Key Points
Mysm1 is required to maintain the quiescence and pool size of HSC, and its deletion severely impairs the survival and function of HSC.
Mysm1 controls HSC homeostasis by regulating Gfi1 expression via modulating histone modifications and transcriptional factors recruitment.
Abstract
Epigenetic histone modifications play critical roles in the control of self-renewal and differentiation of hematopoietic stem cells (HSCs). Mysm1 is a recently identified histone H2A deubiquitinase with essential and intrinsic roles for maintaining functional HSCs. In this study, in addition to confirming this function of Mysm1, by using Mysm1-deficient (Mysm1−/−) mice, we provide more evidence for how Mysm1 controls HSC homeostasis. Mysm1 deletion drives HSCs from quiescence into rapid cycling and increases their apoptotic rate, resulting in an exhaustion of the stem cell pool, which leads to an impaired self-renewal and lineage reconstituting abilities in the Mysm1-deficient mice. Our study identified Gfi1 as one of the candidate genes responsible for the HSC defect in Mysm1-deficient mice. Mechanistic studies revealed that Mysm1 modulates histone modifications and directs the recruitment of key transcriptional factors such as Gata2 and Runx1 to the Gfi1 locus in HSCs. We found that Mysm1 directly associates with the Gfi1 enhancer element and promotes its transcription through Gata2 and Runx1 transactivation. Thus, our study not only elaborates on the initial reports of Mysm1 association with HSC homeostasis but also delineates a possible epigenetic mechanism through which Mysm1 carries out this function in the HSCs.
Introduction
Hematopoietic stem cells (HSCs) are responsible for giving rise to all lineages of blood cells. To sustain blood cell production throughout the lifetime of an individual, a steady-state condition is established during postnatal life in which HSC self-renewal and differentiation are carefully regulated to maintain the HSC pool.1,2 Numerous genes and signaling pathways, including HoxB4, Notch1, growth factor independence 1 (Gfi1), Bmi-1, and the Wnt signaling pathway, have been implicated in this process.3-10 Recently, accumulating evidence indicates that molecules involved in epigenetic and chromatin modifications are also critical for HSC self-renewal and differentiation.11-14 However, although increasing numbers of key players in regulating HSC fate decision have been identified, their roles in HSC biology and their interplay remain poorly understood.
Histone modification is one of the major covalent modifications that occurs at histone tails. Among the histone octamer, H2A is the most abundant ubiquitinated protein and polycomb repressor complex 1 (PRC1) is the main H2A ubiquitin (H2Aub) ligase.15,16 PRC1 plays an important role in transcriptional repression of target genes and is required for the maintenance of both embryonic as well as a broad range of adult stem cells, including HSCs. As observed in knockout mouse lines for Ring1B,17 Bmi1,8 Rae28,18 and Mel18,19 mice harboring mutations in the components of PRC1 commonly develop hematopoietic abnormalities. Once localized to the chromatin, Ring1B and Bmi1, the core components of PRC1, can ubiquitinate histone H2A at lysine 119,15,16,20 which is thought to contribute to transcriptional repression by inhibiting transcription initiation21 or by restraining RNA pol II from elongation.22,23 Although some reports indicate that PRC1 functions as a transcriptional repressor independent of its H2Aub ligase catalytic activity,24 more studies support that H2AK119ub plays a central role in PRC1-mediated gene repression.16,20,25
Despite its important function and early discovery, histone ubiquitination remains the least understood compared with other histone modifications. In particular, the physiologic functions for the increasing number of histone H2A deubiquitinases remain unknown. Recently, ubiquitin-specific protease 16, ubiquitin-specific protease 21, BRCA1-associated protein 1, and the Myb-like, SWIRM, and MPN domains containing protein 1 (Mysm1) were identified as H2A-specific deubiquitinases.21,26-28 They play important roles in H2Aub-mediated HOX gene silencing, X chromosome inactivation, cell-cycle progression, DNA damage or repair, and liver regeneration.21,26,27,29,30
We and others have found that Mysm1 is required for bone marrow (BM) hematopoiesis and lymphocyte differentiation, especially in B-cell development.31,32 Nijnik et al32 have reported the role of Mysm1 in BM hematopoiesis and function. However, the detailed role of Mysm1 exclusively in HSC’s biology and the mechanisms associated with it has not been investigated. In the present study, we enumerate the critical role of Mysm1 in HSC maintenance, self-renewal, differentiation, and function in a more-detailed manner and provide a possible mechanism by which Mysm1 performs this function.
Materials and methods
Mice
Mysm1-deficient mice were generated as described previously.31 In summary, they were generated by crossing Mysm1 mRNA truncation-first floxed mice (Mysm1tm1a/tm1a) with MMTV-cre mice in the B6129F1 background or Tek-cre in the BL/6 background for complete deletion of the floxed Mysm1 exon without any possible transcriptional leakage of the splice acceptor-capture and RNA poly(A) termination strategy designed in the Mysm1-targeted vector.31 MMTV-Cre mice and Tek-Cre have a widespread pattern of Cre expression in various cells, including hematopoietic cells, B and T cells, and their progenitors. In all experiments, wild-type (WT) littermates (+/+) were used for controls. Mice were maintained in a pathogen-free barrier facility, and all experiments were performed in accordance with the University of Southern California Institutional Animal Care and Use Committee.
Cell proliferation and cell-cycle studies
In vivo incorporation of 5-bromo-2′-deoxyuridine (BrdU) into Lin−Sca-1+c-Kit+ (LSK) cells was assessed by the FITC BrdU Flow kit (BD Pharmingen, San Diego, CA). Mice were intraperitoneally injected with 2 mg mL−1 of BrdU for 5 days and then euthanized. BM cells were prepared and stained with antibodies and analyzed by flow cytometry. In some experiments, LSK cells were first sorted and infected with lentivirus or retrovirus constructs for indicated time. Cells were then incubated with BrdU at a final concentration of 10 µM for 30 minutes before staining and analysis.
For cell-cycle analysis, mice received a single intraperitoneal injection of BrdU at the dose of 2 mg per recipient. Then, 1 hour later, mice were euthanized, and BM cells were stained and analyzed by flow cytometry.
Pyronin and Hoechst staining
The quiescence of the freshly isolated HSC was determined by staining with Hoechst 33342 (Molecular Probes, Eugene, OR) and pyronin Y (Sigma-Aldrich, St. Louis, MO). BM cells were resuspended in phosphate-buffered saline containing 2% (v/v) fetal calf serum and 10 µM Hoechst 33342. Cells were then incubated for 30 minutes at 37°C, then washed and subsequently resuspended in phosphate-buffered saline supplemented with 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4; glucose (1 mg/mL); 10% (v/v) fetal calf serum; 10 µM Hoechst 33342; and pyronin Y (1 µg/mL). Cells were incubated for an additional 15 minutes at 37°C, and then were washed and stained for analysis by flow cytometry. Pyronin Y fluorescence was detected at 575 nm in the linear range.
Chromatin immunoprecipitation
Chromatin was immunoprecipitated according to the manufacturer’s instructions (Cell Signaling Technology, Danvers, MA). In brief, cells were crosslinked with 1% (v/v) formaldehyde. Chromatin was isolated, digested by micrococcal nuclease, sheared by sonication, and immunoprecipitated with antibodies. Immunoprecipitated DNA was washed and eluted according to the manufacturer’s instructions. Eluted DNA and sheared input material was analyzed by polymerase chain reaction (PCR). The following antibodies were used: anti-Mysm1 was custom-made; anti-Gata2 (sc-9008x) and anti-Scl (sc-12984x) were both from Santa Cruz Biotechnology (Dallas, TX); anti-PU.1 (2258) was from Cell Signaling Technology (Danvers, MA); and anti-Runx1 (ab23980), anti-H3K4me (ab8895), anti-H3K4me3 (ab1012), anti- H3K27me3 (ab6002), anti-H3K27Ac (ab4329), and anti-H3K9Ac (ab4441) were from Abcam (Cambridge Science Park, Cambridge, United Kingdom). More materials and methods are available in supplemental File 1; see the Blood Web site.
Results
Mysm1 controls HSC differentiation during its transition to MPPs
Nijnik et al32 recently reported the role of Mysm1 in BM hematopoiesis and function. In this study, we attempted to investigate the importance of Mysm1 in HSC differentiation and function in a more detailed manner. To do this, we used Mysm1 knockout-first (Mysm1−/−) mice, as described previously.31 As reported previously, Mysm1−/− mice showed a reduction in the absolute numbers of many hematopoietic lineage cells by various degrees (supplemental Figure 1),31,32 hence, we interrogated whether this reduction in cell numbers was attributable to some defects in the HSC compartment of Mysm1−/− BM. We first checked Mysm1 expression level in BM and sorted lineage-negative (Lin−: CD11b−Gr-1−B220−CD3ε−Ter119−), lineage-positive (Lin+: CD11b+/Gr-1+/B220+/CD3ε+/Ter119+), LSK, long-term HSC (LT-HSC; CD150+CD48−LSK cells), short-term HSC (ST-HSC; CD150+CD48+LSK cells), and multipotent progenitors (MPPs; CD150−CD48+LSK cells) from the BM of C57BL/6 WT mice. Real-time PCR analysis showed that Mysm1 mRNA expression was much greater in the stem cell and progenitor compartments, especially in LT-HSC and ST-HSC than in BM and Lin+ cells (supplemental Figure 1A), indicating the importance of Mysm1 in HSC maintenance and function.
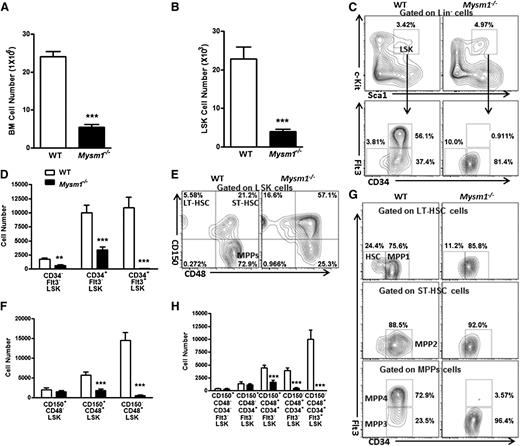
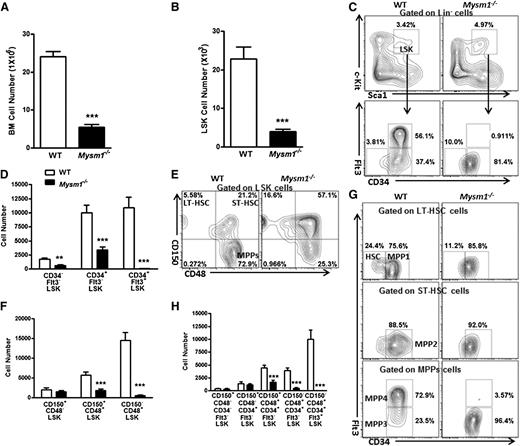
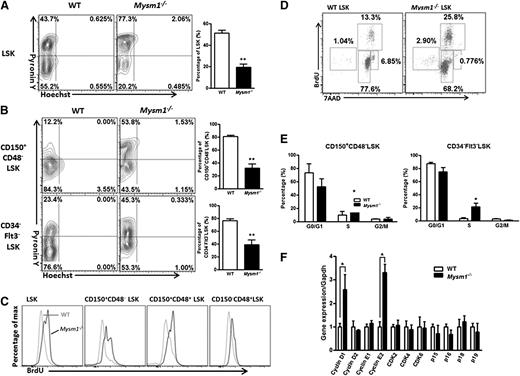
We then analyzed and found that Mysm1-deficient mice had a more than fourfold decrease in the total BM cell numbers and a fivefold reduction in the Lin− BM cells (supplemental Figures 1A and 2B). In Mysm1-deficient mice, a distinct reduction in the absolute numbers of LSK cells, which include the HSC and their earliest progenitors, was observed (Figure 1B). Further analysis of the LSK compartment indicated that Mysm1−/− mice had reduced absolute numbers of CD34−Flt3−, CD34+Flt3−, and CD34+Flt3+LSK cells despite an increase in the proportion of CD34−Flt3− and CD34+Flt3−LSK subsets. Strikingly, the more mature multipotent progenitors, CD34+Flt3+LSK cells, were severely decreased both in proportion and numbers and almost absent in the Mysm1−/− Lin− compartment (Figure 1C-D). We observed similar trend by CD150- and CD48-based immunophenotyping of LT-HSC, ST-HSC, and MPPs, except for an insignificant alteration in the cell numbers of LT-HSC (Figure 1E-F).
Mysm1 deficiency results in a reduction of HSC and its progenitors. Absolute number of (A) BM cells and (B) LSK cells per femur (hind leg) of 8∼12-week-old WT and Mysm1−/− mice; n = 9 mice per group. (C) Distribution of LSK cells and various LSK subsets in the WT and Mysm1−/− BM: Lin negative-gated cells were assessed on the basis of their expression of Sca-1 and c-Kit (top); LSK-gated cells were further defined on the basis of the expression of CD34 and Flt3 (bottom). Numbers adjacent to outlined areas indicate frequency. (D) Absolute number of LSK subsets per femur in the WT and Mysm1−/− mice on the basis of the gates in panel C; n = 8-10 mice per group. (E) Distribution of cells in LSK subsets in the WT and Mysm1−/− mice, assessed by the expression of CD150 and CD48. (F) Absolute number of LSK subsets per femur in the WT and Mysm1−/− mice on the basis of the gates in panel E; n = 8-10 mice per group. (G) Expression of Flt3 and CD34 in the WT and Mysm1−/− BM LSK subsets gated as in panel E. (H) Absolute number of LSK subsets per femur in WT and Mysm1−/− mice (n = 8-10 per group), stained as in panel G. (C,E,G) Data are representative of 3 independent experiments. *P < .05; **P < .01; ***P < .001.
Mysm1 deficiency results in a reduction of HSC and its progenitors. Absolute number of (A) BM cells and (B) LSK cells per femur (hind leg) of 8∼12-week-old WT and Mysm1−/− mice; n = 9 mice per group. (C) Distribution of LSK cells and various LSK subsets in the WT and Mysm1−/− BM: Lin negative-gated cells were assessed on the basis of their expression of Sca-1 and c-Kit (top); LSK-gated cells were further defined on the basis of the expression of CD34 and Flt3 (bottom). Numbers adjacent to outlined areas indicate frequency. (D) Absolute number of LSK subsets per femur in the WT and Mysm1−/− mice on the basis of the gates in panel C; n = 8-10 mice per group. (E) Distribution of cells in LSK subsets in the WT and Mysm1−/− mice, assessed by the expression of CD150 and CD48. (F) Absolute number of LSK subsets per femur in the WT and Mysm1−/− mice on the basis of the gates in panel E; n = 8-10 mice per group. (G) Expression of Flt3 and CD34 in the WT and Mysm1−/− BM LSK subsets gated as in panel E. (H) Absolute number of LSK subsets per femur in WT and Mysm1−/− mice (n = 8-10 per group), stained as in panel G. (C,E,G) Data are representative of 3 independent experiments. *P < .05; **P < .01; ***P < .001.
LSK cells can be further subcategorized on the basis of their expression of CD150, CD48, CD34, and Flt3 into the following five subsets: a most primitive HSC subset (CD34−Flt3−CD150+CD48−LSK), and the increasingly differentiated MPP1 subset (CD34+Flt3−CD150+CD48−LSK), MPP2 subset (CD34+Flt3−CD150+CD48+LSK), MPP3 subset (CD34+Flt3−CD150−CD48+LSK), and MPP4 subset (CD34+Flt3+CD150−CD48+LSK).33,34 Our analysis showed that although there was not a significant change in the most primitive HSC and MPP1 subsets, the absolute numbers of MPP2, MPP3, and MPP4 dramatically decreased (Figure 1G-H). Specifically, the reduction was severe in the most mature MPP4 compartment, with a 20-fold reduction in the percentages and a 750-fold reduction in the cells numbers of Mysm1−/− mice compared with their WT littermates (Figure 1G-H; supplemental Figure 2E). We further noticed that the reduction of MPP4 resulted from a distinct decrease of the Flt3+LSK cells (supplemental Figure 2F). Flt3’s expression enhances lymphoid repopulating potential but not of that of the long-term repopulating potential.35 Reduced Flt3 expression in this compartment may signify an early lesion in lymphoid development. Consistently, Mysm1−/− mice have been reported to show impaired lymphopoiesis.31,32 Together, these data add more to the finding of Nijnik et al32 by demonstrating that Mysm1 is required for differentiation of HSC, specifically during the transition to MPPs.
Mysm1 controls HSC’s quiescence and apoptosis and intrinsically regulates the stem cell differentiation and function
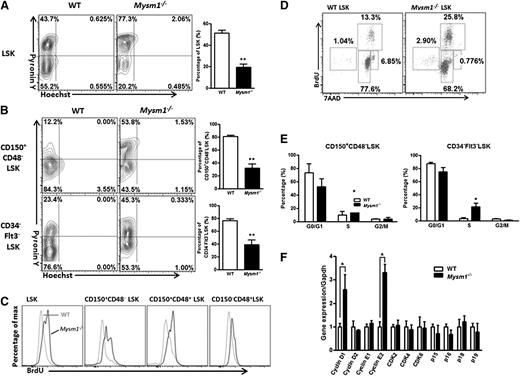
In steady state, most cells in the HSC pool are quiescent, and maintenance of HSC’s quiescence is critical for controlling the stem cell pool. When we measured total RNA/DNA contents in the LSK cells and its subsets by Pyronin Y/Hoechst staining, we found that Mysm1 deficiency significantly reduced, but not increased, the proportion of LSK cells in the G0 phase, whereas there was a significant increase in the proportion of LSK cells in the G1 phase (Figure 2A-B; supplemental Figure 3). To further verify these data, we measured the rate of HSC proliferation by the incorporation of BrdU. Figure 2C-E show that 5 days after BrdU injection, more LSK cells in the Mysm1−/− mice incorporated BrdU, a finding that was more evident in the most primitive Mysm1−/− CD150+CD48−LSK LT-HSC. Consistent with these data, we also noticed an increase in the mRNA levels of cell-cycle genes, cyclin D1 and cyclin E2, and a decrease in the mRNA levels of p15 and p16 in the Mysm1−/− LSK cells in comparison with that of WT cells (Figure 2F). Collectively, our data indicate that Mysm1 maintains HSC in the quiescent stage, preventing them from entering the cycling phase, and that Mysm1’s deletion drives HSC from quiescence to rapid cell cycling.
Loss of Mysm1 drives HSC from quiescence to rapid proliferation. (A-B) Quiescence of HSC was evaluated with Hoechst 33258/Pyronin Y staining in the BM of WT and Mysm1−/− mice. WT or Mysm1−/− BM cells were stained for HSC surface antigens followed by Hoechst 33258/Pyronin Y staining. Representative FACS plots (left) of cells depicting G0 (bottom left quadrant), G1 (top left quadrant), and S/G2/M (top right quadrant) in (A) LSK cells, and (B) LSK subsets. Bar graph (right) shows the percentage of cells in G0 phase for each individual subpopulation. (C) WT, Mysm1−/− mice received 2 mg of BrdU intraperitoneally daily for 5 days. Incorporation of BrdU was analyzed by FACS in BM LSK and its subsets. (D) FACS plots of cell cycle kinetics of WT and Mysm1−/− LSK cells. (E) Cell-cycle analysis in CD150+CD48−LSK and CD34−Flt3−LSK cells. (F) Real-time PCR analysis of cyclins, cyclin-dependent kinases (Cdks), and Cdk inhibitors in LSK cells. The Ct values are normalized to glyceraldehydes-3-phosphate dehydrogenase and are presented relative to WT (value set as 1). (A-F) Data are representative of three independent experiments. *P < .05; **P < .01.
Loss of Mysm1 drives HSC from quiescence to rapid proliferation. (A-B) Quiescence of HSC was evaluated with Hoechst 33258/Pyronin Y staining in the BM of WT and Mysm1−/− mice. WT or Mysm1−/− BM cells were stained for HSC surface antigens followed by Hoechst 33258/Pyronin Y staining. Representative FACS plots (left) of cells depicting G0 (bottom left quadrant), G1 (top left quadrant), and S/G2/M (top right quadrant) in (A) LSK cells, and (B) LSK subsets. Bar graph (right) shows the percentage of cells in G0 phase for each individual subpopulation. (C) WT, Mysm1−/− mice received 2 mg of BrdU intraperitoneally daily for 5 days. Incorporation of BrdU was analyzed by FACS in BM LSK and its subsets. (D) FACS plots of cell cycle kinetics of WT and Mysm1−/− LSK cells. (E) Cell-cycle analysis in CD150+CD48−LSK and CD34−Flt3−LSK cells. (F) Real-time PCR analysis of cyclins, cyclin-dependent kinases (Cdks), and Cdk inhibitors in LSK cells. The Ct values are normalized to glyceraldehydes-3-phosphate dehydrogenase and are presented relative to WT (value set as 1). (A-F) Data are representative of three independent experiments. *P < .05; **P < .01.
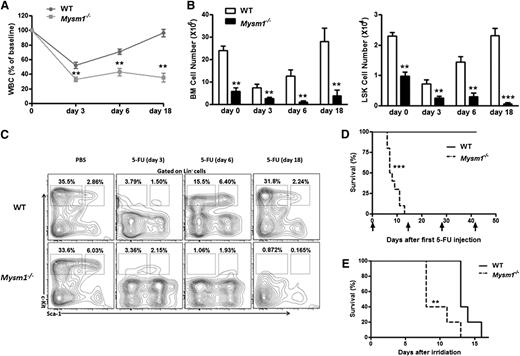
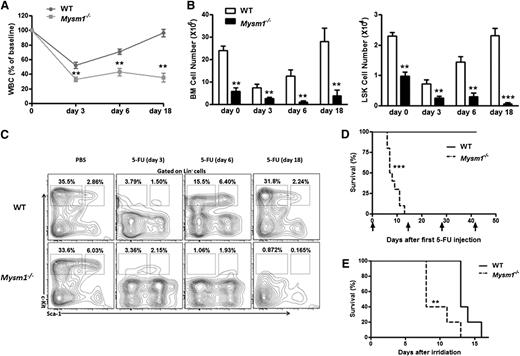
To test whether Mysm1−/− HSCs are likely more actively cycling and thus resulting in an exhaustion of the quiescent stem cell reservoir, we used 5-fluorouracil (5-FU), a chemical sensitive to cycling cells but not to the quiescent HSC. We administered a single dose of 5-FU (75 mg/kg intraperitoneally) to 8-week-old WT and Mysm1−/− mice and performed serial peripheral bleeds to monitor for leucopenia. Compared with WT controls, severe leucopenia was observed in 5-FU−treated Mysm1−/− mice (Figure 3A). Although the BM cells and LSK cells from WT mice exhibited complete recovery 18 days after the administration of 5-FU, cells of 5-FU−treated Mysm1−/− mice remained constantly reduced (Figure 3B). Examination by fluorescence-activated cell-sorting (FACS) analysis revealed that in response to 5-FU, LSK cells derived from WT mice exhibited hyper proliferation on day 6, reverted back to normal, and showed complete recovery on day 18; in contrast, cells derived from Mysm1−/− mice were constantly proliferating and exhausting the stem cell reserves, leading to a steady reduction in the LSK and more mature Lin−Sca-1−c-kit+ cell counts (Figure 3C). We repeated the same experiment with 5-FU injections (150 mg/kg intraperitoneally) administered biweekly to 8-week-old WT and Mysm1−/− mice. Interestingly, 100% of Mysm1−/− mice died before the second 5-FU treatment; in contrast, all WT mice survived after 4 cycles of 5-FU injections (Figure 3D). Similarly, when mice were exposed to a lethal dose of 8 Gy of irradiation, all of the Mysm1−/− mice died within 13 days, in contrast to the WT mice, who survived much longer (Figure 3E). Taken together, these results showed that in the absence of Mysm1, HSC proliferate rapidly and exhaust the stem cell reserves, leading to a defective stem cell renewal capacity, BM exhaustion, and early death.
Exhaustion of HSCs in the absence of Mysm1. (A-C) 8-week-old WT and Mysm1−/− mice were injected with a single dose of 5-FU (75 mg/kg intraperitoneally). (A) At each indicated time, peripheral blood and BM were obtained to monitor hematopoiesis recovery. The counts of white blood cells (WBC) are shown as percentages of initial baseline values for each group of mice; n = 5 mice group. (B) Absolute number of BM (left) and LSK cells (right) of WT and Mysm1−/− mice. (C) Distribution of LSK cells in WT and Mysm1−/− BM. (D) Survival outcome of WT and Mysm1−/− mice after sequential 5-FU treatment (150 mg/kg intraperitoneally) (log-rank nonparametric test) and presented as a Kaplan-Meier survival curve; n = 10 mice per group. (E) Survival outcome of WT and Mysm1−/− mice after a lethal dose of 8 Gy of irradiation; n = 5 mice per group. (A-E) Data are representative of two independent experiments. **P < .01; ***P < .001.
Exhaustion of HSCs in the absence of Mysm1. (A-C) 8-week-old WT and Mysm1−/− mice were injected with a single dose of 5-FU (75 mg/kg intraperitoneally). (A) At each indicated time, peripheral blood and BM were obtained to monitor hematopoiesis recovery. The counts of white blood cells (WBC) are shown as percentages of initial baseline values for each group of mice; n = 5 mice group. (B) Absolute number of BM (left) and LSK cells (right) of WT and Mysm1−/− mice. (C) Distribution of LSK cells in WT and Mysm1−/− BM. (D) Survival outcome of WT and Mysm1−/− mice after sequential 5-FU treatment (150 mg/kg intraperitoneally) (log-rank nonparametric test) and presented as a Kaplan-Meier survival curve; n = 10 mice per group. (E) Survival outcome of WT and Mysm1−/− mice after a lethal dose of 8 Gy of irradiation; n = 5 mice per group. (A-E) Data are representative of two independent experiments. **P < .01; ***P < .001.
We then reexamined whether a reduction in the HSC pool size of Mysm1−/− mice is also caused by their increased apoptotic rate, as reported by Nijnik at al.32 Indeed, we observed an average of 22.57% apoptosis in the Mysm1−/− LSK cells, which was more than sixfold greater than that of the WT mice (3.35%; supplemental Figure 4A-C). Strikingly, in the more mature CD34+Flt3+LSK progenitors, we observed >80% cell death, which was consistent with the sharp reduction in both the frequencies and cell numbers of these cells in the Mysm1−/− mice (Figure 1C; supplemental Figure 2C). In addition, our data displayed clear signs of increased apoptosis, DNA damage, and oxidative stress in the Mysm1−/− LSK and Lin− cells, such as the obvious reduction in the levels of antiapoptotic proteins, a clear increase in the levels of proapoptotic proteins, an increase in the activity of caspase 3/7, a reduction in the mitochondrial potential, an increase in the rate of apoptosis postirradiation, upregulation in the levels of reactive oxygen species, and an increase in the levels rH2AX marks (supplemental Figure 4D-H). Taken together, our data suggest that besides controlling the quiescence and driving them into cycling, Mysm1 also balances the rate of apoptosis of HSC and other progenitors for maintenance of the HSC pool size.
To test whether, besides a severe quantitative reduction in HSC, these cells were also affected in their functions, we sorted LSK cells and assessed the colony-forming units on an in vitro differentiating medium. Mysm1−/− LSK cells overall generated very few colonies (supplemental Figure 5A) compared with WT cells and preferentially generated colonies of granulocytes and macrophages and lost their ability to generate erythroid colonies (supplemental Figure 5B-C). Consistent with this finding, we observed greater frequencies of granulocytes and macrophages in the Mysm1-deficient mice (supplemental Figure 5D). We next used competitive transplantation assays to compare the in vivo functions of Mysm1−/− LSK cells. Each groups of lethally irradiated mice (CD45.1) received 1 × 103 sorted LSK cells from WT or Mysm1−/− mice (CD45.2) along with 2 × 105 competitor BM cells (CD45.1). The chimerism was followed in peripheral blood at 4, 8, 12, and 16 weeks after transplantation. Loss of Mysm1 did not affect the homing of HSC after BM transplantation (supplemental Figure 5E) but markedly affected their hematopoietic-reconstitution ability not only in peripheral blood and BM but also in thymus, spleen, and other peripheral tissues (supplemental Figures 5F-I and 6). These data demonstrate that normal HSC activity is critically dependent on Mysm1, and Mysm1-deficient HSC display intrinsic functional defects such as defective differentiation potential and impaired engraftment.
Gfi1 gene expression is regulated by Mysm1 in HSCs
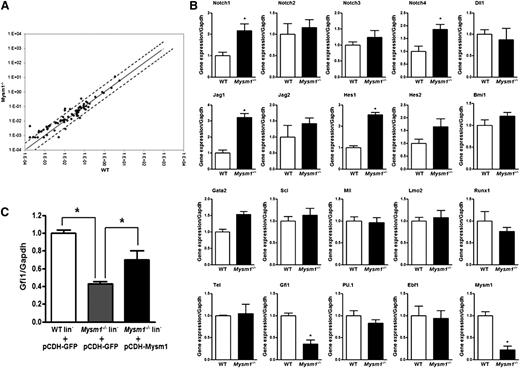
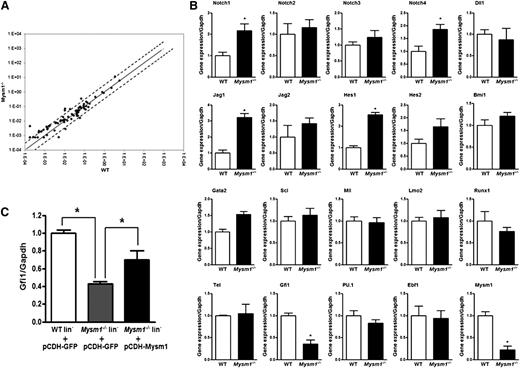
To gain an understanding of how Mysm1 functions in HSC, we performed a PCR array analysis of 84 genes related to the development of blood-cell lineages from HSCs in sorted WT and Mysm1−/− LSK cells. There were relatively limited differences in the gene expression profile between WT and Mysm1−/− LSK cells (Figure 4A). However, we noticed some differential gene expression in some cytokines (Pf4, Csf2), transcription factors (Cbfb, Cebpg), matrix metalloproteinase 9, Notch signaling molecule (Notch4), Wnt signaling molecule (Apc), and surface antigen Cd14 in the Mysm1−/− LSK cells compared with that of WT cells (supplemental Figure 5; supplemental Table 1). Because of limited set of genes profiled on our PCR array, we checked the expression of additional genes associated with HSC maintenance and differentiation by quantitative PCR. Our analysis on selected genes confirmed the increase in mRNA levels of components of Notch-signaling pathway and identified >60% decrease in the mRNA levels of Gfi1 in the Mysm1−/− LSK cells (Figure 4B). Notch is a crucial signaling pathway involved in stem cell maintenance and survival.36 Gfi1 is a zinc finger transcriptional repressor originally recognized for its role in T-cell differentiation and lymphomas.37 Later studies have shown that Gfi1 is also an intrinsic regulator of HSC function.10,38 Loss of Gfi1 causes defective ST-HSC and MPP development, an increase in cycling cells within the HSC pool, and an increase in the apoptotic rate of HSC.9,10,39 Notably, Gfi1−/− HSCs lose the ability to maintain long-term hematopoiesis because of an increase in proliferation and eventual exhaustion of the stem cell pool.9,10 Our analysis here, in which we used Mysm1-deficient HSC, has identified many phenotypes similar to that of Gfi1-deficient HSC. In addition, when Mysm1 was overexpressed in Lin− cells, an increase in the mRNA levels of Gfi1 was detected (Figure 4C). On the basis of these observations coupled with a marked decrease in the mRNA levels of Gfi1 in the Mysm1−/− mice, we hypothesized that Mysm1 may possibly regulate Gfi1 and that its deficiency may be one of many reasons for the defective HSC homeostasis in the Mysm1−/− mice.
Differential gene expression between WT and Mysm1−/− LSK cells. (A) Mouse RT2 Profiler PCR array was performed with sorted LSK cells from WT and Mysm1−/− mice. Solid line indicates no change in gene expression of 1. Dotted lines indicate fold-change (2^(−DCt)) of greater than or equal to 2. (B) Real-time PCR analysis of Notch signaling molecules and important HSC transcriptional factors in WT and Mysm1−/− LSK cells. (C) Sorted LSK cells from WT and Mysm1−/− mice were infected with lentivirus encoding GFP (pCDH-GFP) or Mysm1 (pCDH-Mysm1). Gfi1 expression was assessed 72 hours later by real-time PCR. The Ct values are normalized to glyceraldehydes-3-phosphate dehydrogenase and are presented relative to WT (value set as 1). (B-C) Data are representative of 3 independent experiments. *P < .05.
Differential gene expression between WT and Mysm1−/− LSK cells. (A) Mouse RT2 Profiler PCR array was performed with sorted LSK cells from WT and Mysm1−/− mice. Solid line indicates no change in gene expression of 1. Dotted lines indicate fold-change (2^(−DCt)) of greater than or equal to 2. (B) Real-time PCR analysis of Notch signaling molecules and important HSC transcriptional factors in WT and Mysm1−/− LSK cells. (C) Sorted LSK cells from WT and Mysm1−/− mice were infected with lentivirus encoding GFP (pCDH-GFP) or Mysm1 (pCDH-Mysm1). Gfi1 expression was assessed 72 hours later by real-time PCR. The Ct values are normalized to glyceraldehydes-3-phosphate dehydrogenase and are presented relative to WT (value set as 1). (B-C) Data are representative of 3 independent experiments. *P < .05.
Gfi1 is a direct transcriptional target of Mysm1 and is regulated by Mysm1 and its coordinated action with Gata2 and Runx1
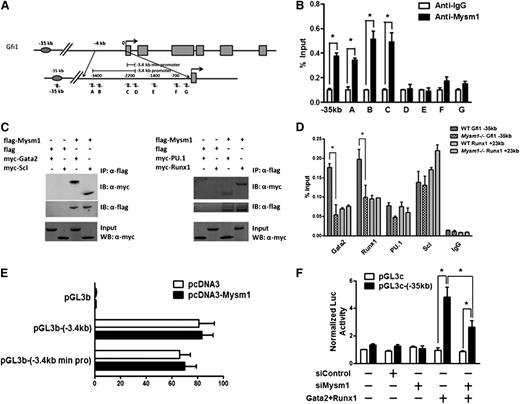
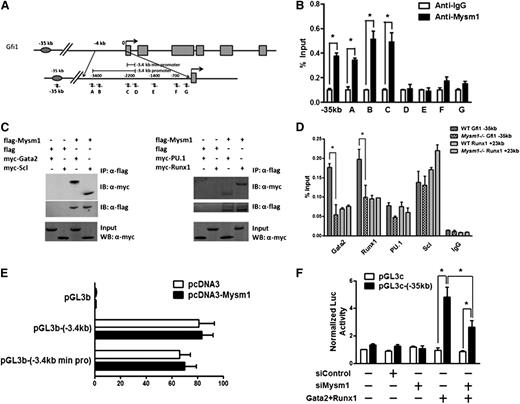
To investigate whether Mysm1 regulates Gfi1 expression by binding to the regulatory elements of Gfi1 in HSC, we performed chromatin immunoprecipitation (ChIP) assays along the Gfi1 regulatory elements. A panel of PCR primers to encompass the −3.4-kb promoter and −35-kb enhancer of the Gfi1 locus was designed (Figure 5A). Immunoprecipitation with the Mysm1-specific antibody, but not with negative-control IgG, enriched the sequences located at the Gfi1 promoter and enhancer regions in WT Lin− progenitors (Figure 5B), demonstrating the direct association of Mysm1 with the Gfi1 regulatory elements.
Gfi1 is a target of Mysm1. (A) Schematic diagram of Gfi1 locus encompassing the promoter and enhancer region; arrows indicate positions of the primers used for ChIP assays. (B) ChIP assays of WT Lin− cells using anti-Mysm1 and anti-IgG. The precipitated DNA was analyzed by real-time PCR using primers along the Gfi1 promoter and enhancer sequence. The relative amount of immunoprecipitated DNA is presented as a percentage of input DNA. (C) In vitro Co-IP experiment in 293T cells. A flag-tagged Mysm1-encoding plasmid was cotransfected with Myc tagged Gata2, Runx1, PU.1, or Scl in 293T cells. Cell lysates were immunoprecipitated using anti-Flag antibody. The immunoprecipitates were examined by western blotting using anti-Myc and anti-Flag antibody. 10% of cell lysates was used as input. (D) ChIP assays in WT and Mysm1−/− Lin− cells using antibodies against Gata2, Runx1, PU.1, Scl, and IgG antibodies. The precipitated DNA was analyzed by real-time PCR using primers along the −35 kb Gfi1 enhancer region. Primers along the +23 kb Runx1 regulatory sequence were used as a positive control. The relative amount of immunoprecipitated DNA is presented as a percentage of input DNA. (E) The −3.4 kb Gfi1 promoter and its minimal core region (−3.4 kb min pro) were cloned into pGL3 basic luciferase vector. Promoter activity was examined in the presence of pcDNA or pcDNA-Mysm1 48 hours after transfection in 293T cells. (F) pGL3-enhancer control (pGL3c) luciferase vector or pGL3c-(−35 kb) luciferase vector which contains the −35 kb Gfi1 enhancer was cotransfected with siRNA control (siControl), siMysm1, or vectors encoding Gata2 and Runx1 into 293T cells. Luciferase activity was recorded 48 hours after transfection. (B,D-F) Data are representative of three independent experiments. (D) Data are representative of two independent experiments. *P < .05.
Gfi1 is a target of Mysm1. (A) Schematic diagram of Gfi1 locus encompassing the promoter and enhancer region; arrows indicate positions of the primers used for ChIP assays. (B) ChIP assays of WT Lin− cells using anti-Mysm1 and anti-IgG. The precipitated DNA was analyzed by real-time PCR using primers along the Gfi1 promoter and enhancer sequence. The relative amount of immunoprecipitated DNA is presented as a percentage of input DNA. (C) In vitro Co-IP experiment in 293T cells. A flag-tagged Mysm1-encoding plasmid was cotransfected with Myc tagged Gata2, Runx1, PU.1, or Scl in 293T cells. Cell lysates were immunoprecipitated using anti-Flag antibody. The immunoprecipitates were examined by western blotting using anti-Myc and anti-Flag antibody. 10% of cell lysates was used as input. (D) ChIP assays in WT and Mysm1−/− Lin− cells using antibodies against Gata2, Runx1, PU.1, Scl, and IgG antibodies. The precipitated DNA was analyzed by real-time PCR using primers along the −35 kb Gfi1 enhancer region. Primers along the +23 kb Runx1 regulatory sequence were used as a positive control. The relative amount of immunoprecipitated DNA is presented as a percentage of input DNA. (E) The −3.4 kb Gfi1 promoter and its minimal core region (−3.4 kb min pro) were cloned into pGL3 basic luciferase vector. Promoter activity was examined in the presence of pcDNA or pcDNA-Mysm1 48 hours after transfection in 293T cells. (F) pGL3-enhancer control (pGL3c) luciferase vector or pGL3c-(−35 kb) luciferase vector which contains the −35 kb Gfi1 enhancer was cotransfected with siRNA control (siControl), siMysm1, or vectors encoding Gata2 and Runx1 into 293T cells. Luciferase activity was recorded 48 hours after transfection. (B,D-F) Data are representative of three independent experiments. (D) Data are representative of two independent experiments. *P < .05.
Previous studies have characterized the enhancer 35 kb upstream of Gfi1 for its activity in the early hematopoietic cells.40 This enhancer is bound and controlled by key HSC regulators, including Scl/Tal1, PU.1/Sfpi1, Runx1, and Gata2.40 We tested whether Mysm1 interacts with these transcriptional factors to regulate Gfi1 enhancer and its expression. We performed coimmunoprecipitation assays in 293T cells and we were surprised to detect direct interactions between Mysm1 and all of these transcriptional factors in 293T cells (Figure 5C). However, when we further examined the association of these transcriptional factors along the Gfi1 regulatory elements in WT and Mysm1−/− Lin− cells by ChIP assays, significant reductions only in the binding of Gata2 and Runx1 and not of Scl and PU.1 to the −35-kb enhancer of Gfi1 locus were observed in Mysm1−/− cells. No differences were detected in transcriptional factor binding in the +23-kb control region of Runx1 locus (Figure 5D). Because no obvious changes in the mRNA expression of Gata2 and Runx1 were found in Mysm1−/− cells (Figure 4C), this reduction of transcription factor binding indicates reduced recruitment of these transcriptional factors to the Gfi1 enhancer in the Mysm1−/− Lin− cells.
To further confirm the direct binding of Mysm1 to Gfi1 regulatory elements and its control of Gfi1 expression, we performed reporter assay in 293T cells. Mysm1 alone did not enhance Gfi1 expression through its promoter region (Figure 5E). However, Gata2 and Runx1 transactivated Gfi1 expression via its −35-kb enhancer, and Mysm1 knockdown significantly reduced this transactivation (Figure 5F). These data, coupled with the interaction of Gata2 and Runx1 with Mysm1, may indicate that Mysm1 regulates Gfi1 expression via interaction and coordination with Gata2 and Runx1. However, the fact that Gata2 and Runx1 association with Gfi1 regulatory elements were reduced in the absence of Mysm1 indicates the existence of a concerted molecular-mechanism between Mysm1 and these transcriptional factors. Although Mysm1 requires the interaction with Gata2 and Runx1 for transactivation of Gfi1, Gata-2 and Runx-1 in turn require Mysm1 for its stable association with the Gfi1 regulatory elements, likely as the result of localized nucleosomic alterations created through its histone-deubiquitinase actity on the Gfi1 locus.
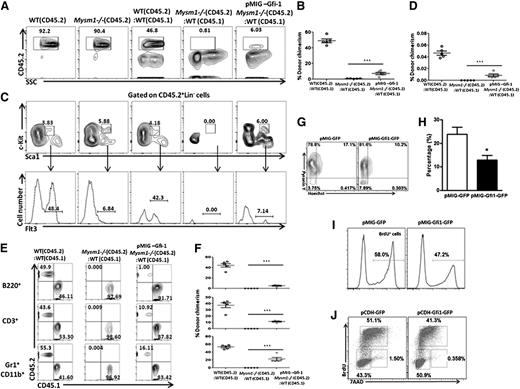
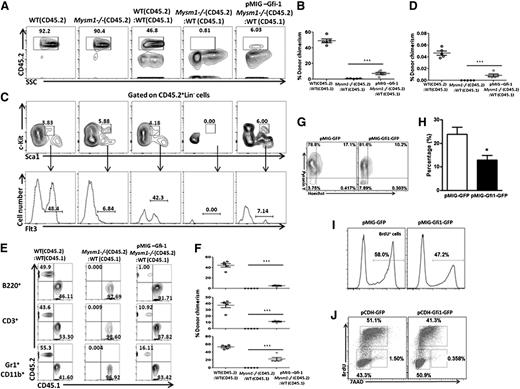
Gfi1 partially restores the function of Mysm1−/− HSC
If this hypothesis was true, forced expression of Gfi1 should restore the impaired function of HSC. To test this, we transplanted LSKFlt3− (CD45.2) cells from WT and Mysm1−/− mice, and LSKFlt3− cells (CD45.2) transduced with Gfi1 (pMIG–Gfi1) from Mysm1−/− mice along with WT CD45.1 BM cells into lethally irradiated CD45.1 recipient mice. Donor chimerism was followed 6 weeks after transplantation. Mysm1−/− donor cells were not detectable after transplantation, whereas the donor Mysm1−/− cells expressing Gfi1 were present, indicating a rescue of engraftment and self-renewal of Mysm1−/− HSC on forced expression of Gfi1 (Figure 6A-B). Not only that, these cells developed and expressed Flt3 in the recipient mice, indicating a rescue of Flt3 levels on forced expression of Gfi1 in contrast to Mysm1−/− cells with defective Gfi1 expression (Figure 6C-D). Furthermore, although Mysm1−/− LSK cells could not reconstitute into different lineages in the recipient mice after transplantation, Mysm1−/− cells expressing Gfi1 were able to do so, indicating that forced Gfi1 expression in the Mysm1−/− cells at least partially restores the function of Mysm1−/− HSC (Figure 6E-F), very likely by protecting against apoptosis and stem cell exhaustion caused by excessive proliferation. We tested this in vitro and consistent to our speculation, the proportion of cells in the G0 phase increased and the apoptotic rate decreased when Mysm1−/− LSK cells were transduced with pMIG-Gfi1-GFP but not with control retroviruses (Figure 6G-H). A lower proliferation rate and a reduction in the proportion of cells in the S phase were also detected in the Gfi1-transduced Mysm1−/− LSK cells compared with GFP-transduced control cells (Figure 6I-J). Collectively, our data demonstrate that Mysm1-mediated Gfi1 transcription is a cell-intrinsic requirement for HSC survival, proliferation, and differentiation.
Forced expression of Gfi1 partially restores the function of Mysm1-deficient HSC. (A-F) 1 × 103 LSKFlt3− cells sorted from WT or Mysm1−/− mice (CD45.2) and Mysm1−/− LSKFlt3− cells transduced with pMIG-Gfi1 were transplanted into lethally irradiated recipients (CD45.1) together with 2 × 105 competitor BM cells (CD45.1). (A) Flow cytometric analyses of WT, Mysm1−/− CD45.2 cells, and donor-derived chimerism (CD45.2) 6 weeks after transplantation in the recipient mice transplanted with WT, Mysm1−/− cells, and Mysm1−/− cells expressed with Gfi1 (pMIG-Gfi1 Mysm1−/−). (B) Percentages of donor-derived cells (CD45.2) in BM. Each dot indicates an individual recipient mouse. (C-D) Flow cytometric analyses and percentages of donor-derived LSKFlt3+ cells in the recipient mice 6 weeks after transplantation. Each dot indicates an individual recipient mouse. (E-F) Flow cytometric analyses and percentages of donor-derived CD45.2 or CD45.1 cells gated on B220+ B cells, CD3+ T cells, and CD11b+Gr1+ myeloid cells in the recipient mice 6 weeks after transplantation. Each dot indicates an individual recipient mouse. (A-F) Data are representative of two independent experiments with n = 5 mice per group in the first experiment and n = 3 mice per group in the second experiment. Shown are means ± SD of 1st experiment with n = 5 mice per group, ***P < .001. (G-J) Sorted Mysm1−/− LSK cells were infected with retroviral vectors encoding either Gfi1 and GFP (pCDH-Gfi1-GFP) or GFP alone (pCDH-GFP) in vitro. Hoechst 33258/Pyronin Y staining (G), annexin V staining (H), BrdU incorporation assay (I) or cell-cycle analysis (J) were performed 48 hours after infection to monitor G0 phase, apoptosis, proliferation, and cell-cycle profile, respectively. (G-J) Data are representative of three independent experiments, *P < .05.
Forced expression of Gfi1 partially restores the function of Mysm1-deficient HSC. (A-F) 1 × 103 LSKFlt3− cells sorted from WT or Mysm1−/− mice (CD45.2) and Mysm1−/− LSKFlt3− cells transduced with pMIG-Gfi1 were transplanted into lethally irradiated recipients (CD45.1) together with 2 × 105 competitor BM cells (CD45.1). (A) Flow cytometric analyses of WT, Mysm1−/− CD45.2 cells, and donor-derived chimerism (CD45.2) 6 weeks after transplantation in the recipient mice transplanted with WT, Mysm1−/− cells, and Mysm1−/− cells expressed with Gfi1 (pMIG-Gfi1 Mysm1−/−). (B) Percentages of donor-derived cells (CD45.2) in BM. Each dot indicates an individual recipient mouse. (C-D) Flow cytometric analyses and percentages of donor-derived LSKFlt3+ cells in the recipient mice 6 weeks after transplantation. Each dot indicates an individual recipient mouse. (E-F) Flow cytometric analyses and percentages of donor-derived CD45.2 or CD45.1 cells gated on B220+ B cells, CD3+ T cells, and CD11b+Gr1+ myeloid cells in the recipient mice 6 weeks after transplantation. Each dot indicates an individual recipient mouse. (A-F) Data are representative of two independent experiments with n = 5 mice per group in the first experiment and n = 3 mice per group in the second experiment. Shown are means ± SD of 1st experiment with n = 5 mice per group, ***P < .001. (G-J) Sorted Mysm1−/− LSK cells were infected with retroviral vectors encoding either Gfi1 and GFP (pCDH-Gfi1-GFP) or GFP alone (pCDH-GFP) in vitro. Hoechst 33258/Pyronin Y staining (G), annexin V staining (H), BrdU incorporation assay (I) or cell-cycle analysis (J) were performed 48 hours after infection to monitor G0 phase, apoptosis, proliferation, and cell-cycle profile, respectively. (G-J) Data are representative of three independent experiments, *P < .05.
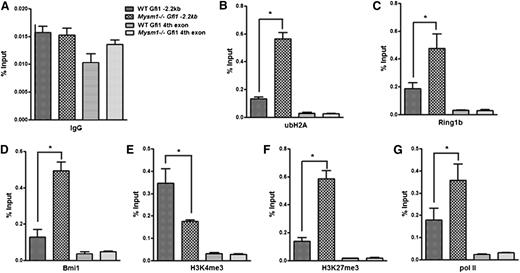
Mysm1 orchestrates histone modifications at the Gfi1 locus
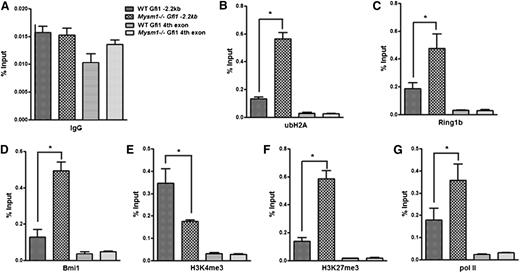
To investigate how Mysm1 activates Gfi1 transcription, we first examined the levels of ubH2A at the Gfi1 enhancer. Our data showed that ubH2A levels, as well as the recruitment of histone-ubiquitinases Ring1b and Bmi1, was increased in the Gfi1 −3.4-kb region of Mysm1−/− Lin− progenitors compared with that of the WT cells (Figure 7A-D). We then analyzed additional histone modifications at the Gfi1 locus that may be impacted by loss of Mysm1 in the Lin− progenitors. ChIP results showed that although the permissive H3K4me3 levels were decreased, the repressive H3K27me3 levels were significantly increased at the Gfi1 enhancer region of Mysm1−/− Lin− progenitors (Figure 7E-F). Notably, the histone modifications were enriched in the same region in which ubH2A, Ring1b, and Bmi1 were enriched, along the Gfi1 promoter locus. Interestingly, the recruitment of paused RNA polymerase (pol) II was also increased in Mysm1−/− Lin− progenitors (Figure 7G). This finding indicates that Gfi1 gene is poised for future activation in the absence of Mysm1-mediated deubiquitinatinase activity at its enhancer region. Collectively, these data indicate that Mysm1 orchestrates histone modifications and facilitates the recruitment of critical transcription factors to the Gfi1 locus. Mysm1 deficiency results in repressive histone modifications and impaired recruitment of key transcription factors causing an inhibition of Gfi1transcription activation.
Mysm1 orchestrates histone modifications at the Gfi1 locus. ChIP assays were performed in WT and Mysm1−/− Lin− cells using (A) anti-IgG, (B) anti-ubH2A, (C) anti-Ring1b, (D) anti-Bmi1, (E) anti-H3K4me3, (F) anti-H3K27me3, and (G) anti-RNA pol II antibodies. The precipitated DNA was analyzed by real-time PCR using primers along the Gfi1 promoter region (primer set C in Figure 5A). Primer amplifying the coding region (fourth exon) of Gfi1 was used as a negative control. The relative amount of immunoprecipitated DNA was presented as a percentage of input DNA. Data are representative of three independent experiments. *P < .05.
Mysm1 orchestrates histone modifications at the Gfi1 locus. ChIP assays were performed in WT and Mysm1−/− Lin− cells using (A) anti-IgG, (B) anti-ubH2A, (C) anti-Ring1b, (D) anti-Bmi1, (E) anti-H3K4me3, (F) anti-H3K27me3, and (G) anti-RNA pol II antibodies. The precipitated DNA was analyzed by real-time PCR using primers along the Gfi1 promoter region (primer set C in Figure 5A). Primer amplifying the coding region (fourth exon) of Gfi1 was used as a negative control. The relative amount of immunoprecipitated DNA was presented as a percentage of input DNA. Data are representative of three independent experiments. *P < .05.
Discussion
Nijnik et al32 recently reported a role for Mysm1 in BM hematopoiesis with an H2A-specific, deubiquitinase−targeted mouse line Mysm1tm1a/tm1a. In this study, we investigated the role of Mysm1, specifically in HSC homeostasis, in a more detailed-manner and found the impairment in Mysm1-controlled Gfi1 regulation as one possible cause for the defective HSC function in Mysm1-deficient mice.
Nijnik et al32 showed an increased proportion of LSK subsets in the total and Lin− BM cells, which also was evident in our results. However, inconsistent with Nijnik et al’s observation that there was no obvious change in the absolute numbers between WT and Mysmtm1a/tm1a LSK cells, we found a significant reduction in Mysm1−/− LSK cells; this reduction was most conspicuous in the more mature CD150−CD48+ or CD34+Flt3+LSK cells in our mouse line. This finding may be attributable to variations in the genetic background of mice or different ages of mice used in experiments. Both Nijnik et al and our group detected an increased apoptosis in the Mysm1−/− LSK cells and its subsets; this finding was regarded as a consequence of elevated genetic instability and oxidative stress. In addition to this finding, we found that Mysm1−/− LSK cells were more sensitive to external stress, such as irradiation and deregulation of Gfi1 and its downstream targets such as bax,41 may have contributed to this abnormality in Mysm1−/− HSC. Not only that, our data provide more rigorous evidence for the defects observed in HSC quiescence, self-renewal, and maintenance of its pool size in the Mysm1−/− mice. On the basis of our experimental evidences and phenotypic similarities between Mysm1−/− and Gfi1−/− mice, we also present here a mechanism of Gfi1 repression as a possible cause for the defective HSC biology in the Mysm1−/− mice.
Quiescence is critical for the maintenance, survival, and self-renewal of HSCs. Published studies of mice deficient in p21, p53, Gfi1, PTEN, Foxos, Pbx1, Mi2β, TSC1, PML, or Fbw7 have shown that a loss of quiescence and unscheduled HSC proliferation results in the loss of self-renewal ability or stem cell exhaustion.2,42,43 We found that Mysm1-deficient HSC had hyperproliferative properties because of their inability to remain quiescent and, unlike c-Cbl−deficient mice,44 Erg1-deficient mice,45 and Itch-deficient mice,34 in which HSC continuous and excessive proliferation resulted in a larger HSC pool but without “stem cell exhaustion,” Mysm1-deficient mice had a reduced pool of HSC and failed to recover in response to stresses such as 5-FU and irradiation because of premature proliferative exhaustion. Importantly, Mysm1-deficient HSC was incompetent in repopulating the hematopoietic system in competitive BM transplantation assays, indicating that the self-renewal of these HSC was impaired.
HSC quiescence, self-renewal, and differentiation are well coordinated by transcription factors,33,42 of which the zinc finger protein Gfi1 is one example. Gfi1 was regarded as one of the most critical regulators in maintaining functional HSC.9,10,38 In searching for the molecular role of Mysm1 in HSC using gene expression analysis, we identified Gfi1 as a possible target of Mysm1. Importantly, Gfi1 rescued the defective function of Mysm1−/− HSC, at least to a partial level. In addition to our experimental evidence that proves the Gfi1 regulation by Mysm1, we noticed many phenotypic similarities between Mysm1- and Gfi1-deficient HSC. In both Gfi1−/− and Mysm1−/− mice, the absolute numbers of LSK cells and Flt3-positive progenitors were significantly reduced, HSC were unable to maintain quiescence, and increased apoptosis was evident in HSCs. Moreover, both deficient mice failed in long-term hematopoiesis.9,10,38 Our further mechanistic investigation revealed the concerted action between Mysm1, Gata2, and Runx1 at the Gfi1 enhancer element to promote its transcription. Mysm1 association with the Gfi1 locus correlated with active histone modifications.
Collectively, these results clearly illustrated that Mysm1 regulates Gfi1 transcription by orchestrating histone modifications and transcription factor recruitment in/to the Gfi1 locus. Although Gfi1 repression may be one possible reason for the defective HSCs in Mysm1−/− mice, one cannot exclude the Mysm1-dependent contribution of other genes in this function, such as the signaling molecules and other cytokine genes identified through our gene-expression analysis. Further studies are needed to elucidate the complete molecular mechanism associated with the Mysm1 function in HSC. Nevertheless, our study elaborates on the initial reports of Nijnik et al32 by numerating the critical role of Mysm1 in HSC homeostasis and illustrates a possible mechanism associated with it.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Y. Chou, L.-F. Wang, H. Won, P. Yates, Q. Nguyen, and other members of the S.-Y. Chen. laboratory for valuable assistance and suggestions.
This work was supported by grants from the National Institutes of Health, Research Grant Program [R01CA090427], National Institute of Allergy and Infectious Diseases [AI084811 and AI068472], and National Cancer Institute [CA116677] to S.-Y.C.; and National Cancer Institute [CA100841] and National Institute of Allergy and Infectious Diseases [AI08185] to X.F.H.
Authorship
Contribution: T.W. and V.N. designed and performed experiments, analyzed data, and wrote the manuscript; X.-X.J., L.J., A.-G.Y., and X.F.H. performed experiments and analyzed data; and S.-Y.C. designed experiments, supervised research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Si-Yi Chen, Keck School of Medicine, University of Southern California, 1450 Biggy St, Los Angeles, CA 90033; e-mail: siyichen@usc.edu.
References
Author notes
T.W. and V.N. contributed equally to this study.