In this issue of Blood, Pozzi et al demonstrate that removing an anionic cage promotes exposure of R169 thereby generating a protein C (PC) that is far more readily activated.1
In the future, it may be possible to deliver anticoagulant function at an injury site with less reliance on cofactor-assisted processes. Physiologically, zymogen PC is subjected to thrombin catalyzed hydrolysis in the presence of thrombomodulin. The resultant PC serves as a serine protease and inactivates Factors Va and VIIIa, 2 key members of the coagulation cascade.2 In addition to its anticoagulant roles, activated PC also exhibits cytoprotective, anti-inflammatory, and profibrinolytic features.2 The mutants described by Pozzi and colleagues may be used to design improved forms of therapeutic PC species whose activation properties can be controlled.
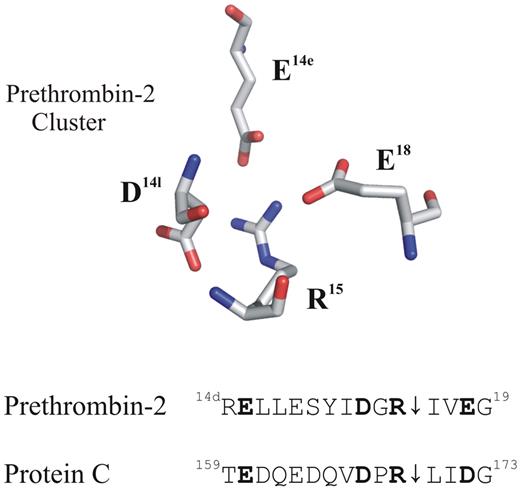
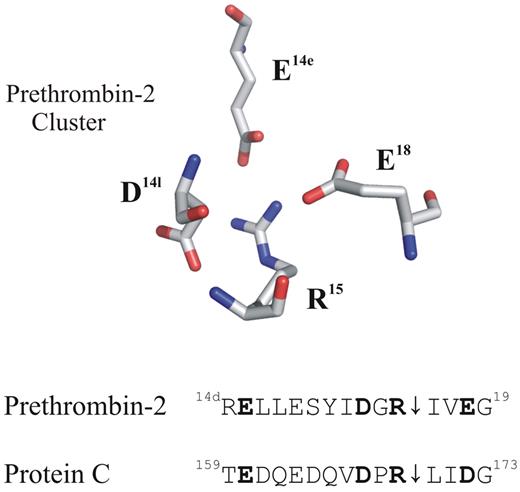
Putative anionic clusters surround the activation regions of prethrombin-2 and protein C (PC). The acidic residues D14l, E14e, and E18 are positioned around the proteolytically sensitive R15 of prethrombin-2 (PDB: 3SQE). PC has comparable residues at E160, D167, and D172 that may also function as an anionic cluster around the reactive R169. For these proteins, cleavage at the 15R-I16 (prethrombin-2) or the 169R-L170 (PC) peptide bond is an important part of the activation process.
Putative anionic clusters surround the activation regions of prethrombin-2 and protein C (PC). The acidic residues D14l, E14e, and E18 are positioned around the proteolytically sensitive R15 of prethrombin-2 (PDB: 3SQE). PC has comparable residues at E160, D167, and D172 that may also function as an anionic cluster around the reactive R169. For these proteins, cleavage at the 15R-I16 (prethrombin-2) or the 169R-L170 (PC) peptide bond is an important part of the activation process.
PC is part of the trypsin-like protease family. It is converted to a mature protease after cleavage at the R169-L170 (P1-P1′) peptide bond by thrombin. A new N-terminus is created that reorients its position within the active site region and helps to establish the oxyanion hole and the primary substrate specificity pocket.3 The membrane-bound protein thrombomodulin assists in inducing the conformational changes needed to promote hydrolysis and conversion of the PC into its activated form. Thrombomodulin has been well documented to cause changes to the thrombin structure.4 There are also reports that thrombomodulin plays a role in presenting the appropriate PC conformation for thrombin cleavage.5 Another example of such a cofactor-assisted system is coagulant Factor VIIa that takes advantage of binding interactions with tissue factor to achieve full activation.6
The R169 residue at the P1 position of PC is not freely accessible and needs to become exposed for proteolytic cleavage. Moreover, there have been reports that acidic thrombin residues neighboring the critical R169 promote electrostatic clash thus hindering the availability of R169.7,8 Interestingly, PC mutants containing D167F (P3 position) and D172N (P3′ position) exhibit a 30-fold increase in thrombin-dependent PC activation in the absence of thrombomodulin.5 In a separate report, R169W was shown to generate a PC species that could be activated by chymotrypsin at levels comparable with that of a thrombin-thrombomodulin complex.9 These different studies suggest that selective amino acid substitutions can be used to help expose the R169.
A review of the PC sequence reveals that this protein may contain an anionic cage surrounding the R169 similar to that found in prethrombin-2. The figure displays the acidic residues that are strategically positioned around the R15 of prethrombin-2. Located below this X-ray crystal structure are some sequence alignments for these 2 proteins. Residues E160, D167, and D172 of PC occupy the same positions as E14e, D14l, and E18 of prethrombin-2. An earlier project by Pozzi et al revealed that alanine mutants at the E14e, D14l, and E18 positions generated a prethrombin-2 species that could spontaneously convert to the active serine protease thrombin.10 In this mutant prethrombin-2 environment, the critical R15 residue was becoming exposed for cleavage and subsequent activation. A similar strategy to control activation of PC might be revealed by probing its anionic E160, D167, and D172 network and testing if the R169 residue could be liberated. So far, no X-ray crystal structures have been reported for PC. The current article by Pozzi and colleagues provides the first view of how a portion of PC (162QEDVDPR169) binds to the extended thrombin active site region.1 The VDPR portion makes strong contacts with the thrombin active site region and includes 1 member of the anionic cluster.
An important goal of the current work by Pozzi et al was to express the triple PC mutant E160A/D167A/D172A (EDD) and test its ability to be activated.1 Intriguingly, this PC mutant was found to undergo slow autoactivation over a period of 72 hours. With such an autoactivation system, the delicate balance between coagulation and anticoagulation would still need to be kept in check. Also of great interest from the latest project by Pozzi and colleagues was the observation that the thrombin-dependent activation of the triple mutant PC (EDD) was now 63-fold enhanced over that of wild-type PC. The addition of thrombomodulin only led to a further 3-fold improvement. These results strongly suggest that removal of the putative anionic cage has promoted conformational changes to PC that mimic those of the thrombomodulin effect. Therapeutic PC species could now be designed that were no longer restricted to environments containing this cofactor protein.
The advantages of liberating an R residue from its anionic cage have now been effectively demonstrated using both prethrombin-2 and Protein C. A valuable next step would be to initiate large-scale production of selective therapeutic proteins and examine their contributions to procoagulant and anticoagulant events. Much knowledge has been gathered on the steps involved in zymogen activation. The current studies make us better poised to selectively control enzyme activation for desired therapeutic applications.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■