To the editor:
Chronic lymphocytic leukemia (CLL) may be driven by antigen recognition through the B-cell receptor (BCR).1-7 A recent paper in Nature8 suggested a new mechanism for such antigenic drive by functionally characterizing an epitope previously identified by our group as being recognized by virtually all CLL BCRs within a large cohort of CLL patients.9 This epitope has now been shown to be part of the CLL BCR itself, thus conferring apparently autonomous signaling of BCRs within the cell membrane that may promote growth and survival of the leukemic cell. This provides an entirely new view on CLL pathobiology and its mechanism of disease that appears to apply to the majority of CLL patients.8
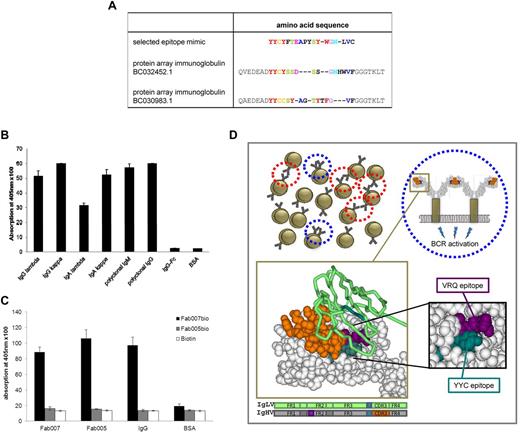
We describe herein an additional epitope involved in self-recognition of CLL BCRs. We screened random phage display peptide libraries for peptides specifically binding to the CLL BCR (expressed as a Fab fragment) of a randomly chosen patient (Fab007). We identified peptides mimicking the epitope recognized by Fab007 and validated their strong binding to this BCR (data not shown). The consensus amino acid sequence of this epitope (YYC) is homologous to Ig heavy and light chains of different gene families used by Igs of the IgG, IgM, IgA, IgE, and IgD isotype, including the respective CLL Fab007 itself (data not shown). Binding of the CLL Fab007 to the Fab part of Igs was verified in a protein array (Figure 1A) and in ELISAs (Figure 1B). Further binding assays indicated that CLL Fab007 recognizes itself and other CLL BCRs even in patients (eg, CLL Fab005) in whom the recently described epitope VRQ8 is not present (Figure 1C). A large binding study on primary CLL samples indicated that approximately 50% of CLL cases interact with the YYC motif, most of them showing also considerable reactivity with the VRQ motif,9 suggesting redundant recognition of alternative epitopes. Interestingly, our new epitope, which is located in framework region 3 of the variable part of Igs, is sterically adjacent and builds a structural continuum with the VRQ epitope located in framework region 2 that was recently suggested as driving autonomous BCR signaling in most CLL patients8 (Figure 1D bottom right panel). This remarkable colocalization of different epitopes mediating self-recognition of CLL BCRs may explain how autonomous signaling can occur even at a single-cell level8 (Figure 1D blue circles) and not just in the context of 1 CLL cell recognizing receptors in adjacent cells (Figure 1D red circles). This is because the region containing both described epitopes is (1) structurally exposed on the surface of the protein and (2) approximately the same distance from the cell surface as the CDR3 region mediating this autorecognition (Figure 1D). This allows interaction of adjacent BCRs within the membrane of the same CLL cell. Our data support the recently established theory of autostimulatory mechanisms in CLL pathobiology and point out redundant recognition profiles and structural explanations that provide the basis for self-recognition of the BCR and self-antigen binding despite the low-affinity typically attributed to CLL BCRs.
BCR self-recognition in CLLviaalternative epitopes. (A) Alignment of the insert sequence of the selected epitope mimicking phage YYCYFTEAPYSYWGNLVC with 2 species of Ig identified by protein array screening. Fab007 bound specifically to 2 distinct Igs, BC032452.1 and BC030983.1, on a protein array (ProtoArray human protein microarray; Invitrogen). Sequence homology of the phage insert sequence YYCYFTEAPYSYWGNLVC selected on Fab007 with an epitope within the variable region of Igs BC032452.1 and BC030983.1 is shown. Homologous sequences (single-letter amino acid code) are colored. (B) CLL BCR Fab007 binds to immobilized Igs of the IgG, IgA, and IgM isotype. All Igs and control BSA were coated and incubated with Fab007. Binding of Fab007 was detected by ELISA with an HRP-conjugated anti–His-tag antibody. Data are shown as means from triplicate experiments (± SEM). Sources of Igs: monoclonal Igs of the IgG (κ and lambda) and IgA (κ and lambda) isotype were purified from the sera of multiple myeloma patients by protein-A chromatography for IgG and jacalin chromatography for IgA. Polyclonal IgG and IgM were from Octapharma or USB products, respectively; the Fc fragment of human IgG1 (IgG-Fc) was from R&D Systems (P-Selectin/Fc-gamma, #137-PS). (C) CLL BCR Fab007 displays “self-reactivity.” Fab007 was labeled with biotin and tested for its binding to immobilized unlabeled receptor in an ELISA assay. Bound biotinylated Fab007 (Fab007bio) was detected using alkaline phosphatase–conjugated avidin. Unrelated Fab005 was used as a control (note that this BCR contained the YYC epitope presented herein but not the VRQ epitope described in Dühren-von Minden et al8 ). Data are shown as means from triplicate experiments (± SEM). (D) Illustration of the structural basis of potential autostimulatory mechanisms in CLL. Red circles mark potential interactions between BCRs on 2 different CLL cells; blue circles mark potential interactions between BCRs on the same CLL cell. The bottom panels show the structure of Ig heavy chains (gray space-filling model) and Ig light chains (green tube model) with the spatially related VRQ epitope (depicted in violet) and YYC epitope (depicted in dark green). The heavy chain CDR3 region is depicted in orange. Below, the exact positions of both epitopes are shown within the framework regions of Ig heavy (IgHV) and Ig light (IgLV) chains (VRQ epitope in violet; YYC epitope in dark green). For the structural Ig model, a random crystallized Fab fragment from the RCSB database (www.rcsb.org) was chosen (identifier 3GBM).
BCR self-recognition in CLLviaalternative epitopes. (A) Alignment of the insert sequence of the selected epitope mimicking phage YYCYFTEAPYSYWGNLVC with 2 species of Ig identified by protein array screening. Fab007 bound specifically to 2 distinct Igs, BC032452.1 and BC030983.1, on a protein array (ProtoArray human protein microarray; Invitrogen). Sequence homology of the phage insert sequence YYCYFTEAPYSYWGNLVC selected on Fab007 with an epitope within the variable region of Igs BC032452.1 and BC030983.1 is shown. Homologous sequences (single-letter amino acid code) are colored. (B) CLL BCR Fab007 binds to immobilized Igs of the IgG, IgA, and IgM isotype. All Igs and control BSA were coated and incubated with Fab007. Binding of Fab007 was detected by ELISA with an HRP-conjugated anti–His-tag antibody. Data are shown as means from triplicate experiments (± SEM). Sources of Igs: monoclonal Igs of the IgG (κ and lambda) and IgA (κ and lambda) isotype were purified from the sera of multiple myeloma patients by protein-A chromatography for IgG and jacalin chromatography for IgA. Polyclonal IgG and IgM were from Octapharma or USB products, respectively; the Fc fragment of human IgG1 (IgG-Fc) was from R&D Systems (P-Selectin/Fc-gamma, #137-PS). (C) CLL BCR Fab007 displays “self-reactivity.” Fab007 was labeled with biotin and tested for its binding to immobilized unlabeled receptor in an ELISA assay. Bound biotinylated Fab007 (Fab007bio) was detected using alkaline phosphatase–conjugated avidin. Unrelated Fab005 was used as a control (note that this BCR contained the YYC epitope presented herein but not the VRQ epitope described in Dühren-von Minden et al8 ). Data are shown as means from triplicate experiments (± SEM). (D) Illustration of the structural basis of potential autostimulatory mechanisms in CLL. Red circles mark potential interactions between BCRs on 2 different CLL cells; blue circles mark potential interactions between BCRs on the same CLL cell. The bottom panels show the structure of Ig heavy chains (gray space-filling model) and Ig light chains (green tube model) with the spatially related VRQ epitope (depicted in violet) and YYC epitope (depicted in dark green). The heavy chain CDR3 region is depicted in orange. Below, the exact positions of both epitopes are shown within the framework regions of Ig heavy (IgHV) and Ig light (IgLV) chains (VRQ epitope in violet; YYC epitope in dark green). For the structural Ig model, a random crystallized Fab fragment from the RCSB database (www.rcsb.org) was chosen (identifier 3GBM).
Authorship
Acknowledgments: The authors thank Dr Nicholas Chiorazzi and Dr Charles Chu (The Feinstein Institute for Medical Research, Manhasset, NY) for kindly supplying BCRs as recombinant proteins, and Barbara Gösch, Mareile Cordes, and Janina Rahlff for expert technical assistance. This work was supported by the José Carreras Leukemia Foundation (grant R10/36f to M.T. and M.B.) and the German Research Foundation (Deutsche Forschungsgemeinschaft, grant TR448/6-1 to M.T.).
Contribution: M.B. and M.T. designed the study; M.B., F.M., M.F., C.W., and F.S. acquired the data; M.B., F.M., B.L., H.V., R.M., and M.T. analyzed and interpreted the data; and M.B. and M.T. wrote the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin Trepel, Department of Oncology and Hematology, Hubertus Wald Cancer Center, University Medical Center Hamburg- Eppendorf, Martinistr 52, 20246 Hamburg, Germany; e-mail: m.trepel@uke.de.