Key Points
Histones migrate into the cytoplasm of normal erythroblasts during maturation, leading to extruded nuclei largely depleted of protein.
Loss of nuclear exportin Xpo7 inhibits normal erythroid nuclear condensation and enucleation; histones remain in Xpo7-knockdown nuclei.
Abstract
Global nuclear condensation, culminating in enucleation during terminal erythropoiesis, is poorly understood. Proteomic examination of extruded erythroid nuclei from fetal liver revealed a striking depletion of most nuclear proteins, suggesting that nuclear protein export had occurred. Expression of the nuclear export protein, Exportin 7 (Xpo7), is highly erythroid-specific, induced during erythropoiesis, and abundant in very late erythroblasts. Knockdown of Xpo7 in primary mouse fetal liver erythroblasts resulted in severe inhibition of chromatin condensation and enucleation but otherwise had little effect on erythroid differentiation, including hemoglobin accumulation. Nuclei in Xpo7-knockdown cells were larger and less dense than normal and accumulated most nuclear proteins as measured by mass spectrometry. Strikingly, many DNA binding proteins such as histones H2A and H3 were found to have migrated into the cytoplasm of normal late erythroblasts prior to and during enucleation, but not in Xpo7-knockdown cells. Thus, terminal erythroid maturation involves migration of histones into the cytoplasm via a process likely facilitated by Xpo7.
Introduction
Terminal differentiation of red cell progenitors into mature erythrocytes is a complex and highly regulated process including the survival of committed progenitors, a set number of terminal cell divisions, the accumulation of hemoglobin and other erythroid-specific proteins, and global nuclear condensation culminating in enucleation only in mammals.1 Transcription ceases as late erythroblasts terminally exit the cell cycle, after which the nucleus condenses to about 1/10th of its original volume with highly condensed chromatin2 and culminates in enucleation. Multiple cellular pathways (reviewed in Ji et al2 ) regulate erythroid enucleation, including histone deacetylation, actin polymerization, vesicle trafficking and cytokinesis, cell-matrix interactions, and even erythroid-specific micro RNAs, but the preceding features of global nuclear condensation remain poorly understood.
Our limited understanding of erythroid nuclear condensation comes from extensive work in chickens and involves linker histone H5 or the nonhistone myeloid and erythroid nuclear termination protein, neither of which is present in mammals.3 In mammals, only histone acetylation has been linked to erythroid chromatin condensation; mechanisms common to enucleation of other cell types such as caspase activation/DNA degradation in lens cells and keratinocytes, chromosome condensation (condensin), and nuclear condensation in sperm (replacement of histones with transitional proteins and protamines) are seemingly irrelevant in red cells. Treatment of human erythroid precursors4 or murine spleen erythroblasts5 with histone deacetylase inhibitors blocked enucleation, whereas either inhibition or short hairpin RNA (shRNA) knockdown of histone deacetylase 26 blocked both chromatin condensation and enucleation of mouse fetal liver erythroblasts in vitro. Deacetylation of histones generates a positive charge on the corresponding lysine residue; 1 plausible hypothesis is that these new positive charges interact with the negative charges on the phosphodiester bonds of DNA to facilitate chromatin condensation.
Our examinations of nuclear components during red blood cell development combined with the most highly abundant transcripts in late erythroid development7 led us to the subsequent study of a highly induced and erythroid-specific nuclear export protein, Exportin 7 (Xpo7) or RanBP16, revealing that normal erythroid chromatin condensation involves a heretofore-unconnected process—the export of many nuclear proteins including core histones. Active transport across the nuclear membrane is carried out on proteins larger than 20 to 40 kDa by members of the Importin-β nuclear transport receptor family in response to a nucleocytoplasmic RanGTP/GDP gradient.8 Exportins recruit their cargo at the 1000+-fold higher levels of RanGTP found in the nucleus, whereas importins bind their cargo at high RanGDP levels found in the cytoplasm; both receptors then release their cargo in the opposite compartment.9 Eight export mediators (Exportins 1-7 and -t) have been identified in higher eukaryotes with varying substrate specificity. Exportin 2 (Xpo2, also CAS) exports adaptor importin-α; Exportin-t (Xpo-t) exports tRNA; and Exportin 6 (Xpo6) exports actin.10 In contrast, some exportins such as Exportin 1 (Xpo1 or CRM1) have a versatile cargo-binding site allowing it to transport many structurally unrelated cargoes sharing only a specific leucine-rich nuclear export signal.11 Although evolutionarily conserved across several species, Xpo7 and its cargo are poorly understood. Xpo7 appears to have broad substrate specificity as only 3 cargoes have been elucidated in detail that share very little structural composition other than a predominance of basic amino acid residues12 and do not contain the leucine-rich nuclear export signal common to CRM1 cargo proteins.
Methods
Erythroid progenitor isolation and culture
Total fetal liver cells were isolated from E14.5 wild-type C57BL/6 embryos (Jackson Laboratory) and then stained with biotin-labeled anti-TER119 and antihematopoietic lineage antibodies to obtain early erythroid progenitors by streptavidin-conjugated magnetic bead separation as previously described.13 TER119-negative erythroid progenitors were then cultured in erythropoietin-containing medium on fibronectin-coated plates for 48 hours as previously described.14 All animal studies were approved by the Massachusetts Institute of Technology/Whitehead Institute for Biomedical Research and Yale University Institutional Animal Use and Care Committees. Details on isolation are in the supplemental Experimental Procedures available on the Blood Web site.
Retroviral constructs
Retroviruses for shRNA were constructed using the murine stem cell retroviral vector (MSCV)-pgk-green fluorescent protein (GFP)-U3-U6P-Bbs vector (MSCV-pgk promoter-GFP-U6 promoter-shRNA) as described previously.14 The control shRNA construct was designed against the firefly luciferase gene.
H2A-Turquoise fusion experiments used the construct composed of MSCV-MIGR1 vector where the GFP gene was replaced by the fusion gene mTurquoise-H2A, an enhanced cyan fluorescent protein fused with histone H2A (obtained from Addgene #36207).15 Details on shRNA and construct design are in the supplemental Experimental Procedures.
Generation of retroviral supernatants and infection of primary cells
The retrovirus-packaging cell line 293T was maintained and transfected with retroviral plasmid as previously described.14 For infection of purified TER119-negative fetal liver cells, 2 to 6 × 105 cells were resuspended in 1 mL thawed viral supernatant containing 4 μg/mL Polybrene (Sigma) and centrifuged at 1200 rpm for 1 hour at 25°C as previously described.14 Aliquots of each sample were harvested at 24 and 48 hours of culture for further molecular biology and biochemical studies described below.
Western blotting, proteasome inhibitor, and silver staining
Approximately 3 × 106 GFP+ cells were sorted from each retrovirally infected sample after 24, 36, or 48 hours of retroviral infection, or normal cells were isolated from similar time points during the 48-hour culture, and cytoplasmic and nuclear fractions were separated using the PARIS isolation kit (Life Technologies) and then lysed in high salt (300 mM) radioimmunoprecipitation assay buffer. The nuclear fraction was further disrupted by sonication for 3 minutes using a Bioruptor test tube sonicator (Diagenode) and then boiled for 10 minutes, followed by centrifugation at 14 000 rpm for 10 minutes. Lysates were then separated using Novex 4% to 12% electrophoresis gels (Life Technologies) and wet transferred onto polyvinylidene fluoride membranes (Immobilon P; Millipore), followed by immunoblotting with the indicated antibodies. Further details and proteasome inhibitor experiments are in the supplemental Experimental Procedures.
Antibodies
Xpo7 antibody made against either the C terminus of Xpo7 (published in Mingot et al12 ) or against the different exon 1a and 1b isoforms was obtained from Dr Görlich. Details on their construction are in the supplemental Experimental Procedures. Histone-specific antibodies used were rabbit polyclonal histone H2A (#ab18255; Abcam), rabbit polyclonal histone H3 (#06-755; EMD-Millipore), calf thymus conjugate histone H1 conjugated to Alexa-488 (H13188; Life Technologies), rabbit monoclonal pan-methyl-histone H3K9 (#44735; Cell Signaling), rabbit polyclonal macro-histone H2A.1 (#ab37264; Abcam), and rabbit polyclonal histone H3-acetyl K27 (ab4729; Abcam). Mouse monoclonal glyceraldehyde-3-phosphate dehydrogenase antibody (clone 6C5) and goat polyclonal Lamin B antibody (M20) were obtained from Santa Cruz Biotechnologies. Lysine methyltransferase rabbit polyclonal antibodies against ESET (#2196), G9a (#3306), and SUV39H1 (#8729) were obtained from Cell Signaling Technology.
Secondary antibodies used for immunoblotting were as follows: TrueBlot horseradish peroxidase anti-mouse, anti-rabbit, or anti-goat immunoglobulin (Ig)G at 1:2000 (eBioscience). Secondary antibody used for immunofluorescence was Alexa Fluor 594 donkey anti-rabbit (#A21207; Life Technologies). Details of the immunofluorescence staining protocol are included in the supplemental Experimental Procedures.
Flow cytometric analysis of erythroid differentiation and enucleation and fluorescence-activated cell sorter
Cells were immunostained with allophycocyanin-conjugated anti-TER119 antibody at 1:200 (BD Pharmingen), phycoerythrin-conjugated anti-CD71 antibody at 1:200 (BD Pharmingen), and 10 µg/mL Hoechst 33342 (Sigma) for 15 minutes at room temperature. Propidium iodide (Sigma) was added to exclude dead cells from analysis. Flow cytometry and fluorescence-activated cell sorter (FACS) were carried out as previously described.14 GFP+ cells were sorted for additional biochemical and molecular studies described below.
Hemoglobin quantification
Mouse fetal liver cells infected with control and Xpo7 shRNA retroviruses as described above were sorted for GFP at 48 hours of culture, and then 2 × 106 GFP+ cells were lysed in 200 µL Drabkin’s reagent according to the manufacturer’s instructions (Sigma). Spectrophotometric analysis was then performed at 540 nm.
Cell cycle and apoptosis assays
Cells were sorted for GFP+ and then prepared for apoptosis and cycle analysis as previously described.14
RNA isolation and quantitative real-time polymerase chain reaction
RNA was isolated using the RNAeasy Micro kit (Qiagen) from cultured cells 48 hours after retroviral infection, sorted GFP+ cells from each knockdown sample, or from populations R2-R5 from the fetal liver. DNA was obtained via reverse transcription (Stratagene), and quantitative polymerase chain reaction (qPCR) was performed using SYBR green real-time PCR (Applied Biosciences/Life Technologies) as previously described.14 Primers against erythroid-specific genes are from Hattangadi et al.14
Microarray sample preparation, hybridization, and analysis
Total RNA was isolated from the control and Xpo7-knockdown samples as described. Each samples’ cDNA was prepared and hybridized to a mouse whole genome oligo expression microarray (Agilent), as previously described.14 Analysis was performed using global mean normalization utilizing R and Bioconductor16 as previously described.14 Significantly changed genes were defined as at least a twofold increase or decrease from control and with P < .05 and are included in supplemental Table 2.
4′,6-Diamidino-2-phenylindole staining, confocal microscopy, and nuclear volume calculation
After sorting, 5 × 104 to 2 × 106 GFP+ cells were spun onto slides for 3 minutes at 800 rpm (Cytospin 3) and air dried. Cells were fixed with 4% paraformaldehyde, and then nuclei were stained with 0.1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI, Dilactate; Invitrogen) for 2 minutes prior to mounting with coverslips using ProLong Antifade Reagent (Invitrogen).
Details on image acquisition and analysis are in the supplemental Experimental Procedures.
Immunofluorescence
Cells were spun onto poly-l-lysine–coated coverslips in 24-well plates at 1200 rpm for 5 minutes, fixed in fresh 4% paraformaldehyde for 20 minutes, and permeabilized by incubation in 0.25% TritonX100 for 10 minutes. Coverslips were then blocked, stained, and mounted as described in the supplemental Experimental Procedures. Slides were imaged using ZEN 2010 software (Zeiss) on a laser scanning confocal microscope (Zeiss LSM 700) and then analyzed and processed using the Fiji17 package of ImageJ imaging software (http://fiji.sc/wiki/index.php/Fiji).
Results
Nuclear proteins including histones are exported into the cytoplasm during normal terminal erythroid differentiation
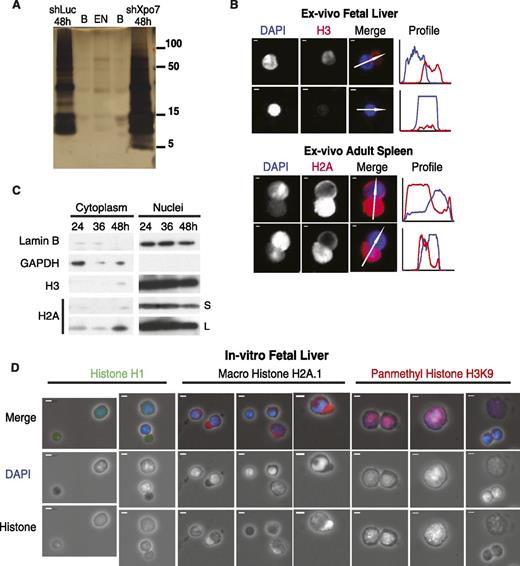
To evaluate the process of terminal erythroid nuclear condensation, we examined the composition of isolated erythroblast nuclei during development using silver stain of polyacrylamide gel protein analysis followed by mass spectrometry proteomics (Figure 1A). We discovered that extruded nuclei are largely depleted of protein (Figure 1A; supplemental Figure 1B), including the major histone species, compared with cells just prior to enucleation in cells at 48 hours of culture when transcription is still active. We then examined the most abundant transcripts present at this stage and found that an abundant nuclear export protein, Exportin 7, or Xpo7, was the only exportin present in late erythroblasts. We then compared nuclei from control cultured erythroblasts at 48 hours, Xpo7-knockdown (Xpo7-KD) erythroblasts at 48 hours, and pycnotic extruded nuclei. Silver stain of equal numbers of nuclei from each sample (Figure 1A) revealed that nuclei from Xpo7-KD cells showed a moderate accumulation of all nuclear proteins (confirmation that equal numbers of nuclei were used is included as qPCR of genomic DNA for the HPRT gene in supplemental Figure 1A). Mass spectrometry analysis of the silver stained bands revealed that Xpo7-KD nuclei accumulated nuclear proteins of almost all functions (supplemental Table 4) and that the relative distribution of histones was largely preserved through erythropoiesis (supplemental Table 3). In other words, there was no specific histone variant that replaced core histones in the condensed extruded nucleus, nor any specific nuclear protein that accumulated after Xpo7 knockdown. We repeated these experiments (silver stain and mass spectrometry) with similar results (data not shown).
Nuclear proteins such as histones are exported into the cytoplasm during normal erythroid differentiation. (A) Silver stain of nuclei isolated from cultured erythroblasts across differentiation (from left, lanes are control shRNA at 48 hours, blank, extruded nuclei, blank, and Xpo7-KD nuclei at 48 hours). Protein sizes in kilodaltons are shown on right. Mass spectrometry of each lane was performed; a complete dataset uploaded to the proteomics identification database (PRIDE) is at http://www.ebi.ac.uk/pride under accession no. 1-20140306-107700. (B) Immunofluorescence (IF) of mouse tissues (fetal liver and adult spleen) isolated ex vivo from C57/Bl6 mice showing migration of histones into the reticulocyte before and during enucleation. Red staining is against either histone 3 or histone 2A using Alexa-594–conjugated secondary antibody, and blue staining is against DNA using DAPI. Profiles on right reflect intensities of each channel (corresponding colors) across the axis of the white arrow shown in the merge micrograph. In all panels, scale bar = 10 μm. (C) Erythroblasts cultured in erythropoietin-containing media were harvested after 24, 36, and 48 hours and fractionated into cytoplasmic and nuclear extracts using the PARIS Isolation Kit (Life Technologies). Immunoblotting was performed against a cytoplasmic protein (glyceraldehyde-3-phosphate dehydrogenase), nuclear membrane protein (Lamin B), and histones H3 and H2A. S, short exposure of blot, 5 seconds; L, long exposure of blot, 15 minutes. (D) IF of cultured erythroblasts at 48 hours after culture showing migration of histones into the cytoplasm before and during enucleation. Three primary antibodies against nuclear proteins are shown: histone 1, macro histone H2A.1, and pan-methyl histone H3 on lysine 9; blue staining is against DNA using DAPI. Secondary antibodies used were Alexa-594–conjugated secondary antibody (macro H2A.1 and pan-methyl H3K9) or Alexa-488–conjugated secondary antibody (H1). In all panels, scale bar = 10 μm.
Nuclear proteins such as histones are exported into the cytoplasm during normal erythroid differentiation. (A) Silver stain of nuclei isolated from cultured erythroblasts across differentiation (from left, lanes are control shRNA at 48 hours, blank, extruded nuclei, blank, and Xpo7-KD nuclei at 48 hours). Protein sizes in kilodaltons are shown on right. Mass spectrometry of each lane was performed; a complete dataset uploaded to the proteomics identification database (PRIDE) is at http://www.ebi.ac.uk/pride under accession no. 1-20140306-107700. (B) Immunofluorescence (IF) of mouse tissues (fetal liver and adult spleen) isolated ex vivo from C57/Bl6 mice showing migration of histones into the reticulocyte before and during enucleation. Red staining is against either histone 3 or histone 2A using Alexa-594–conjugated secondary antibody, and blue staining is against DNA using DAPI. Profiles on right reflect intensities of each channel (corresponding colors) across the axis of the white arrow shown in the merge micrograph. In all panels, scale bar = 10 μm. (C) Erythroblasts cultured in erythropoietin-containing media were harvested after 24, 36, and 48 hours and fractionated into cytoplasmic and nuclear extracts using the PARIS Isolation Kit (Life Technologies). Immunoblotting was performed against a cytoplasmic protein (glyceraldehyde-3-phosphate dehydrogenase), nuclear membrane protein (Lamin B), and histones H3 and H2A. S, short exposure of blot, 5 seconds; L, long exposure of blot, 15 minutes. (D) IF of cultured erythroblasts at 48 hours after culture showing migration of histones into the cytoplasm before and during enucleation. Three primary antibodies against nuclear proteins are shown: histone 1, macro histone H2A.1, and pan-methyl histone H3 on lysine 9; blue staining is against DNA using DAPI. Secondary antibodies used were Alexa-594–conjugated secondary antibody (macro H2A.1 and pan-methyl H3K9) or Alexa-488–conjugated secondary antibody (H1). In all panels, scale bar = 10 μm.
Corroborating this observation, imaging of erythroblasts isolated ex vivo from fresh murine fetal liver and spleen (Figure 1B) revealed that during enucleation, histone staining appears in the cytosol and separates from DAPI staining, with very little histone remaining in extruded nuclei (Figure 1B, second panel from top). Western blot analysis of cultured erythroblasts confirmed these findings: core histones H2A and H3 appear in the cytoplasm near the end of our 48-hour erythroid differentiation culture when many of the cells have begun to enucleate (Figure 1C). Of note, migration of nuclear protein to the cytoplasm appears to be affected by the activity of the proteasome as proteasome inhibition with MG132 increases the level of cytoplasmic histone H2A even further (supplemental Figure 2A).
Examination of 48-hour cultured erythroblasts by immunofluorescence (Figure 1D; supplemental Figure 2B) revealed that other core histones such as histone H1 and histone variants such as macro histone H2A.1 also migrate into the cytoplasm late during red cell development. In contrast, histones containing modifications that are associated with heterochromatin such as histone H3-methyl K9 remain in the nucleus until the nucleus is highly condensed or during enucleation, at which point this histone accumulates in the cytoplasm (Figure 1D; supplemental Figure 2B, far right panels). Given this finding, we then examined the localization of 3 histone methyltransferases that catalyze this modification and found the same phenomenon: that these nuclear proteins are found throughout the nucleus of larger cells, but that as the nuclei condense, the histone methyltransferases migrate to the cytoplasm (supplemental Figure 2C).
Xpo7 is erythroid-specific, highly induced, and uses an erythroid-specific start site
Given this novel phenomenon of nuclear protein export to the cytoplasm during terminal erythroid development, we went on to examine the role that the nuclear export protein Xpo7 plays in this process. We previously showed7 that the majority of genes induced during terminal murine erythroid differentiation including the master regulators are upregulated simultaneously with induction of the glycoprotein Ter119.14 Briefly, we isolated RNA from progressive stages of erythroid development using 5 distinctly stained populations of mouse fetal liver cells corresponding to progressive (albeit somewhat mixed) stages of definitive erythroid development (Figure 2A) and then analyzed the mRNAs in each population by high-throughput sequencing. On filtering for genes involved in nuclear structure and organization such as heterochromatin formation, we found that Exportin 7 or Xpo7 (also known as RANBP16) was 1 of the 1500 most abundant transcripts in terminal erythropoiesis. Xpo7 was actually induced ∼150-fold in late erythroblasts and accumulated even further later in erythropoiesis after transcription has presumably ended (Figure 2B). In contrast, the 5 other known murine exportins and 6 murine importins (5 importin-α subunits and importin β-1) all decreased to negligible levels (Figure 2B). We confirmed this expression pattern of Xpo7 transcript by qPCR (Figure 2C) and found that it closely matched the pattern of induction of erythroid-important regulators such as GATA1 and erythroid-specific proteins such as β-globin,14 suggesting that Xpo7 expression is also tightly regulated during erythropoiesis.
Xpo7 is highly induced during terminal erythroid differentiation, highly erythroid specific, and uses an erythroid-specific start site. (A) FACS of in vivo fetal liver erythropoiesis. Briefly, mouse fetal liver was sorted by flow cytometry into 5 separate populations corresponding to progressive stages of definitive erythroid development using CD71/TER119 staining patterns (regions R1-R5). (B) RNA-seq levels (from Wong et al7 ) of all nuclear exportins during murine definitive erythropoiesis. (C) qPCR of Xpo7 mRNA transcript during terminal erythropoiesis. (D) Tissue expression (measured by qPCR) of Xpo7 transcript in several different mouse tissues. (E) Browser depicting the genomic structure of the murine Xpo7 gene, which consists of 28 exons spanning over 112 kb, transcribed from the reverse strand, from right to left. Boxed area shows close-up of (F), with black bars depicting previously annotated exons; gray bar shows an alternative first exon transcribed only in late erythroblasts. (F) RNA-seq data (numbers on left are log2[RPKM values]) from the earliest committed erythroid precursors (BFU-Es) through early precursors (CFU-Es) to TER119-positive late erythroblasts (from Flugare et al13 ), showing reads corresponding to each exon of Xpo7. Note the alternative exon depicted in the University of California, Santa Cruz gene prediction tracks (arrow) is transcribed only in TER119+ late erythroblasts. The sequence of the alternative exons 1a and 1b is included in supplemental Table 1. BFU-E, erythroid burst forming unit; CFU-E, erythroid colony forming unit.
Xpo7 is highly induced during terminal erythroid differentiation, highly erythroid specific, and uses an erythroid-specific start site. (A) FACS of in vivo fetal liver erythropoiesis. Briefly, mouse fetal liver was sorted by flow cytometry into 5 separate populations corresponding to progressive stages of definitive erythroid development using CD71/TER119 staining patterns (regions R1-R5). (B) RNA-seq levels (from Wong et al7 ) of all nuclear exportins during murine definitive erythropoiesis. (C) qPCR of Xpo7 mRNA transcript during terminal erythropoiesis. (D) Tissue expression (measured by qPCR) of Xpo7 transcript in several different mouse tissues. (E) Browser depicting the genomic structure of the murine Xpo7 gene, which consists of 28 exons spanning over 112 kb, transcribed from the reverse strand, from right to left. Boxed area shows close-up of (F), with black bars depicting previously annotated exons; gray bar shows an alternative first exon transcribed only in late erythroblasts. (F) RNA-seq data (numbers on left are log2[RPKM values]) from the earliest committed erythroid precursors (BFU-Es) through early precursors (CFU-Es) to TER119-positive late erythroblasts (from Flugare et al13 ), showing reads corresponding to each exon of Xpo7. Note the alternative exon depicted in the University of California, Santa Cruz gene prediction tracks (arrow) is transcribed only in TER119+ late erythroblasts. The sequence of the alternative exons 1a and 1b is included in supplemental Table 1. BFU-E, erythroid burst forming unit; CFU-E, erythroid colony forming unit.
The expression pattern of the importins and exportins was confirmed by western blotting for the importins and exportins that showed the highest transcript levels (supplemental Figure 3A): Xpo5, Kpna2, and Kpnb1. Of note, the most abundant exportin in definitive erythropoiesis was the erythroid-specific isoform of Xpo7 (Xpo7b in supplemental Figure 3A, using antibody raised against the protein with a novel erythroid-specific first exon), whereas the nonerythroid isoform (Xpo7a in supplemental Figure 3A, raised against the nonerythroid isoform of Xpo7) decreased similarly to the other murine importins and exportins. The sequence of the alternative exons 1a and 1b is included in supplemental Table 2. Interestingly, the components of the RanGTPase system (including Ran, RanGAP, and Rcc1) decreased considerably but not to negligible levels (supplemental Figure 3B), supporting that the process of ongoing nuclear export can still proceed in the cell via Xpo7 as needed, despite the fact that all other exportins and importins are missing. The Xpo7 transcript was not present in any adult mouse tissue other than TER119-positive late erythroblasts (Figure 2D); this expression pattern is conserved in humans, as seen in the Novartis tissue expression database (supplemental Figure 4).
Analyzing our previously published RNA-seq analysis,13 we uncovered an erythroid-specific first exon of Xpo7 that is expressed in TER119-positive erythroblasts, but not in earlier (TER119-negative) erythroid progenitors (Figure 1E-F). This alternative, erythroid-specific first exon (exon 1b) is located in the 50-kb intronic region between the previously annotated exon 1 (here, exon 1a) and exon 2. This alternative start site likely contains an erythroid-specific promoter (supplemental Figure 5) because analysis of published histone modification data7 reveals that the modification H3-trimethyl K4 (normally associated with active promoter regions and not introns) is present near exon 1b in TER119+ erythroblasts but not earlier erythroid progenitors (data not shown). Data from the ENCODE project18 support this hypothesis, as in late erythroblasts there are GATA1 and TAL1 binding sites within 5 kb of exon 1b (red box, supplemental Figure 5), but not near exon 1a (blue box), which instead shows bivalent H3-trimethyl K4 and H3-trimethyl K27 modifications that when found together signify that this promoter was likely active either in earlier cells or in nonerythroid tissues.19 Finally, location analysis of H3-trimethyl K36—signifying RNA polymerase transcript elongation (light blue arrow, supplemental Figure 5)—reveals that elongation of the Xpo7 transcript extends downstream of the exon 1b (red box) but does not include the region immediately downstream of exon 1a (blue box), supporting the notion that erythroid exon 1 indeed contains an alternative promoter and is not merely a splice variant.
Published chromatin immunoprecipitation data show that this alternative start site for Xpo7 is bound by erythroid master regulators GATA1 in G1E-ER4 cells18 (supplemental Figure 5, bottom 4 tracks) and EKLF in fetal liver late erythroblasts.18,20 The induction of a GATA1 peak at the 1b promoter binding site in erythroblasts 24 hours after reintroduction of GATA1 into the GATA1-null hematopoietic cell line G1-ER (bottom 2 tracks on supplemental Figure 5, red box) further supports the tight, erythroid-specific regulation of Xpo7 expression.
Xpo7 knockdown blocks enucleation but does not affect other important features of terminal erythroid differentiation
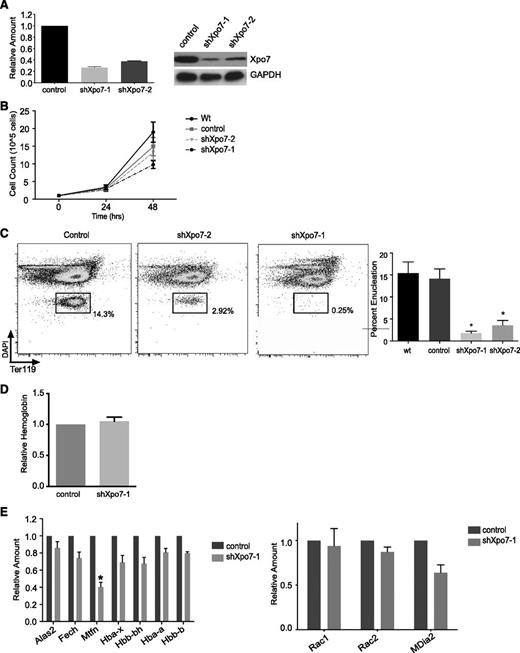
We knocked down Xpo7 in primary murine fetal liver erythroblast cultures using retroviral shRNA infection14 ; ∼70% knockdown of Xpo7 transcript was achieved with the shRNA construct shXpo7-1 (Figure 3A). Xpo7 knockdown resulted in at most a twofold inhibition in proliferation during the last 24 hours of culture (Figure 3B), although this proliferation defect was not due to an increase in apoptosis or significant cell cycle disruption (supplemental Figure 6). Xpo7 knockdown did not affect normal induction of cell surface TER119 (supplemental Figure 7) but did block enucleation (using Hoechst and TER119 staining as in Ji et al21 ), as quantified for 5 biological replicates (Figure 3C; supplemental Figure 8A). Of note, additional enucleation does not occur in our erythroid culture after 48 hours (supplemental Figure 8B). There was no effect on the total amount of hemoglobin accumulated per cell (Figure 3D). We also performed microarray analysis on erythroblasts 36 hours after Xpo7 knockdown and found that the vast majority of the transcriptional profile was unchanged (based on the Student t test performed on the intensity values across the transcriptome). The complete dataset has been uploaded to the Gene Expression Omnibus database under accession no. GSE54457; significantly changed transcripts (greater than twofold increase or decrease) are shown in supplemental Table 2. Using qPCR, we confirmed that the transcript levels of key genes involved in heme biosynthesis were largely unaffected by Xpo7 knockdown, and there was no significant change in the direct regulators of enucleation Rac1 or mDia2 (Figure 3E). Taken together, these data support the hypothesis that the major function of Xpo7 occurs in the nucleus after transcription of the erythroid-specific program has largely ended and erythroblasts have left the cell cycle.
Xpo7 knockdown inhibits enucleation but does not affect important aspects of erythroid differentiation such as hemoglobin accumulation or the erythroid expression program. (A) (Left) qPCR showing specific knockdown of Xpo7 transcript and (right) western blot showing a decrease in Xpo7 protein for 2 different shRNA constructs. (B) Cell counts counted at 24-hour intervals during in vitro culture of shRNA-infected erythroblasts. (C) Enucleation (left) measured by FACS of cultured erythroblasts using DAPI and TER119 staining (as in Ji et al21 ) and (right) quantified for 5 independent experiments. (D) Hemoglobin quantification per cell by spectrometry using Drabkin’s reagent (as in Dessypris1 ). (E) Transcript levels of erythroid-specific genes as measured by qPCR. Genes chosen are required for (left) hemoglobin production or (right) the process of enucleation. *P < .01 only for mitoferrin-1 (Slc25a37). Complete dataset has been uploaded to the GEO database under accession no. GSE54457; significantly changed transcripts (greater than twofold increase or decrease) are shown in supplemental Table 2.
Xpo7 knockdown inhibits enucleation but does not affect important aspects of erythroid differentiation such as hemoglobin accumulation or the erythroid expression program. (A) (Left) qPCR showing specific knockdown of Xpo7 transcript and (right) western blot showing a decrease in Xpo7 protein for 2 different shRNA constructs. (B) Cell counts counted at 24-hour intervals during in vitro culture of shRNA-infected erythroblasts. (C) Enucleation (left) measured by FACS of cultured erythroblasts using DAPI and TER119 staining (as in Ji et al21 ) and (right) quantified for 5 independent experiments. (D) Hemoglobin quantification per cell by spectrometry using Drabkin’s reagent (as in Dessypris1 ). (E) Transcript levels of erythroid-specific genes as measured by qPCR. Genes chosen are required for (left) hemoglobin production or (right) the process of enucleation. *P < .01 only for mitoferrin-1 (Slc25a37). Complete dataset has been uploaded to the GEO database under accession no. GSE54457; significantly changed transcripts (greater than twofold increase or decrease) are shown in supplemental Table 2.
Xpo7 knockdown severely disrupts terminal erythroid chromatin condensation
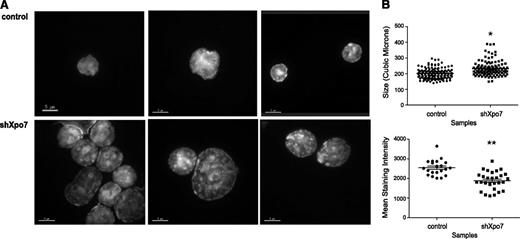
Analysis by flow cytometry revealed that Xpo7-KD erythroblasts showed higher forward- and side-scatter values, consistent with immature erythroblasts (data not shown), suggesting that the nucleus was not able to progress normally through differentiation. To more closely quantify the defects in chromatin condensation after Xpo7 knockdown, 48-hour cultured cells were sorted for GFP (retroviral expression) and examined using confocal microscopy with 3-dimensional Z-stacks after DAPI staining. Although control cells showed normal nuclear condensation, Xpo7-KD cells showed less homogenous staining of their nuclei (Figure 4A). Nuclear volume and DAPI density were quantified for >100 cells after DAPI staining (as in Ji et al21 ), revealing that Xpo7-KD nuclei were significantly larger and less dense than those of control cells (Figure 4B). These data support the hypothesis that Xpo7 plays a role in erythroid nuclear condensation.
Xpo7 knockdown significantly disrupts erythroid terminal nuclear condensation. (A) Erythroblasts infected with control or Xpo7 shRNA imaged at 48 hours of culture. Cells were stained with DAPI and imaged at 100× using confocal fluorescent microscopy. (B) (Upper) Quantification of volume of nuclei, measured by calculation of area for each Z-stack slice of confocal images in A, and (Lower) average DAPI intensity per combined deconvoluted 2-dimensional image.
Xpo7 knockdown significantly disrupts erythroid terminal nuclear condensation. (A) Erythroblasts infected with control or Xpo7 shRNA imaged at 48 hours of culture. Cells were stained with DAPI and imaged at 100× using confocal fluorescent microscopy. (B) (Upper) Quantification of volume of nuclei, measured by calculation of area for each Z-stack slice of confocal images in A, and (Lower) average DAPI intensity per combined deconvoluted 2-dimensional image.
Migration of nuclear proteins such as histones into the cytoplasm during normal terminal erythroid differentiation is disrupted by Xpo7 knockdown
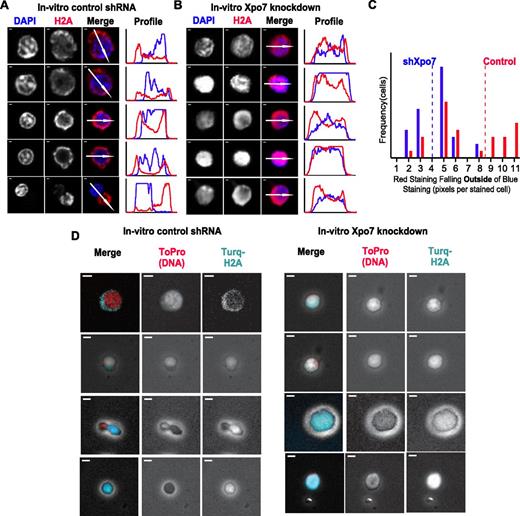
Examination of 48-hour cultured erythroblasts with either control or Xpo7 shRNA revealed that knockdown of Xpo7 disrupts the movement of histones, as histone staining was present in the cytoplasm of control cells (Figure 5A) yet remained high in density over Xpo7-KD nuclei (Figure 5B). This is most clearly illustrated by the fluorescence intensity profiles to the right of each panel of cell micrographs: in control enucleating cells (Figure 5A), histone staining (red line) accumulates outside the trace line of DAPI staining (blue line), whereas in Xpo7-KD cells (Figure 5B), H2A staining and DAPI staining remain colocalized. The area of cytoplasmic H2A staining that is no longer colocalized with DAPI staining was quantified for cells with positive staining in both channels by creating a binary mask of each channel (supplemental Figure 9A) and then measuring how much red staining occurs outside of blue staining by subtraction of the 2 binary images (supplemental Figure 9B). A histogram of the amounts of nonnuclear (cytoplasmic) histone staining (Figure 5C) revealed a significant paucity of cytoplasmic H2A staining after Xpo7 knockdown; there was almost twice as much cytoplasmic histone staining in control cells on average than in Xpo7-KD cells. To illustrate this point further, we used a fluorescent-tagged histone H2A construct and coinfected fetal liver erythroid progenitors with both shRNA vectors as described, as well as an mTurquoise-H2A fusion construct. More cytoplasmic turquoise staining in control than Xpo7-KD cells supports the notion that export of histone H2A is disrupted by Xpo7 knockdown (Figure 5D).
The migration of nuclear proteins such as histones out of the nucleus during normal erythroid differentiation is inhibited by Xpo7 knockdown. (A-B) Immunofluorescence of cultured erythroblasts after 48 hours of culture containing shRNA against either (A) control or (B) Xpo7. Red staining is against histone 2A using an Alexa-594 secondary antibody and blue staining is against DNA (DAPI). Note that there is less red staining overlying the nuclei in the control shRNA-infected cells and increased accumulation in the cytoplasm, but in contrast, after Xpo7 knockdown, histone staining remains colocalized with the nucleus. In all panels, scale bar = 10 μm. (C) Histogram of amount (pixels per cell) of cytoplasmic, or noncolocalized with DAPI, red staining in cultured control (red) or Xpo7-KD (blue) cells at 48 hours. Mean noncolocalized red staining (pixels per cell) for 35 cells is depicted as a dotted line (control, 8.49; shXpo7, 4.00). There is significantly more staining outside of the nucleus in control cells compared with Xpo7-KD cells (Student t test, P < .001). (D) Immunofluorescence of cultured erythroblasts after 48 hours of culture containing shRNA against either control or Xpo7 and a plasmid containing a histone H2A-mTurquoise fusion protein. Red staining shows nuclear DNA using ToPro dye, and turquoise depicts localization of the fusion protein. There is less turquoise staining over the nucleus in control shRNA-infected cells (and more in the cytoplasm), whereas in Xpo7-KD cells, red and turquoise staining colocalize.
The migration of nuclear proteins such as histones out of the nucleus during normal erythroid differentiation is inhibited by Xpo7 knockdown. (A-B) Immunofluorescence of cultured erythroblasts after 48 hours of culture containing shRNA against either (A) control or (B) Xpo7. Red staining is against histone 2A using an Alexa-594 secondary antibody and blue staining is against DNA (DAPI). Note that there is less red staining overlying the nuclei in the control shRNA-infected cells and increased accumulation in the cytoplasm, but in contrast, after Xpo7 knockdown, histone staining remains colocalized with the nucleus. In all panels, scale bar = 10 μm. (C) Histogram of amount (pixels per cell) of cytoplasmic, or noncolocalized with DAPI, red staining in cultured control (red) or Xpo7-KD (blue) cells at 48 hours. Mean noncolocalized red staining (pixels per cell) for 35 cells is depicted as a dotted line (control, 8.49; shXpo7, 4.00). There is significantly more staining outside of the nucleus in control cells compared with Xpo7-KD cells (Student t test, P < .001). (D) Immunofluorescence of cultured erythroblasts after 48 hours of culture containing shRNA against either control or Xpo7 and a plasmid containing a histone H2A-mTurquoise fusion protein. Red staining shows nuclear DNA using ToPro dye, and turquoise depicts localization of the fusion protein. There is less turquoise staining over the nucleus in control shRNA-infected cells (and more in the cytoplasm), whereas in Xpo7-KD cells, red and turquoise staining colocalize.
Discussion
Our study not only uncovered a new regulator of erythroid nuclear condensation, Xpo7, but it also revealed that the migration of some nuclear proteins—including core histones H2A and H3—to the cytoplasm occurs prior to enucleation and may be facilitated by the exporter Xpo7.
The highly erythroid specific expression of Xpo7, its induction to very high levels, and its abundance even late in erythroid development after the nucleus has shut down all support a unique erythroid-specific function for this nuclear export protein. In addition, we uncovered an erythroid-specific promoter for Xpo7; previously published work18 has revealed direct binding of this promoter by the GATA1-Tal1 complex in erythroid cells. Previous work22 showed that many important erythroid-specific genes use erythroid-specific promoters during development, and more generally, that intergenic enhancers act as unidirectional alternative promoters for tissue-specific regulators.23 Taken together, these data support the hypothesis that Xpo7 is a highly regulated, critical erythroid-specific regulator.
Our functional analysis of Xpo7 via retroviral knockdown in our well-studied in vitro primary fetal liver erythroid cell culture system revealed its major function in the erythroid nucleus after transcription has ended and after erythroblasts have left the cell cycle: following Xpo7 knockdown, nuclear condensation and enucleation were severely inhibited, whereas major features of the erythroid expression program—hemoglobin production, induction of many erythroid-important genes, and erythroid cell surface marker acquisition—remained intact. Presumably, this is because Xpo7’s role in facilitating nuclear condensation occurs entirely after transcription has ended.
We recognize the weakness of our in vitro culture system: unlike in vivo in the mouse, in our in vitro culture system, enucleation is not complete, although it is measureable and consistent from 15% to 30%. In addition, after enucleation is measured at 48 hours, unlike reticulocytes in the mouse, the retics (and extruded nuclei) in culture are not stable, so we do not see additional enucleation after the end of our cultures.
The key finding of our study is that during the final stages prior to enucleation, some nuclear proteins such as histones migrate out of the condensed erythroid precursor nucleus into the cytoplasm. This process is disrupted by Xpo7 knockdown. Our findings suggest a basic process underlying chromatin condensation that is unique to erythroid development: the removal of some nuclear proteins including histones from the late erythroblast nucleus. We speculate that some of these nuclear proteins may be replaced by smaller basic proteins—as occurs during spermatogenesis— or by a multicharged cation such as Ca2+, given that chromatin cannot condense without neutralization of its overall negative charge and anticipate that we will uncover this molecule with our ongoing studies.
These data also imply the reason why Xpo7 might be such a tightly regulated, abundant, and erythroid-specific nuclear export protein: that helping to facilitate the removal of vital nuclear proteins such as histones may only occur once the nucleus is no longer necessary for erythroid progenitor survival and function. Ours is the first study linking the process of export of some critical nuclear proteins such as core histones with erythroid chromatin condensation and suggesting that the highly specialized nuclear export protein Xpo7 plays an important role in this function. Given that active nuclear transport via importins and exportins is confined to proteins >20 to 40 kDa and histone chaperones are known to be responsible for chaperoning translated histones into the nucleus and during mitosis during nucleosome assembly and disassembly, we do not propose that Xpo7 exports histones directly. We speculate that Xpo7 may export a regulator of this process that may maintain nucleosome stability or perhaps histone chaperone affinity for core DNA binding proteins, that when removed from the nucleus, allows nucleosomes to disassemble and nuclear proteins to migrate out of the nucleus. We hypothesize that the exported nuclear proteins may be degraded by other processes such as autophagy or the ubiquitin-proteasome system based on our experiments using the proteasome inhibitor MG132. Although the original identification of the role of ubiquitin in ATP-dependent proteolytic degradation made use of reticulocyte cell-free systems,24,25 in erythropoiesis, the resultant free amino acids could be used by the reticulocyte to synthesize hemoglobin and other major red cell proteins that continue to be translated even after the nucleus is extruded.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge Dr Dorus Gadella for the mTurquoise-H2A plasmid (obtained from Addgene). The authors thank Eliza Vasile (MIT) and Wendy Salmon (Whitehead Institute) for superb expertise in confocal and fluorescent microscopy and Chad Araneo from the Whitehead FACS Core facility and Prat Thiru from the Whitehead Bioinformatics Core facility for assistance.
This work was supported by National Institutes of Health National Heart, Lung, and Blood Institute grant P01 HL32262 (to H.F.L.), the National Institute of General Medicine grant K08 GM102718 (to H.C.P.), and the National Institute of Diabetes, Digestive, and Kidney Diseases grant K08 DK076848 (to S.M.H.).
Authorship
Contribution: S.M.H. designed experiments, performed research, analyzed data, and wrote and revised the manuscript; H.C.P., M.M.-H., and D.G. performed experiments and reviewed the manuscript; J.S., K.B., A.A.-F., K.P., S.V., S.M.-M., and J.W. performed experiments; and H.F.L. designed experiments and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.M.H. is Division of Pediatric Hematology-Oncology, Yale University School of Medicine, New Haven, CT.
The current affiliation for K.B. is Harvard Medical School, Boston, MA.
The current affiliation for M.M.-H. is Institute for Stem Cell Biology and Regenerative Medicine (InStem), National Center for Biological Sciences, Bangalore, India.
Correspondence: Shilpa M. Hattangadi, Pediatric Hematology-Oncology, Yale University School of Medicine, 333 Cedar St, LMP 2073, New Haven, CT 06520; e-mail: shilpa.hattangadi@yale.edu.

![Figure 2. Xpo7 is highly induced during terminal erythroid differentiation, highly erythroid specific, and uses an erythroid-specific start site. (A) FACS of in vivo fetal liver erythropoiesis. Briefly, mouse fetal liver was sorted by flow cytometry into 5 separate populations corresponding to progressive stages of definitive erythroid development using CD71/TER119 staining patterns (regions R1-R5). (B) RNA-seq levels (from Wong et al7) of all nuclear exportins during murine definitive erythropoiesis. (C) qPCR of Xpo7 mRNA transcript during terminal erythropoiesis. (D) Tissue expression (measured by qPCR) of Xpo7 transcript in several different mouse tissues. (E) Browser depicting the genomic structure of the murine Xpo7 gene, which consists of 28 exons spanning over 112 kb, transcribed from the reverse strand, from right to left. Boxed area shows close-up of (F), with black bars depicting previously annotated exons; gray bar shows an alternative first exon transcribed only in late erythroblasts. (F) RNA-seq data (numbers on left are log2[RPKM values]) from the earliest committed erythroid precursors (BFU-Es) through early precursors (CFU-Es) to TER119-positive late erythroblasts (from Flugare et al13), showing reads corresponding to each exon of Xpo7. Note the alternative exon depicted in the University of California, Santa Cruz gene prediction tracks (arrow) is transcribed only in TER119+ late erythroblasts. The sequence of the alternative exons 1a and 1b is included in supplemental Table 1. BFU-E, erythroid burst forming unit; CFU-E, erythroid colony forming unit.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/12/10.1182_blood-2013-11-537761/4/m_1931f2.jpeg?Expires=1769256395&Signature=woQHuie6rA-1jtB7hEtOq3w7pdWlk9ecp9uGlNFoBz488aUw~CaBf0tI7XGRTnudI-kvi0T9YwQMtu0Wuu-LXfY6hWCMn~-t5O8HISysSaNcwzzja84ODTDPCtkmuMGpS5V4FCc3pf65bQtuVKYFS9FTeW5VAi6jlhelDZoii1SkeSwebOIXC9kHYbjVGoocUUtNBOvAx1vqZQhCht8E8P4vlEEeKEahUBbSpOPryvcGon6Z7KqXBWCwkEqH2JuevNNfrRXndaLqCEx-zXlqtHQF0pvXnXm656-G6d9xoc8faKOkDDQV3SyCbkOD4HNXBtevB~WRPDddWwxNx0Fzpw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)