from groups 1, 2, 3, or 4 (Figure 1E-F) demonstrated that CD166+CD150+ cells (group 3) provide approximately 20-fold higher levels of chimerism than other groups examined. The number of repopulating units17 (RU) per 10 cells (Figure 1G) illustrated that LDA and RU calculations generated very similar results, and ascertained that repopulating HSC among LSK48−CD166+CD150+ cells are approximately sixfold higher than among LSK48−CD166−CD150+ cells.
Characterization of the functional properties of murine HSC fractionated with CD166. (A) C57BL/6 low-density BM cells were analyzed for the expression of lineage markers and CD48 (left dot plot). Lineage-CD48− (Lin-48−) cells were analyzed for the expression of Sca-1 and cKit (middle dot plot) to identify Lin-Sca1+cKit+CD48− (LSK48−) cells (group 1 in middle dot plot). LSK48− cells were analyzed with CD150 and CD166 (right dot plot) to define groups 2, 3, and 4. Percentages next to each group represent the relative size of each population within the dot plot. A total of 100, 50, and 25 cells from groups 1 through 4 were transplanted into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). (B) Chimerism 4-months PT for the 25-cells/mouse group. *P < .05 vs group 3. (C) Engraftment kinetics for groups 1, 2, 3, and 4 over 4 months measured in the PB of recipients of 100-cells/mouse group. *P < .05 vs corresponding values from LSK48−CD166+CD150+ cells. These data are from 1 of 3 independent experiments with similar results. (D) LDA analysis from BM engraftment at 4-months PT in mice receiving 25, 50, and 100 cells (5 mice/group). P = .01 for the overall test for differences in HSC frequencies between any of the groups. Table shows the estimated frequencies and range of repopulating cells within each group tested. (E) Chimerism at 4-months PT in primary recipients after the competitive transplantation of 10 cells/mouse from groups 1 to 4 identified in (A), along with 10e5 BoyJ competitor cells. (F) Chimerism at 4-months PT in secondary recipients of the equivalent of one half femur from primary recipients (BM cells were collected 4-months postprimary transplant) shown in (E) without competitor cells [4-5 mice/group in (E) and (F)]. (G) RU per 10 cells calculated from data shown in (E) as described in Harrison and Astle.17 *P < .05 vs group 3. (H) Multilineage differentiation 4-months PT of groups 1 through 4 in mice receiving 100 cells/animal. *P < .05 between LSK48−CD166+CD150+ cells and LSK48−, CD166+CD150−, and CD166−CD150+ cells within each group. (I) Expression of CD166 on LSK48−CD150+CD9+ cells (in the 3 dot plots from L to R). (J) Chimerism at 4-months PT in mice receiving 10 cells per mouse from groups I, II, and III transplanted competitively into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). *P < .05 vs group III. (K) Expression of CD166 on the low (L), medium (M), and high (H) fractions of murine BM side population (SP) cells. Data shown in (K) are from 1 representative experiment from 4 independent analyses with similar results.
Characterization of the functional properties of murine HSC fractionated with CD166. (A) C57BL/6 low-density BM cells were analyzed for the expression of lineage markers and CD48 (left dot plot). Lineage-CD48− (Lin-48−) cells were analyzed for the expression of Sca-1 and cKit (middle dot plot) to identify Lin-Sca1+cKit+CD48− (LSK48−) cells (group 1 in middle dot plot). LSK48− cells were analyzed with CD150 and CD166 (right dot plot) to define groups 2, 3, and 4. Percentages next to each group represent the relative size of each population within the dot plot. A total of 100, 50, and 25 cells from groups 1 through 4 were transplanted into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). (B) Chimerism 4-months PT for the 25-cells/mouse group. *P < .05 vs group 3. (C) Engraftment kinetics for groups 1, 2, 3, and 4 over 4 months measured in the PB of recipients of 100-cells/mouse group. *P < .05 vs corresponding values from LSK48−CD166+CD150+ cells. These data are from 1 of 3 independent experiments with similar results. (D) LDA analysis from BM engraftment at 4-months PT in mice receiving 25, 50, and 100 cells (5 mice/group). P = .01 for the overall test for differences in HSC frequencies between any of the groups. Table shows the estimated frequencies and range of repopulating cells within each group tested. (E) Chimerism at 4-months PT in primary recipients after the competitive transplantation of 10 cells/mouse from groups 1 to 4 identified in (A), along with 10e5 BoyJ competitor cells. (F) Chimerism at 4-months PT in secondary recipients of the equivalent of one half femur from primary recipients (BM cells were collected 4-months postprimary transplant) shown in (E) without competitor cells [4-5 mice/group in (E) and (F)]. (G) RU per 10 cells calculated from data shown in (E) as described in Harrison and Astle.17 *P < .05 vs group 3. (H) Multilineage differentiation 4-months PT of groups 1 through 4 in mice receiving 100 cells/animal. *P < .05 between LSK48−CD166+CD150+ cells and LSK48−, CD166+CD150−, and CD166−CD150+ cells within each group. (I) Expression of CD166 on LSK48−CD150+CD9+ cells (in the 3 dot plots from L to R). (J) Chimerism at 4-months PT in mice receiving 10 cells per mouse from groups I, II, and III transplanted competitively into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). *P < .05 vs group III. (K) Expression of CD166 on the low (L), medium (M), and high (H) fractions of murine BM side population (SP) cells. Data shown in (K) are from 1 representative experiment from 4 independent analyses with similar results.
Analysis of multilineage differentiation at 4-months PT suggested that differentiation of CD150+ cells (regardless of CD166 expression status) is skewed toward the myeloid lineage (Figure 1H),whereas that of CD150− cells, especially the CD166+CD150− cells, is skewed toward the lymphoid lineage, as previously reported for both subgroups.18 Skewed lineage differentiation of CD150+ and CD150− cells was also observed in mice receiving different size grafts (supplemental Figure 2). Recently, Karlsson et al19 reported a new phenotypic characterization of murine HSC using the tetraspanin CD9. We examined whether CD9+ HSC19 express CD166. Approximately one-third of LSKCD48−CD150+ cells expressed CD9 (Figure 1I) (FMO controls shown in supplemental Figure 1B). Only CD166+CD9+ cells sustained significant in vivo engraftment (Figure 1J), suggesting that CD166 expression is obligatory for the complete identification of functional HSC. That CD166 marks HSC was also gleaned from coexpression of CD166 and side population cells20 (Figure 1K).
The majority of Lin−CD34+CD38− cells in human umbilical CB expressed CD166 (Figure 2A). The repopulating potential of Lin−CD34+CD38−CD166+ cells in NSG mice (Figure 2B) was >4-fold higher than that of equal numbers of Lin−CD34+CD38−CD166− cells and far exceeded chimerism sustained by Lin−CD34+CD38− cells. Recently, CD49f (along with lineage markers, CD34, CD38, and CD90) was used to identify robust human HSC capable of engrafting in a xenotransplantation model at the single cell level.21 A modified staining protocol (Figure 2C; FMO controls shown in supplemental Figure 1C) demonstrated that CD166 and CD49f fractionate CB Lin−CD34+CD38− cells into 4 distinct subgroups (Figure 2C). Repopulating cells in NSG mice were predominantly CD166+ (Figure 2D; groups 2 and 3), regardless of the status of CD49f, suggesting that CD166+, but not CD166− cells within the CD34+Lin−CD38− fraction contains the majority of long-term engrafting cells. Long-term engraftment of CD49f+ cells was principally restricted to cells coexpressing CD166, demonstrating the importance of CD166 in identifying human LT-HSC. Serial transplantation studies (cells isolated as described in Figure 2C) confirmed that CD166+, but not CD166− cells sustained durable long-term engraftment (groups 2 and 3) (Figure 2E-F). CD166 identified similar subgroups in adult human BM (supplemental Figure 3) illustrating that CD166 identifies putative HSC from different hematopoietic tissues at different stages of ontogeny.
Characterization of the functional properties of human CB-derived HSC fractionated with CD166. (A) Human CB cells were analyzed with a combination of lineage markers, CD34 and CD38 and Lin−CD34+CD38− cells (group 1) were analyzed for the expression of CD166 to identify CD34+CD38−CD166− (group 2) and CD34+CD38−CD166+ (group 3) cells. Percentages next to each group represent the relative size of each population within the dot plot. (B) A total of 5000 cells from groups 1, 2, and 3 were transplanted into NSG mice and chimerism was assessed 4-months later (n = 3-4 per group). *P < .05, CD34+CD38−CD166+ vs other groups. (C) Fractionation of human CB cells with CD34 and a combination of lineage markers and CD38 to select Lin−CD34+CD38− cells (group 1 in left dot plot). Lin−CD34+CD38− cells were analyzed with CD166 and CD49f (right dot plot) to define groups 2, 3, 4, and 5. Percentages next to each group represent the relative size of each population within the dot plot. A total of 1000 cells from groups 1 through 5 were transplanted into NSG mice. (D) BM chimerism 4-months PT |(n = 3-4 per group). *P < .05, groups 2 and 3 vs other groups. Similar results were obtained in another independent experiment (4000 cells/group; n = 4/group). The 2 data sets were not pooled. (E-F) Chimerism at 4-months PT (E) sustained by 1000 cells per mouse from groups 1 to 4, as shown in panel (C) and in secondary recipients (F) transplanted with BM cells collected from primary recipients (one-half a femur equivalent from pooled BM cells per recipient) shown in (E) (n = 4-5 per group in [E] and [F]). *P < .05, groups 2 and 3 vs other groups. (F) Flow cytometric analysis of representative mice from (F) are shown in supplemental Figure 10.
Characterization of the functional properties of human CB-derived HSC fractionated with CD166. (A) Human CB cells were analyzed with a combination of lineage markers, CD34 and CD38 and Lin−CD34+CD38− cells (group 1) were analyzed for the expression of CD166 to identify CD34+CD38−CD166− (group 2) and CD34+CD38−CD166+ (group 3) cells. Percentages next to each group represent the relative size of each population within the dot plot. (B) A total of 5000 cells from groups 1, 2, and 3 were transplanted into NSG mice and chimerism was assessed 4-months later (n = 3-4 per group). *P < .05, CD34+CD38−CD166+ vs other groups. (C) Fractionation of human CB cells with CD34 and a combination of lineage markers and CD38 to select Lin−CD34+CD38− cells (group 1 in left dot plot). Lin−CD34+CD38− cells were analyzed with CD166 and CD49f (right dot plot) to define groups 2, 3, 4, and 5. Percentages next to each group represent the relative size of each population within the dot plot. A total of 1000 cells from groups 1 through 5 were transplanted into NSG mice. (D) BM chimerism 4-months PT |(n = 3-4 per group). *P < .05, groups 2 and 3 vs other groups. Similar results were obtained in another independent experiment (4000 cells/group; n = 4/group). The 2 data sets were not pooled. (E-F) Chimerism at 4-months PT (E) sustained by 1000 cells per mouse from groups 1 to 4, as shown in panel (C) and in secondary recipients (F) transplanted with BM cells collected from primary recipients (one-half a femur equivalent from pooled BM cells per recipient) shown in (E) (n = 4-5 per group in [E] and [F]). *P < .05, groups 2 and 3 vs other groups. (F) Flow cytometric analysis of representative mice from (F) are shown in supplemental Figure 10.
CD166−/− HSC do not engraft in WT recipients
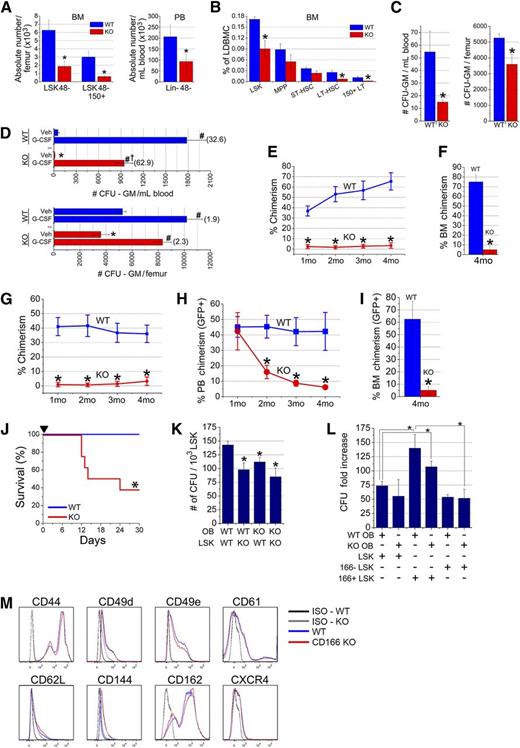
Because CD166 is expressed by HSC (Figure 1) and OB is critical for their function,2,11 we reasoned it may play an important role in the niche via CD166-mediated HSC–OB interactions. To investigate the role of CD166 in hematopoiesis, we examined the functional properties of HSC from CD166 knockout (KO) mice. As expected, BM cells from the central region, considered here the vascular niche, and from flushed and then digested bones, considered here the endosteal region of KO mice did not express CD166 (supplemental Figure 4A) nor did OB from these mice (supplemental Figure 4B). Absolute numbers of LSK48− and LSK48−CD150+ cells in the BM and Lin−CD48− cells in the PB of CD166−/− mice were significantly reduced (Figure 3A). Lineage composition in the central region revealed that KO mouse BM contained a slightly, but significantly, higher percentage of lymphoid cells and a slightly, but significantly, lower percentage of myeloid cells (supplemental Figure 5A). However, cell composition in the endosteal fraction did not show significant differences between WT and KO mice (supplemental Figure 5B). The frequency of HSPC including LSK, LT-HSC (LSKCD34−CD135−), and CD150+LT-HSC was significantly lower in the BM of KO mice (Figure 3B), although the frequencies of multipotent progenitors (MPP) (LSKCD34+CD135+) and ST-HSC (LSKCD34+CD135−) were not significantly reduced in KO mice (Figure 3B). The clonogenic potential of steady state BM- and PB-derived progenitors was significantly reduced in the BM and PB of KO mice (Figure 3C). However, CD166−/− mice mobilized normally in response to granulocyte colony-stimulating factor (G-CSF), although the overall number of circulating clonogenic progenitors was significantly lower than in WT mice (Figure 3D), suggesting that CD166 may play a role in G-CSF–induced mobilization of HSPC. The size and cellularity of the spleens of CD166−/− mice were not different than those of WT mice.
Hematopoietic parameters of CD166−/−mice. (A) Absolute numbers of LSK48− and LSK48−150+ cells in the BM and Lin-48− cells in the PB of WT and KO mice (6 mice per group). *P < .05 compared with WT. (B) Percentages of HSPC classes in the marrow of WT and KO mice (6 mice/group). Classes of HSPC were identified as such: LSK (Lin−Sca1+Kit+); MPP (LSKCD34+CD135+); ST-HSC (LSKCD34+CD135−); LT-HSC (LSKCD34−CD135−); 150+LT (CD150+LSKCD34−CD135−). *P < .05 compared with WT. (C) Numbers of colony forming unit-granulocyte/macrophage (CFU-GM) in 1 mL of PB and in BM contained in 1 femur from WT and KO mice (n = 5-6 mice per group). *P < .05 compared with WT. (D) Numbers of CFU-GM in 1 mL of PB and in BM contained in 1 femur from WT and KO mice mobilized with G-CSF (2 daily injections of 1 µg/mouse for 4 days). Cells were collected on day 5 (n = 4-5 mice per group). *P < .05 between KO and WT vehicle (Veh) control. #P < .05 compared with Veh in each set. †P < .05 between G-CSF KO and WT. Values in parentheses are fold-increase in CFU-GM numbers between Veh and G-CSF in each tissue. (E) Lethally irradiated F1 mice (n = 13 per group from 3 independent experiments) received 1000 LSK cells from the central region of CD166−/− mice plus 10e5 CD45.1 competitor cells. *P < .05 compared with WT at corresponding time point. (F) BM chimerism at 4-months PT of LSK cells from 1 of the 3 experiments in (E). *P < .05 compared with WT. (G) Lethally irradiated F1 mice (n = 5 per group) received 1000 LSK cells from the endosteal region of CD166−/− mice plus 10e5 CD45.1 competitor cells. *P < .05 compared with WT at corresponding time point. (H) Engraftment of 1000 GFP+ LSK cells from GFP+ WT mice transplanted with 10e5 CD45.1 competitor cells into lethally irradiated WT or KO mice (n = 5 per group). *P < .05 compared with WT at corresponding data points. Chimerism is reported as a percentage of GFP+ cells in PB. (I) BM chimerism at 4-months PT of GFP+ LSK cells in mice in (H). *P < .05 compared with WT. (J) Survival of lethally irradiated recipients (700 and 400 cGy split dose) over a 30-day period after transplantation with 10e5 low-density BM cells from WT or KO donors. *P < .05 compared with WT group. (K) Numbers of colony-forming units per 10e3 sorted LSK cells from the BM of WT or KO donors retained on WT or KO OB after 16 hours of coculture of both cell types. *P < .05 compared with WT OB-LSK coculture. (L) Fold increase in the number of colony-forming unit produced after 7 days of coculture combinations as shown in the table. Each culture received 1000 LSK cells on day 0. Data are reported as fold-increase relative to the colony-forming unit number obtained from freshly isolated 1000 LSK cells on day 0. *P < .05 compared with WT OB and CD166+ LSK coculture (third bar from the left). (M) Expression of different adhesion markers on BM cells from WT and CD166−/− donors. All antibodies were obtained from Becton-Dickinson or Pharmingen.
Hematopoietic parameters of CD166−/−mice. (A) Absolute numbers of LSK48− and LSK48−150+ cells in the BM and Lin-48− cells in the PB of WT and KO mice (6 mice per group). *P < .05 compared with WT. (B) Percentages of HSPC classes in the marrow of WT and KO mice (6 mice/group). Classes of HSPC were identified as such: LSK (Lin−Sca1+Kit+); MPP (LSKCD34+CD135+); ST-HSC (LSKCD34+CD135−); LT-HSC (LSKCD34−CD135−); 150+LT (CD150+LSKCD34−CD135−). *P < .05 compared with WT. (C) Numbers of colony forming unit-granulocyte/macrophage (CFU-GM) in 1 mL of PB and in BM contained in 1 femur from WT and KO mice (n = 5-6 mice per group). *P < .05 compared with WT. (D) Numbers of CFU-GM in 1 mL of PB and in BM contained in 1 femur from WT and KO mice mobilized with G-CSF (2 daily injections of 1 µg/mouse for 4 days). Cells were collected on day 5 (n = 4-5 mice per group). *P < .05 between KO and WT vehicle (Veh) control. #P < .05 compared with Veh in each set. †P < .05 between G-CSF KO and WT. Values in parentheses are fold-increase in CFU-GM numbers between Veh and G-CSF in each tissue. (E) Lethally irradiated F1 mice (n = 13 per group from 3 independent experiments) received 1000 LSK cells from the central region of CD166−/− mice plus 10e5 CD45.1 competitor cells. *P < .05 compared with WT at corresponding time point. (F) BM chimerism at 4-months PT of LSK cells from 1 of the 3 experiments in (E). *P < .05 compared with WT. (G) Lethally irradiated F1 mice (n = 5 per group) received 1000 LSK cells from the endosteal region of CD166−/− mice plus 10e5 CD45.1 competitor cells. *P < .05 compared with WT at corresponding time point. (H) Engraftment of 1000 GFP+ LSK cells from GFP+ WT mice transplanted with 10e5 CD45.1 competitor cells into lethally irradiated WT or KO mice (n = 5 per group). *P < .05 compared with WT at corresponding data points. Chimerism is reported as a percentage of GFP+ cells in PB. (I) BM chimerism at 4-months PT of GFP+ LSK cells in mice in (H). *P < .05 compared with WT. (J) Survival of lethally irradiated recipients (700 and 400 cGy split dose) over a 30-day period after transplantation with 10e5 low-density BM cells from WT or KO donors. *P < .05 compared with WT group. (K) Numbers of colony-forming units per 10e3 sorted LSK cells from the BM of WT or KO donors retained on WT or KO OB after 16 hours of coculture of both cell types. *P < .05 compared with WT OB-LSK coculture. (L) Fold increase in the number of colony-forming unit produced after 7 days of coculture combinations as shown in the table. Each culture received 1000 LSK cells on day 0. Data are reported as fold-increase relative to the colony-forming unit number obtained from freshly isolated 1000 LSK cells on day 0. *P < .05 compared with WT OB and CD166+ LSK coculture (third bar from the left). (M) Expression of different adhesion markers on BM cells from WT and CD166−/− donors. All antibodies were obtained from Becton-Dickinson or Pharmingen.
LSK cells from KO mice collected from the central (Figure 3E-F) or endosteal regions (Figure 3G) almost failed to engraft in lethally irradiated recipients. Given the low chimerism levels in primary recipients, secondary transplants were not performed. Multilineage differentiation analysis demonstrated a significant increase in lymphoid and a significant decrease in myeloid reconstitution in recipients of KO cells (supplemental Figure 6) reflecting the same skewness in KO mice (supplemental Figure 5). When used as recipients of LSK cells from GFP C57BL/6 mice, only short-term repopulating cells engrafted efficiently in KO mice (Figure 3H). Chimerism in WT recipients at 4-months PT was more than 12-fold higher than in KO hosts (Figure 3I), demonstrating that the CD166−/− HN cannot support LT-HSC engraftment. In addition, BM cells from KO mice also had a compromised radioprotective function (Figure 3J).
Homophilic interactions between CD166 expressed on OB and LSK cells was evident from adherence studies in which significantly fewer KO clonogenic cells within the LSK fraction adhered to OB when either, or both LSK and OB, were derived from KO mice (Figure 3K). The negative impact of the loss of homophilic CD166 interaction between OB and LSK was evident in the number of clonogenic cells produced in mixed cocultures (Figure 3L). The highest level of loss of hematopoietic function was evident between cocultures of WT OB plus CD166+ LSK cells and KO OB plus CD166− LSK cells (bars 3 and 6 in Figure 3L). These data demonstrate that selection of CD166+ LSK cells enriches hematopoietic progenitor cell activity and suggests that other HSC-OB compensatory adhesion interactions may be operable. However, we did not detect any significant difference in the expression patterns of several adhesion molecules on WT or KO marrow cells (Figure 3M).
Stress response and homing of hematopoietic cells from CD166 KO mice
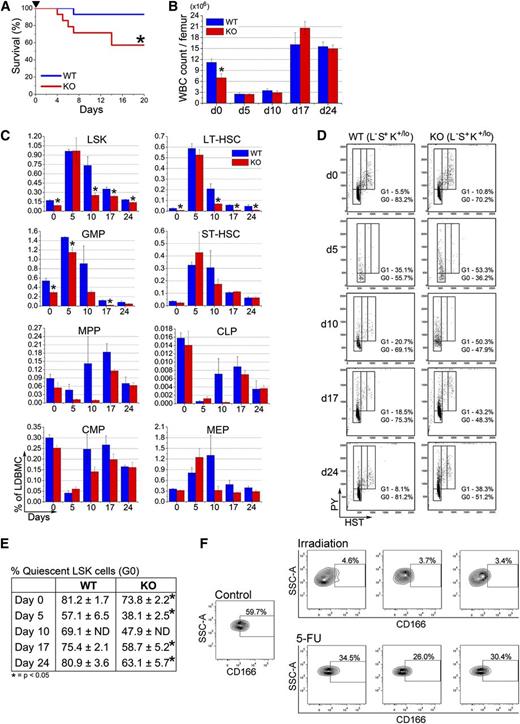
The behavior of CD166−/− LSK cells and the compromised competence of the HN of KO mice prompted us to examine the response of KO HSPC to stress. Following a single 150 mg/kg BW injection of 5-fluorouracil (5-FU), survival of CD166−/− mice was significantly reduced compared with WT mice (Figure 4A). The BM cellularity of surviving KO mice 5, 10, 17, and 24 days after 5-FU injection was unchanged from that of WT mice (Figure 4B). At these time points, some classes of HSPC,22,23 specifically LSK, LT-HSC (LSKCD34−CD135−), and granulocyte-macrophage progenitors (Lin−Sca1−CD117+CD34+CD16/32w+), were significantly reduced in KO relative to WT mice (Figure 4C), whereas recovery of ST-HSC, MPP, common lymphoid progenitor, common myeloid progenitor, and megakaryocyte-erythroid progenitor was not different between the 2 genotypes (Figure 4C). A significantly larger fraction of day 0, day 5, day 10, day 17, and day 24 KO LSK cells were in G1 and S/G2+M phases of the cell cycle (Figure 4D-E), demonstrating the cell cycle-dependent sensitivity of KO HSPC to 5-FU.
Behavior of CD166−/− HSPC under hematopoietic stress. (A) Survival of WT and KO mice after a single intraperitoneal injection of 5-FU (150 mg/kg, arrowhead) (n = 14 per genotype). Survival was monitored daily. *P < .05; Kaplan-Meyer curves were analyzed with a log-rank (Mantel-Cox) nonparametric test. (B) Total white blood cell count per femur before (day 0) and on days 5, 10, 17, and 24 after treatment with 5-FU (n = 4 per genotype per time point). *P < .05 compared with WT. (C) Phenotypic analysis of HSPC from BM of WT and KO mice 5, 10, 17, and 24 days post 5-FU (n = 4 per genotype per time point). *P < .05 compared with WT. (D) Representative analysis of LSK cells from 1 mouse per genotype per time point with Hoechst 33342 and Pyronin Y (n = 4 per genotype per time point). (E) Percentage of LSK cells in the G0 phase of cell cycle on days 5, 10, 17, and 24 post-5-FU treatment analyzed as shown in (D) (n = 4 per genotype per time point). *P < .05 compared with WT. (F) Expression of CD166 on BM-derived LSK cells from 1 representative control mouse or from each of 3 separate mice irradiated with 400 cGy or treated with 150 mg/kg 5-FU 10 days earlier.
Behavior of CD166−/− HSPC under hematopoietic stress. (A) Survival of WT and KO mice after a single intraperitoneal injection of 5-FU (150 mg/kg, arrowhead) (n = 14 per genotype). Survival was monitored daily. *P < .05; Kaplan-Meyer curves were analyzed with a log-rank (Mantel-Cox) nonparametric test. (B) Total white blood cell count per femur before (day 0) and on days 5, 10, 17, and 24 after treatment with 5-FU (n = 4 per genotype per time point). *P < .05 compared with WT. (C) Phenotypic analysis of HSPC from BM of WT and KO mice 5, 10, 17, and 24 days post 5-FU (n = 4 per genotype per time point). *P < .05 compared with WT. (D) Representative analysis of LSK cells from 1 mouse per genotype per time point with Hoechst 33342 and Pyronin Y (n = 4 per genotype per time point). (E) Percentage of LSK cells in the G0 phase of cell cycle on days 5, 10, 17, and 24 post-5-FU treatment analyzed as shown in (D) (n = 4 per genotype per time point). *P < .05 compared with WT. (F) Expression of CD166 on BM-derived LSK cells from 1 representative control mouse or from each of 3 separate mice irradiated with 400 cGy or treated with 150 mg/kg 5-FU 10 days earlier.
We also assessed CD166 expression on LSK cells in response to radiation. BM cells from irradiated mice exhibit a persistent engraftment defect postirradiation,24,25 whereas those from mice treated with 5-FU engraft successfully.26 Expression of CD166 on LSK from sublethally irradiated mice was significantly reduced (Figure 4F). However, 5-FU treatment did not significantly reduce CD166 expression, suggesting that the engraftment defect observed in cells exposed to radiation may be associated with decreased CD166 expression.
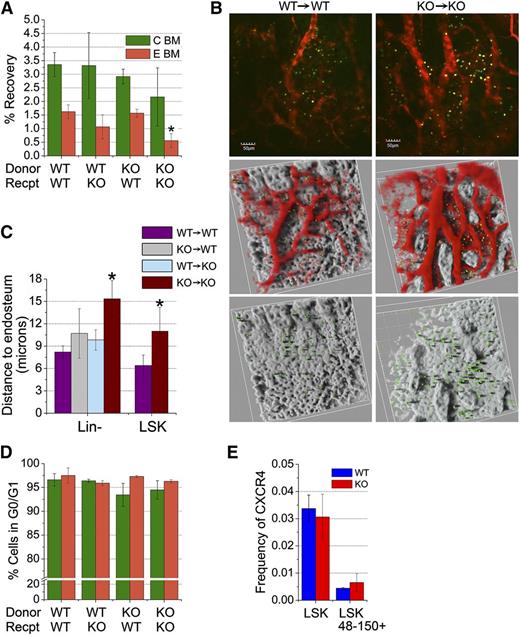
Homing of KO cells to the endosteal or central regions of the marrow of KO recipients was inefficient (Figure 5A). Using intravital imaging, we examined the behavior of BM-homed transplanted cells to assess the impact of CD166 on interactions between transplanted cells and elements of the HN. Figure 5B illustrates that both WT and KO cells home to the marrow of both genotypes. However, distances between WT cells and the endosteum of WT recipients (supplemental Figure 7) were significantly shorter than those measured between KO cells homed to the marrow of KO mice and the endosteum of these recipients (Figure 5C). Interestingly, this defect was not as pronounced (Figure 5C) when CD166 was absent on either transplanted cells or the endosteum of recipient mice only (KO→WT or WT→KO), suggesting that other compensatory adhesion interactions between transplanted HSC and the endosteum are also critical for HSC function. Most importantly, differences in the settings WT→WT vs KO→KO were consistent whether grafts contained low-density BM cells (data not shown), lineage depleted BM cells, or LSK cells (Figure 5C). A smaller percentage of BM-homed KO cells from the central region were in G0/G1 compared with WT cells (Figure 5D), but these differences were not significant, which suggests that increased cell cycle kinetics27,28 are most likely not responsible for the gradual decline in the engraftment of these cells. Interestingly, expression of CXCR4 on KO HSPC cells was not altered (Figure 5E), suggesting that CD166 is not involved in directed homing of HSPC to the BM.
Homing of CD166−/− cells to the hematopoietic niche. (A) Homing of CD166−/− hematopoietic cells to the BM of lethally irradiated recipients. Recovery of BM-homed cells 16-hours PT (corrected for total body BM). Cells were stained with CTV, injected into lethally irradiated recipients and then recovered from flushed central (C BM) or from digested endosteal (E BM) regions (n = 3-4 mice per group). *P < .05 compared with WT. (B) Intravital images of CFSE-stained Lin− cells in the calvariae of nonirradiated recipients 16-hours PT. Panels on the left depict WT cells in WT recipients and those on the right show KO cells in KO recipients. Top 2 panels show CFSE-stained cells (green) in the BM microenvironment. Images of vessels (collected at 605 nm) that were stained with tetramethyl rhodamine isothiocyanate (TRITC)–conjugated dextran appear in red. Images in the middle row depict 2 different areas of the calvarium with the bone rendered silver to facilitate visualization of marrow-homed cells. The bottom row shows the same areas depicted in the middle row after removal of the images of the vasculature. Distances from each cell to the surface of the endosteum was measured using Amira 3D Visualization and Image Analysis software, version 4.1, using a 3-dimensional caliper tool. Additional information can be found in supplemental Figure 7. (C) Average distances between WT or KO Lin− and LSK cells and the endosteum. Distances between cells and the bone surface (reconstructed with a constant threshold value, visible in gold) were measured using a 3-dimensional caliper tool. *P < .05 compared with WT. Data shown are from 1 representative experiment from a total of 3. At least 1 recipient mouse per condition was used in each experiment. In the analysis shown between 8 and 25 cells per condition were measured for the Lin− group and 5 to 7 for the LSK group. Similar data were obtained in the other experiments. (D) Cell cycle analysis of BM-homed cells for 16-hours PT. BM homed CTV+ cells were isolated by cell sorting, stained with propidium iodide, and flow cytometrically analyzed for cell cycle status to determine cells in G0, G1, and G2+M (n = 3-4 per group). (E) Frequencies of WT and KO marrow-derived LSK and LSK48-150+ cells expressing CXCR4 (BD, clone 2B11, cat# 561734).
Homing of CD166−/− cells to the hematopoietic niche. (A) Homing of CD166−/− hematopoietic cells to the BM of lethally irradiated recipients. Recovery of BM-homed cells 16-hours PT (corrected for total body BM). Cells were stained with CTV, injected into lethally irradiated recipients and then recovered from flushed central (C BM) or from digested endosteal (E BM) regions (n = 3-4 mice per group). *P < .05 compared with WT. (B) Intravital images of CFSE-stained Lin− cells in the calvariae of nonirradiated recipients 16-hours PT. Panels on the left depict WT cells in WT recipients and those on the right show KO cells in KO recipients. Top 2 panels show CFSE-stained cells (green) in the BM microenvironment. Images of vessels (collected at 605 nm) that were stained with tetramethyl rhodamine isothiocyanate (TRITC)–conjugated dextran appear in red. Images in the middle row depict 2 different areas of the calvarium with the bone rendered silver to facilitate visualization of marrow-homed cells. The bottom row shows the same areas depicted in the middle row after removal of the images of the vasculature. Distances from each cell to the surface of the endosteum was measured using Amira 3D Visualization and Image Analysis software, version 4.1, using a 3-dimensional caliper tool. Additional information can be found in supplemental Figure 7. (C) Average distances between WT or KO Lin− and LSK cells and the endosteum. Distances between cells and the bone surface (reconstructed with a constant threshold value, visible in gold) were measured using a 3-dimensional caliper tool. *P < .05 compared with WT. Data shown are from 1 representative experiment from a total of 3. At least 1 recipient mouse per condition was used in each experiment. In the analysis shown between 8 and 25 cells per condition were measured for the Lin− group and 5 to 7 for the LSK group. Similar data were obtained in the other experiments. (D) Cell cycle analysis of BM-homed cells for 16-hours PT. BM homed CTV+ cells were isolated by cell sorting, stained with propidium iodide, and flow cytometrically analyzed for cell cycle status to determine cells in G0, G1, and G2+M (n = 3-4 per group). (E) Frequencies of WT and KO marrow-derived LSK and LSK48-150+ cells expressing CXCR4 (BD, clone 2B11, cat# 561734).
Bone phenotype and function in CD166 KO mice
To assess the impact of loss of CD166 on OB phenotype and function, we analyzed multiple properties of OB from KO mice. A significantly smaller fraction of 2-day calvarial OB from KO mice were in G0/G1 during days 1 and 2 of culture (supplemental Figure 8A). Except for alkaline phosphatase, expression of messenger RNA levels of collagen type 1, osteocalcin, osteopontin, and Runx2, were similar between WT and KO OB (supplemental Figure 8B). Expression of HoxB4 and N-cadherin, which have been implicated in maintaining HSC function,29-31 but not stromal cell-derived factor 1 were significantly lower among KO OB (supplemental Figure 8C). Although these changes were significant, they were not substantial suggesting that reduced signaling through HoxB4 and N-cadherin may play a role in the altered function of HSPC from KO mice and the diminished competence of the CD166−/− HN. Expression levels of fibulins, which negatively influence the adhesive properties of the endosteal niche,32 were significantly increased in KO OB (supplemental Figure 8D), suggesting that loss of CD166 upregulates the expression of fibulins and indirectly reduces the adhesive properties of OB, possibly through increased expression of MMP2, a pathway known to be triggered by CD166.33
CD166 regulation
We analyzed the promoter region of CD166 and recognized the presence of 3 consensus motifs for STAT3 binding. Interestingly, using a hematopoietic cell-targeted deletion of STAT3,34 we recently established that the engraftment potential of STAT3−/− BM cells was significantly reduced.35 Chromatin immunoprecipitation with polymerase-II using the leukemia cell line TAIL7,36 which exhibits STAT3 activation and strong expression of CD166 (supplemental Figure 9), showed significant enrichment in GAPDH, but not of the control ORF-free intergenic region IGX1A (Figure 6A). However, immunoprecipitation with STAT3 antibody showed marked enrichment for the CD166 promoter sequence, as well as for Survivin, a well-known transcriptional target of STAT3. To determine whether STAT3 transcriptional activity regulates CD166 expression on HSC, we examined BM cells from WT and STAT3−/− mice. Figure 6B illustrates a substantial decrease in the percentage of CD166+ cells within the LSK and LSK48−CD150+ fractions in STAT3−/− mice relative to controls. Expression of CD166 was significantly reduced on murine BM-derived Lin− cells treated for 24 hours with the STAT3-selective inhibitor STATTIC (Figure 6C). Finally, suppression of STAT3 expression with short hairpin RNA concomitantly reduced CD166 expression on MOLT4 cells (Figure 6D). Collectively, these data suggest that expression of CD166 can be modulated by STAT3 and present the possibility that loss of engraftment potential of STAT3−/− cells35 is most likely linked to decreased CD166 expression on these cells.
STAT3 regulation of CD166 expression. (A) ChiP analysis of the binding of STAT3 to the CD166 promoter. Data are from 1 representative experiment from a total of 3 that showed very similar results in each. (B) CD166 expression on LSK and LSK48−150+ cells from WT and STAT3−/− mice (n = 3 mice per group). Data shown are from 1 mouse/genotype. Identical results were obtained from the other 2 mice. LSK cells were gated from column “A” and LSK48−150+ were gated from column “B.” (C) CD166 expression on BM-derived Lin− cells treated with STATTIC for 24 hours. Percentage of CD166+ cells declined from 53% (solid line plot) to 17% (dotted line plot) after treatment. (D) Expression of CD166 in human MOLT4 cells transduced with scrambled (Src), CD166, or STAT3 short hairpin RNA. Short hairpin RNA was cloned into Age1 and EcoR1 sites of the MISSION vector (Sigma), which was then selected by puromycin.
STAT3 regulation of CD166 expression. (A) ChiP analysis of the binding of STAT3 to the CD166 promoter. Data are from 1 representative experiment from a total of 3 that showed very similar results in each. (B) CD166 expression on LSK and LSK48−150+ cells from WT and STAT3−/− mice (n = 3 mice per group). Data shown are from 1 mouse/genotype. Identical results were obtained from the other 2 mice. LSK cells were gated from column “A” and LSK48−150+ were gated from column “B.” (C) CD166 expression on BM-derived Lin− cells treated with STATTIC for 24 hours. Percentage of CD166+ cells declined from 53% (solid line plot) to 17% (dotted line plot) after treatment. (D) Expression of CD166 in human MOLT4 cells transduced with scrambled (Src), CD166, or STAT3 short hairpin RNA. Short hairpin RNA was cloned into Age1 and EcoR1 sites of the MISSION vector (Sigma), which was then selected by puromycin.
Discussion
Interactions between HSC and cells of the HN, which are vital for stem cell pool maintenance, depend on ligand-receptor molecules coexpressed independently on interacting cells. Among the most characterized interactions in the HN are those between HSC and OB, which sustain HSC functions, especially self-renewal.1,3,6 To date, not a single, functional surface marker has been identified as a common marker on murine and human HSC and on cells of the HN, nor has there been any description of a marker capable of homophilic interactions that is coexpressed on all these cell types.
Using strict phenotypic definitions of murine15 and human21 engrafting cells, we demonstrated that CD166 is a critical and functional common marker that positively identifies repopulating HSC beyond what is currently possible with previous approaches. To our knowledge, this is the first report to describe a common functional marker on both human and murine HSC, and to demonstrate that loss of this marker not only impacts stem cell function, but also compromises the competence of the HN. In addition, we also demonstrate a possible novel regulatory pathway impacting HSC function that involves STAT3-mediated regulation of CD166 expression. STAT3, which plays a significant role in the hematopoietic system, is activated by several factors involved in homeostatic and stress hematopoiesis, including IL-6, G-CSF, erythropoietin, thrombopoietin, and leukemia inhibitory factor. Although STAT3 deletion markedly decreases the hematopoietic repopulating ability,35 little is known about the downstream effectors of this defect. Our studies demonstrate a direct link between loss of STAT3 in HSC and decreased CD166 expression (Figure 6B) and we suggest this pathway as a possible explanation for the decreased repopulating potential of STAT3−/− BM cells and a likely tentative mechanism for the role of STAT3 in hematopoiesis.
In our transplantation studies, the significant majority of both murine and human long-term repopulating cells were CD166+, demonstrating the importance of CD166 in identifying LT-HSC in both species. The degree to which CD166 facilitated the enrichment of long-term repopulating cells was evident from the frequency of these cells among LSK48−CD166+CD150+ cells (1:17). Although the frequency of 1:17 LT- HSC among this phenotype is lower than the 1:3 frequency reported by Kiel et al15 for CD150+ cells, calculation of the frequency of RUs using a different approach17 yielded results (<2 RU per 10 cells, Figure 1G) almost identical to those from Kiel et al.15 These analyses suggest that exact HSC enumeration or quantification may be limited by the sensitivity of the currently available experimental protocols and subtle differences between practices in different laboratories. Similar observations can be made regarding CD49f in the human system. Although our studies do not contradict the findings of Notta et al,21 they suggest that only a subfraction of CD49f+ cells that also expresses CD166 contains bona fide marrow reconstituting cells.
Our current and previous investigations2,4,5,11 illustrate that CD166 is a functional marker on both HSC and OB. Functionality of CD166 was demonstrated by the almost complete inability of CD166−/− LSK cells to engraft in WT recipients and by the failure of the CD166−/− microenvironment to support the engraftment of WT LT-HSC. Furthermore, recent data (B.R.C. and E.F.S., unpublished observations) suggest that human CB cells treated with CD166 short hairpin RNA have a reduced engraftment potential in NSG mice, suggesting again that CD166 is a functional marker of engrafting stem cells. Imaging data suggest that simultaneous expression of CD166 on HSC, as well as cells of the niche, is critical for engraftment and repopulation. Our results are consistent with those reported by Jeannet et al,12 in which the engraftment potential of purified CD166−/− LSK cells in secondary transplantation recipients was significantly reduced.
Although loss of CD166 did not repress the expression of other stem cell markers on putative HSC, homing of CD166−/− cells to the marrow of CD166−/− recipients was significantly compromised (Figure 5A). Jeannet et al12 only examined the homing of KO cells to the BM of WT recipients, and hence no defect in homing was observed. Our data (Figure 5A) demonstrate that overall, the homing of KO cells to the BM is inefficient, and that homing of KO cells to the microenvironment of KO recipients is significantly reduced. In the studies of Jeannet et al,12 the HN of KO mice was not analyzed. Because CXCR4 expression on CD166−/− HSPC is normal (Figure 5E), it suggests that CD166 may not play a critical role in homing and retention of HSPC in the marrow, but it may be critical for HSC–HN interactions that ensure survival and function of these cells. Imaging studies (Figure 5) confirm this premise and lend support to the suggestion that CD166 may be critical for HSC–OB interactions that sustain stem cell survival and engraftment. More definitive studies are required to ascertain the role of CD166 in HSC-niche interactions. Our studies examined lodgment of marrow-homed CD166−/− cells under permutations not examined by Jeannet et al,12 perhaps leading to different conclusions regarding homing of KO cells to the BM of recipient mice.
The extent to which loss of CD166 impacts HSC function and the fact that KO mice are viable with no apparent major defects is interesting. Although the engraftment defect of CD166−/− HSC is severe and almost total, the defect observed in the HN of CD166−/− mice is partial. Collectively, data (in Figure 3H,J) suggest that ST-HSC survive momentarily in KO mice, but are quickly depleted early after transplantation. These defects predict an acute hematopoietic injury in CD166−/− mice that would impact survival. The susceptibility of CD166−/− mice to metabolic stress underscores the abnormalities of their hematopoietic system that are not detected under steady-state hematopoiesis. Our data suggest that loss of CD166 is more critical for HSC function than for the competence of the HN, and highlight OB differentiation and maturation irregularities in CD166−/− mice that may partially contribute to the overall defect of the HN. A more detailed analysis of the bones of KO mice (presented elsewhere) revealed defects that may also partially contribute to loss of niche competence. Nevertheless, our analysis documents significantly higher levels of alkaline phosphatase messenger RNA in CD166−/− OB (supplemental Figure 8B), suggesting that these OB are more mature than their WT counterparts. Because we have already demonstrated that immature CD166+ OB are responsible for the hematopoiesis enhancing activity,2,4,5,11 these data suggest that maturation of OB may be a contributing factor to the inability of the HN of KO mice to support the engraftment of LT-HSC (Figure 3H). Our studies also illustrated that HoxB4 and N-cadherin expression is moderately, but significantly reduced in OB from KO mice. Although the importance of HoxB4 in sustaining HSC function is well-documented,37 the role of N-cadherin in hematopoiesis is controversial.31,38 Whether the observed differences in the expression levels of HoxB4 and N-cadherin contribute to the inability of the KO microenvironment to support LT-HSC engraftment remains to be determined.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the operators of the Indiana University Melvin and Bren Simon Cancer Center Flow Cytometry Resource Facility for their outstanding technical help and support. This work was supported in part by National Heart, Lung, and Blood Institute grant HL55716 (E.F.S.) and the Indiana Center for Excellence in Molecular Hematology (National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK090948). The Flow Cytometry Research Facility is partially funded by National Cancer Institute grant P30 CA082709.
We acknowledge the In Vivo Therapeutics Core of the Indiana University Simon Cancer Center (partially funded by National Cancer Institute grant P30 CA082709 and National Institute of Diabetes and Digestive and Kidney Diseases grant P01 DK090948) as well as the nursing staff and Dr Arthur Baluyut at the St. Vincent Hospital (Indianapolis, IN) for providing umbilical CB samples for this study.
Authorship
Contribution: B.R.C. performed the majority of experimental work and assisted in writing the paper; M.K. performed many animal studies; Y.-H.C. performed all experimental work involving preparation and culturing of OB; H.Z. performed all chromatin immunoprecipitation studies; B.A.P. contributed to ex vivo culture studies and animal work; H.E.B. and L.M.P. contributed to data analysis and interpretation; H.H. contributed to experimental design and data analysis; A.Z. assisted in animal preparation for the imaging studies and performed imaging; M.M.K. performed a substantial volume of the imaging work and analyzed imaging data; N.C. and A.A.C. assisted in experimental design, data interpretation and animal modeling; M.A.K. designed all the work related to OB preparation and culturing, helped in the design of other experiments, and assisted in writing the manuscript; and E.F.S. designed the research, interpreted data, assisted in some experimental work, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edward F. Srour, Indiana University School of Medicine, 980 W Walnut St, R3-C312, Indianapolis, IN 46202; e-mail: esrour@iupui.edu.

![Figure 1. Characterization of the functional properties of murine HSC fractionated with CD166. (A) C57BL/6 low-density BM cells were analyzed for the expression of lineage markers and CD48 (left dot plot). Lineage-CD48− (Lin-48−) cells were analyzed for the expression of Sca-1 and cKit (middle dot plot) to identify Lin-Sca1+cKit+CD48− (LSK48−) cells (group 1 in middle dot plot). LSK48− cells were analyzed with CD150 and CD166 (right dot plot) to define groups 2, 3, and 4. Percentages next to each group represent the relative size of each population within the dot plot. A total of 100, 50, and 25 cells from groups 1 through 4 were transplanted into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). (B) Chimerism 4-months PT for the 25-cells/mouse group. *P < .05 vs group 3. (C) Engraftment kinetics for groups 1, 2, 3, and 4 over 4 months measured in the PB of recipients of 100-cells/mouse group. *P < .05 vs corresponding values from LSK48−CD166+CD150+ cells. These data are from 1 of 3 independent experiments with similar results. (D) LDA analysis from BM engraftment at 4-months PT in mice receiving 25, 50, and 100 cells (5 mice/group). P = .01 for the overall test for differences in HSC frequencies between any of the groups. Table shows the estimated frequencies and range of repopulating cells within each group tested. (E) Chimerism at 4-months PT in primary recipients after the competitive transplantation of 10 cells/mouse from groups 1 to 4 identified in (A), along with 10e5 BoyJ competitor cells. (F) Chimerism at 4-months PT in secondary recipients of the equivalent of one half femur from primary recipients (BM cells were collected 4-months postprimary transplant) shown in (E) without competitor cells [4-5 mice/group in (E) and (F)]. (G) RU per 10 cells calculated from data shown in (E) as described in Harrison and Astle.17 *P < .05 vs group 3. (H) Multilineage differentiation 4-months PT of groups 1 through 4 in mice receiving 100 cells/animal. *P < .05 between LSK48−CD166+CD150+ cells and LSK48−, CD166+CD150−, and CD166−CD150+ cells within each group. (I) Expression of CD166 on LSK48−CD150+CD9+ cells (in the 3 dot plots from L to R). (J) Chimerism at 4-months PT in mice receiving 10 cells per mouse from groups I, II, and III transplanted competitively into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). *P < .05 vs group III. (K) Expression of CD166 on the low (L), medium (M), and high (H) fractions of murine BM side population (SP) cells. Data shown in (K) are from 1 representative experiment from 4 independent analyses with similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2014-03-565721/4/m_519f1.jpeg?Expires=1765093666&Signature=HuT0-kRr-vKZlnZjcN4H9x5hhRCviybAu0Rs4XviUvfmduipol1l-bikuq9gH7HZhJ4UoLVYI8hpE7xHVMynHNuKqP~XGpQuTvMrPtfBdQr2Sw-qKeDACyDQY3qVmU~A5Mn0gdVWqttaEch2At5vfG45c4NhzKQ~POt~Wmubya6PiOj7kKLK8xHED5hPRhXA6pj8fHs6PCWp-li5I4HoQlWSLUrRFgYHGtWqDXDDb54UXAo1oeiq1fgfg0Hw1CAZLjCs9J0AdYNJOzsRXduRG7bFRJWk9XLqpm-7LLNTqlebVxVzjCf1CnlT1Gir8rhcVz7bYwqVNUq3fuVG3hU5ZA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Characterization of the functional properties of human CB-derived HSC fractionated with CD166. (A) Human CB cells were analyzed with a combination of lineage markers, CD34 and CD38 and Lin−CD34+CD38− cells (group 1) were analyzed for the expression of CD166 to identify CD34+CD38−CD166− (group 2) and CD34+CD38−CD166+ (group 3) cells. Percentages next to each group represent the relative size of each population within the dot plot. (B) A total of 5000 cells from groups 1, 2, and 3 were transplanted into NSG mice and chimerism was assessed 4-months later (n = 3-4 per group). *P < .05, CD34+CD38−CD166+ vs other groups. (C) Fractionation of human CB cells with CD34 and a combination of lineage markers and CD38 to select Lin−CD34+CD38− cells (group 1 in left dot plot). Lin−CD34+CD38− cells were analyzed with CD166 and CD49f (right dot plot) to define groups 2, 3, 4, and 5. Percentages next to each group represent the relative size of each population within the dot plot. A total of 1000 cells from groups 1 through 5 were transplanted into NSG mice. (D) BM chimerism 4-months PT |(n = 3-4 per group). *P < .05, groups 2 and 3 vs other groups. Similar results were obtained in another independent experiment (4000 cells/group; n = 4/group). The 2 data sets were not pooled. (E-F) Chimerism at 4-months PT (E) sustained by 1000 cells per mouse from groups 1 to 4, as shown in panel (C) and in secondary recipients (F) transplanted with BM cells collected from primary recipients (one-half a femur equivalent from pooled BM cells per recipient) shown in (E) (n = 4-5 per group in [E] and [F]). *P < .05, groups 2 and 3 vs other groups. (F) Flow cytometric analysis of representative mice from (F) are shown in supplemental Figure 10.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2014-03-565721/4/m_519f2.jpeg?Expires=1765093666&Signature=O3k9uUO5f95tMVfGQ0Ih~0DD3N6jp5I6LLFugZoZm9bpiGkp0Dnzf8U0EdVoEAlvw6QiJVTpAlZp8O01kvDWxpvUOKIw7e~wactNm4L1O2J-PRiozI1nmFZ~R~ECJ3LqXUAsr45Rrfevvd7Iiw2RB2AB--6vh-L5HiW00AF8TzsWZRZuZVTDagr0mIV2EDpZTkOOdzR4bnahJRlkUAHMudyd3993ysHTjmz9NaAlJuFhLZCPlsXiG5mzRKmw7KBNeCTqR2qmarTHjp2Fvms-BHi9pT~kP9G6R2Plao-7q01IpxHLjilwrSY105NDyjXxbfjgWxUIwvNpbDcvS5jdYg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 1. Characterization of the functional properties of murine HSC fractionated with CD166. (A) C57BL/6 low-density BM cells were analyzed for the expression of lineage markers and CD48 (left dot plot). Lineage-CD48− (Lin-48−) cells were analyzed for the expression of Sca-1 and cKit (middle dot plot) to identify Lin-Sca1+cKit+CD48− (LSK48−) cells (group 1 in middle dot plot). LSK48− cells were analyzed with CD150 and CD166 (right dot plot) to define groups 2, 3, and 4. Percentages next to each group represent the relative size of each population within the dot plot. A total of 100, 50, and 25 cells from groups 1 through 4 were transplanted into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). (B) Chimerism 4-months PT for the 25-cells/mouse group. *P < .05 vs group 3. (C) Engraftment kinetics for groups 1, 2, 3, and 4 over 4 months measured in the PB of recipients of 100-cells/mouse group. *P < .05 vs corresponding values from LSK48−CD166+CD150+ cells. These data are from 1 of 3 independent experiments with similar results. (D) LDA analysis from BM engraftment at 4-months PT in mice receiving 25, 50, and 100 cells (5 mice/group). P = .01 for the overall test for differences in HSC frequencies between any of the groups. Table shows the estimated frequencies and range of repopulating cells within each group tested. (E) Chimerism at 4-months PT in primary recipients after the competitive transplantation of 10 cells/mouse from groups 1 to 4 identified in (A), along with 10e5 BoyJ competitor cells. (F) Chimerism at 4-months PT in secondary recipients of the equivalent of one half femur from primary recipients (BM cells were collected 4-months postprimary transplant) shown in (E) without competitor cells [4-5 mice/group in (E) and (F)]. (G) RU per 10 cells calculated from data shown in (E) as described in Harrison and Astle.17 *P < .05 vs group 3. (H) Multilineage differentiation 4-months PT of groups 1 through 4 in mice receiving 100 cells/animal. *P < .05 between LSK48−CD166+CD150+ cells and LSK48−, CD166+CD150−, and CD166−CD150+ cells within each group. (I) Expression of CD166 on LSK48−CD150+CD9+ cells (in the 3 dot plots from L to R). (J) Chimerism at 4-months PT in mice receiving 10 cells per mouse from groups I, II, and III transplanted competitively into F1 mice with 10e5 BoyJ competitor cells (5 mice/group). *P < .05 vs group III. (K) Expression of CD166 on the low (L), medium (M), and high (H) fractions of murine BM side population (SP) cells. Data shown in (K) are from 1 representative experiment from 4 independent analyses with similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2014-03-565721/4/m_519f1.jpeg?Expires=1765195905&Signature=O0B4aHXoGVDdYIjK0PCR1aPrmwwyswB3eJPGhLPX1S-ninKXrrXggs2K4ClQ8EBQKyiDAWjIp9FPfB6DA-3DAI7M90sLoip8VghIN5thJl3cboG~BfXhLA7Djc7X1UBmtck37Q5Of3WIPS~-Tkam3xZKGXcai61WsThpPvDf1jhLdETaa4DZnOT2J-zMYIz8HmILE~Fos2zLSmw9RmnwEpLzzHaSeQ9LmXdEg9lwZqsW5tBwQsv50XetlC39SORT~TraSegRHp14bGuwlGpPgaTC-h7z1y5GDjDSW01NMqT8W8LctJbfaBZL~hTzscvooKeeU94b4-Nc1BZsWxr6pg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Characterization of the functional properties of human CB-derived HSC fractionated with CD166. (A) Human CB cells were analyzed with a combination of lineage markers, CD34 and CD38 and Lin−CD34+CD38− cells (group 1) were analyzed for the expression of CD166 to identify CD34+CD38−CD166− (group 2) and CD34+CD38−CD166+ (group 3) cells. Percentages next to each group represent the relative size of each population within the dot plot. (B) A total of 5000 cells from groups 1, 2, and 3 were transplanted into NSG mice and chimerism was assessed 4-months later (n = 3-4 per group). *P < .05, CD34+CD38−CD166+ vs other groups. (C) Fractionation of human CB cells with CD34 and a combination of lineage markers and CD38 to select Lin−CD34+CD38− cells (group 1 in left dot plot). Lin−CD34+CD38− cells were analyzed with CD166 and CD49f (right dot plot) to define groups 2, 3, 4, and 5. Percentages next to each group represent the relative size of each population within the dot plot. A total of 1000 cells from groups 1 through 5 were transplanted into NSG mice. (D) BM chimerism 4-months PT |(n = 3-4 per group). *P < .05, groups 2 and 3 vs other groups. Similar results were obtained in another independent experiment (4000 cells/group; n = 4/group). The 2 data sets were not pooled. (E-F) Chimerism at 4-months PT (E) sustained by 1000 cells per mouse from groups 1 to 4, as shown in panel (C) and in secondary recipients (F) transplanted with BM cells collected from primary recipients (one-half a femur equivalent from pooled BM cells per recipient) shown in (E) (n = 4-5 per group in [E] and [F]). *P < .05, groups 2 and 3 vs other groups. (F) Flow cytometric analysis of representative mice from (F) are shown in supplemental Figure 10.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2014-03-565721/4/m_519f2.jpeg?Expires=1765195905&Signature=RyaJ0Aqa-9iTQXgRv56m88hUBL6ga-1gkbf0K21JGy7NiEA3HQKSc-1VkfyDPPRl1fFbF8hYmiRVGp~L93TgJzWZe-BqEZQIl3buKp5ry5Gl1IITh1aWJIawxzG1WpXBSwWAm70E8Jv-sYQP8yzmt3sHe2UEpQLv86zooT5iy7soE7k6QIC6jgVevjprILjVUgHbZDzMy5oQfvMI7BkBWhQfGQQcCMeQBOJUPUkcfAekcx-9c6Z-2xlUKRXzCi3RcvUkZlyGRJWLH9p28a7Z8LOTB~J8CbRnWbdNe3V8N0eBVJx7wqxnZP~1VaP0bG6~BsKPyUJvyrotstMyVlfrIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)