Abstract
Primitive erythroid (EryP) progenitors are the first cell type specified from the mesoderm late in gastrulation. We used a transgenic reporter to image and purify the earliest blood progenitors and their descendants from developing mouse embryos. EryP progenitors exhibited remarkable proliferative capacity in the yolk sac immediately before the onset of circulation, when these cells comprise nearly half of all cells of the embryo. Global expression profiles generated at 24-hour intervals from embryonic day 7.5 through 2.5 revealed 2 abrupt changes in transcript diversity that coincided with the entry of EryPs into the circulation and with their late maturation and enucleation, respectively. These changes were paralleled by the expression of critical regulatory factors. Experiments designed to test predictions from these data demonstrated that the Wnt-signaling pathway is active in EryP progenitors, which display an aerobic glycolytic profile and the numbers of which are regulated by transforming growth factor-β1 and hypoxia. This is the first transcriptome assembled for a single hematopoietic lineage of the embryo over the course of its differentiation.
Introduction
The regulation of lineage commitment and differentiation of progenitor cells is a fundamental problem in developmental biology. In the postimplantation mammalian embryo, primitive erythroid (EryPs) or red blood cells are the first cell type to be specified from nascent mesoderm late in gastrulation.1 EryPs emerge in great numbers within the “blood islands” of the yolk sac (YS), and constitute the predominant circulating blood cell until a second wave of definitive, enucleated erythrocytes (EryDs) are produced by the fetal liver.2-4 EryPs are crucial for the transition from rapidly growing embryo to fetus: failure in primitive erythropoiesis is uniformly associated with embryonic lethality. In addition to their function in oxygen delivery to cells within the embryo, EryPs are thought to play a critical role in vascular remodeling during development.5,6 The importance of this lineage is underscored by the fact that primitive erythropoiesis is conserved among vertebrate species.7
In the mouse, EryP progenitors are found in the YS between embryonic day 7.5 (E7.25) and E9.0.4 Their numbers decrease abruptly within the next 12 hours, and by E9.5, when embryonic circulation has begun, they can no longer be detected.4 As EryPs circulate, they continue to mature in a stepwise, essentially synchronous fashion.8 These nucleated erythroblasts undergo a series of dramatic cellular and morphologic changes, including up-regulation of embryonic globin genes, expression of cell adhesion proteins, cytoskeletal reorganization, decreased cell proliferation, nuclear condensation, and, finally, from E12.5-E14.5, nuclear extrusion.8-11 The enucleated EryPs are rapidly outnumbered by adult-type erythrocytes, but remain in the circulation through the end of gestation (our unpublished data and Fraser et al8 ). The transient appearance of EryP progenitors in the embryo and the synchronous, stepwise maturation of their progeny make this lineage an attractive model for cell specification and terminal differentiation.
Early hematopoietic cells remain poorly characterized because of the relative inaccessibility and small size of the embryo, the transient appearance of their progenitors, and the lack of suitable cell surface markers for their isolation. Detailed chronologic expression profiling will be essential for an understanding of the genetic networks that regulate the development and differentiation of the EryP lineage and for comparative analyses of embryonic versus adult erythropoiesis. We have developed a transgenic mouse system in which a nuclear green fluorescent protein (GFP) reporter is expressed specifically in EryPs, allowing the tracking of these cells and their nuclei throughout gestation.11,12 In the present study, we show that the expression of this transgene can be used to mark and prospectively isolate the earliest hematopoietic progenitors of the mouse embryo from their first appearance at ∼E7.5. To identify the processes necessary for commitment, expansion, maturation, and terminal differentiation of progenitors for this first hematopoietic lineage, a global transcriptional analysis was performed for successive stages of development from E7.5-E12.5, and revealed not only well-studied red blood cell genes but also some surprises. Experiments were designed to test predictions based on the expression profiles. We show that the Wnt pathway functions autonomously in EryP progenitors, the numbers of which are regulated by transforming growth factorβ1 (TGFβ1) and hypoxia. Interestingly, EryP progenitors express genes associated with aerobic glucose metabolism (the Warburg effect), a phenotype characteristic of cancer and other rapidly proliferating cells. This study is the first comprehensive, genome-wide expression profiling undertaken for a single lineage from the gastrulating embryo and will provide a valuable resource for understanding early hematopoietic development.
Methods
Detailed experimental procedures are described in the supplemental Materials (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This study was approved by the Mount Sinai School of Medicine institutional animal care and use committee.
Mouse lines and embryo dissection
Primary microarray data acquisition and analyses
The labeled cRNA samples were hybridized to Illumina Mouse WG-6 v1.1 Expression BeadChip genome-wide arrays. The data files generated by the EryP array analyses have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24127) for use by other investigators as accession code GSE24127. The expression level cutoffs were set at 7.2 for E7.5- and E8.5-amplified samples and at 6.0 for E8.5-E12.5 samples (all log2 scale).
Flow cytometry and cell sorting
Cell suspensions were sorted based on GFP fluorescence using an Influx (Cytopeia) or MoFlo (Dako-Cytomation) cell sorter, with dead cells excluded by propidium iodine staining. For multicolor cell sorting, staining with 6-diamidino-2-phenylindole was used to exclude dead cells, and surface fluorescence was analyzed using an LSR II flow cytometer (Becton Dickinson). Data analysis was performed using FlowJo software 8.8.6 (TreeStar). Monoclonal antibodies used in this study are listed in supplemental Table 1.
qRT-PCR analysis and transcript verification
RNA from sorted cell populations was reverse transcribed to cDNA using Superscript III reverse transcriptase and oligo dT(20), as described previously.12 Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using either Taqman primers and probes or standard oligonucleotide primers and SYBR Green PCR master mix. Oligonucleotide primers are listed in supplemental Table 2.
Imaging
Images were acquired using an Axiocam MRc or MRm camera (Zeiss), analyzed with the Axiovision software package (Zeiss), and postprocessed using Adobe Photoshop. Whole embryos were imaged on a Lumar V12 stereomicroscope (Zeiss) fitted with a NeoLumar S 1.5× objective. For time-lapse experiments, laser scanning confocal data were acquired using a Zeiss LSM510 META scan head fitted onto a Zeiss Axiovert 200M, with a plan-apochromat 10×/0.45 NA lens. For fluorophore excitation, a 488-nm argon laser line (3% power) was used. Giemsa-stained cytospun cells were imaged using an inverted Zeiss Axiovert 25 microscope fitted with a 32× (LD-A-Plan/0.4 NA) objective.
Immunohistochemistry
Results
The ϵ-globin::H2B-EGFP transgene labels the first committed EryP progenitors in the developing mouse embryo
The identification and isolation of the first hematopoietic (EryP) progenitors has presented challenges because of a lack of specific cell-surface markers. Whereas CD31 (PECAM1) and Tie-2 expression have been used to enrich for EryP progenitors from E7.5 embryos,14 both of these proteins are also expressed on vascular endothelial and definitive hematopoietic progenitors. Similarly, low-level expression of CD41 has been used to enrich for EryP progenitors.15 CD41 is expressed on definitive hematopoietic but not on endothelial progenitors.15,16 We have developed a transgenic mouse model in which expression of a histone H2B-GFP reporter is driven by human ϵ-globin regulatory elements specifically within the EryP lineage; this line has been validated by a variety of criteria and GFP shown to mark essentially all EryPs but not EryDs.11,12 In addition, the same regulatory elements have been used by us for 2 related transgenic mouse lines.8,17-20 Whereas globin gene expression has traditionally been viewed as a late marker of differentiation, embryonic globins are in fact transcribed in the YS from as early as E7.5.17,18 We therefore investigated whether the ϵ-globin::H2B-EGFP transgene is also expressed at this developmental stage and whether it might permit the identification and enrichment of EryP progenitors.
We first determined the time of onset of GFP fluorescence in ϵ-globin::H2B-EGFP embryos (Figure 1A). The first GFP(+) cells (EryP) were evident in the extraembryonic region of late streak/early bud embryos (∼ E7.5; Figure 1A). From the neural stages on, all embryos displayed a ring of GFP(+) cells encircling the YS. Therefore, ϵ-globin::H2B-EGFP transgene expression is activated toward the end of gastrulation, at the time when the first erythroid cells are specified from nascent mesoderm.1 The emergence and expansion of GFP(+) EryPs were visualized using time-lapse confocal microscopy of cultured embryos (Figure 1B and supplemental Video 1). The static and time-lapse images of GFP-expressing cells in the early postgastrulation embryo further confirms the utility of the ϵ-globin::H2B-EGFP line as a model for primitive erythropoiesis.
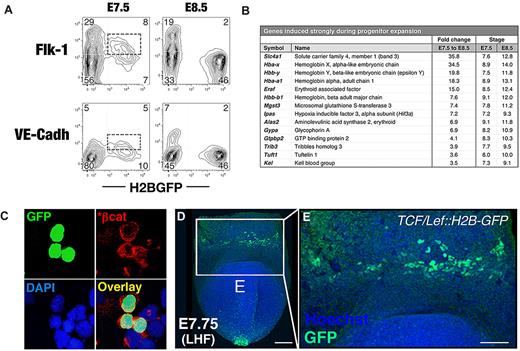
GFP expression from the ϵ-globin::H2B-EGFP transgene marks the primitive erythroid lineage in the blood islands of the YS and can be used to identify and isolate EryP progenitors. (A) GFP expression in ϵ-globin::H2B-EGFP primitive streak to early somite–stage transgenic embryos. To visualize the emergence and expansion of GFP(+) EryP cells, time-lapse videos of cultured ϵ-globin::H2B-EGFP embryos were acquired. GFP(+) cells appear in a narrow band of 3-5 cell diameters in the proximal YS during mid-to-late gastrulation (MS/LS stage). Scale bar, 500 μm. MS, midstreak; LS, late streak; EB, early bud; LHF, late headfold; ss, somite stage; ESom, early somite. (B) Selected snapshots from time-lapse video (supplemental Video 1) of an ϵ-globin::H2B-EGFP embryo cultured in vitro under physiologic conditions from the LS to the ESom stage. Early-bud-stage embryos were imaged over a period of 14 hours, from the time before transgene induction (∼ EB, t = 0) through the ESom stages (t = 860 minutes; supplemental Video 1). Confocal images were acquired as sequential optical x-y sections taken at 4-μm z intervals. Images were taken at 20-minute intervals (total imaging time: 14.5 hours). (C) Flow cytometric histogram profiles of dispersed ϵ-globin::H2B-EGFP transgenic embryos reveals a clearly identifiable GFP(+) population. (D) Cells from whole E7.5 or E8.5 embryos were FACS sorted to GFP(+) and GFP(−) populations. Left panel, Giemsa-stained cytospun cells from FACS sort. Scale bar, 20 μm. EryP-progenitor numbers were measured using a clonogenic assay. Virtually all progenitor activity was recovered in the GFP(+) population. Characteristic EryP colonies (right panels) showed red pigmentation (hemoglobin) and GFP fluorescence. Scale bar, 50 μm. (E) Real-time RT-PCR expression of endogenous embryonic ϵy- and βh1-globin genes in GFP(+) and GFP(−) FACS-sorted cells from E7.5 ϵ-globin::H2B-EGFP transgenic embryos. Expression was normalized to ubiquitin b (Ubb). (F) EryP numbers at the YS stages of development.
GFP expression from the ϵ-globin::H2B-EGFP transgene marks the primitive erythroid lineage in the blood islands of the YS and can be used to identify and isolate EryP progenitors. (A) GFP expression in ϵ-globin::H2B-EGFP primitive streak to early somite–stage transgenic embryos. To visualize the emergence and expansion of GFP(+) EryP cells, time-lapse videos of cultured ϵ-globin::H2B-EGFP embryos were acquired. GFP(+) cells appear in a narrow band of 3-5 cell diameters in the proximal YS during mid-to-late gastrulation (MS/LS stage). Scale bar, 500 μm. MS, midstreak; LS, late streak; EB, early bud; LHF, late headfold; ss, somite stage; ESom, early somite. (B) Selected snapshots from time-lapse video (supplemental Video 1) of an ϵ-globin::H2B-EGFP embryo cultured in vitro under physiologic conditions from the LS to the ESom stage. Early-bud-stage embryos were imaged over a period of 14 hours, from the time before transgene induction (∼ EB, t = 0) through the ESom stages (t = 860 minutes; supplemental Video 1). Confocal images were acquired as sequential optical x-y sections taken at 4-μm z intervals. Images were taken at 20-minute intervals (total imaging time: 14.5 hours). (C) Flow cytometric histogram profiles of dispersed ϵ-globin::H2B-EGFP transgenic embryos reveals a clearly identifiable GFP(+) population. (D) Cells from whole E7.5 or E8.5 embryos were FACS sorted to GFP(+) and GFP(−) populations. Left panel, Giemsa-stained cytospun cells from FACS sort. Scale bar, 20 μm. EryP-progenitor numbers were measured using a clonogenic assay. Virtually all progenitor activity was recovered in the GFP(+) population. Characteristic EryP colonies (right panels) showed red pigmentation (hemoglobin) and GFP fluorescence. Scale bar, 50 μm. (E) Real-time RT-PCR expression of endogenous embryonic ϵy- and βh1-globin genes in GFP(+) and GFP(−) FACS-sorted cells from E7.5 ϵ-globin::H2B-EGFP transgenic embryos. Expression was normalized to ubiquitin b (Ubb). (F) EryP numbers at the YS stages of development.
EryP progenitors are present in the mouse embryo during a narrow developmental window: they first appear at E7.5 and are absent by E9.0.1,4 Because bright GFP expression was easily detected as early as E7.5 in ϵ-globin-H2B-EGFP transgenic embryos, we investigated whether EryP progenitors could be identified by GFP expression. Whole E7.5 or E8.25 ϵ-globin::H2B-EGFP embryos were dispersed and GFP(+) cells were isolated using fluorescence-activated cell sorting (FACS; Figure 1C) and plated in clonogenic assays in methylcellulose. At E7.5 and E8.5, as many as 15% and 48%, respectively, of all cells in the embryo expressed GFP (Figure 1C). The sorted GFP(+) cells (Figure 1D) gave rise to EryP colonies (EryP-CFC; Figure 1D and supplemental Figure 1A) containing green fluorescent erythroblasts. No EryP-CFC formed from the GFP(−) fractions. The number of EryP progenitors per embryo decreased significantly from E7.5-E8.5 and were absent by E9.5 (supplemental Figure 1A and data not shown). GFP(+) cells from E8.5 embryos did not contain progenitors for definitive hematopoiesis (not shown). The endogenous βh1- and ϵ-globin genes were expressed only in the GFP(+) population as early as E7.5 (Figure 1E).
Expansion of the developing EryP lineage is rapid, as indicated by GFP expression (Figure 1A) and cell-doubling times (supplemental Figure 1B). The highest proliferative activity was observed during the 24-hour period when EryPs are first detected in the YS (E7.5) to shortly before the onset of circulation (E8.5), with a > 30-fold increase in cell numbers and a doubling time of ∼ 4 hours (supplemental Figure 1B). By E12.5, when EryP proliferation had essentially ceased, the population had expanded by 30 000-fold (∼ 14-15 cell divisions; supplemental Figure 1B). At present there is no unequivocal method to determine the percentage of GFP(+) cells that are progenitors (ie, give rise to colonies in clonogenic assays). In contrast to the large increase in GFP(+) cell numbers, progenitor numbers increased by only ∼ 3- to 4-fold from E7.5-E8.5 (our estimates from multiple experiments). These differences may indicate that not all of the GFP(+) cells found in the E7.5-E8.5 YS are progenitors. In our best experiments, 25%-30% (E7.5) or 10% (E8.5) of cells plated yielded EryP colonies. However, it is possible that during experimental manipulation (ie, dissection, embryo dispersion to single cells, FACS sorting, and plating in methylcellulose), some cells were lost, damaged, or had begun to differentiate before plating in methylcellulose.
Gene expression profiling of the EryP lineage throughout development
The specific expression of GFP in maturing EryPs and their progenitors allowed us to isolate these cells at distinct stages of differentiation and to define the EryP transcriptome during development. GFP(+) cells were FACS sorted from ϵ-globin::H2B-EGFP embryos at 24-hour intervals during gestation from E7.5-E12.5 (supplemental Figure 2A). Total RNA was extracted from the purified GFP(+) cell populations (3 replicates per stage; supplemental Figure 2A), converted to labeled cRNA, and hybridized to the Illumina Mouse WG-6 BeadChip platform. Because the number of sorted cells obtained from pooled E7.5 embryos was very small (7000-21 000 cells from > 70 embryos per pool), it was necessary to amplify this RNA. RNA was also amplified from pooled E8.5 embryos to permit direct comparison between the E7.5 and E8.5 stages. Thus, 2 complementary EryP datasets were generated: one from amplified samples (E7.5 and E8.5, progenitor stages) and a second from samples that were not amplified (E8.5-E12.5, maturing EryPs). Pairwise comparison between samples revealed high sample-to-sample correlation (> 98% concordance), with tight clustering between replicate samples from the same stage (supplemental Figure 2B-C).
Dynamic gene expression patterns observed during EryP maturation
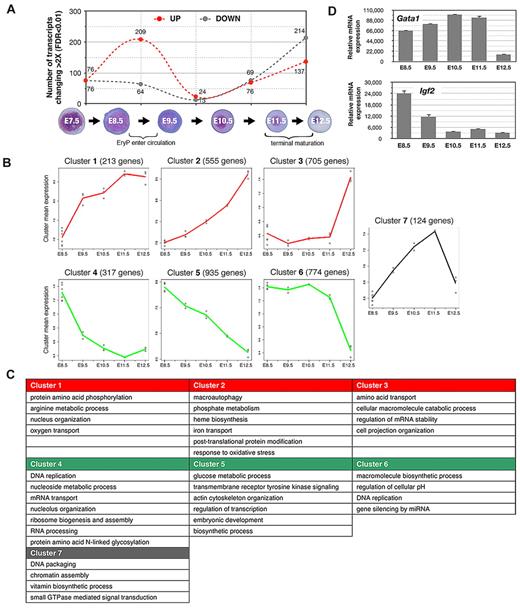
The greatest numbers of increases and decreases in gene expression (> 2-fold) were detected during 2 24-hour windows, from E8.5-E9.5 (transition from YS to circulation) and from E11.5-E12.5 (fetal liver stage; Figure 2A), coinciding with key physiologic changes in the embryo. Interestingly, we observed a nearly reciprocal expression of transmembrane protein genes in E8.5 versus E11.5 EryPs (supplemental Figure 2D), which is suggestive of opposing functions that may reflect the need for these cells to respond to the distinct microenvironmental niches of the YS and circulation.
Global gene expression profiling of the primitive erythroid lineage. Labeled cRNA samples were hybridized to Illumina Mouse WG-6 v1.1 Expression BeadChip genome-wide arrays. Quality control of array data was performed using the Bioconductor lumi R package. The filtered genes were clustered into 7 major patterns using the maSigPro algorithm. We were able to survey a genome-wide probe set representing 46 630 murine transcripts, encompassing the emergence of EryP progenitors in the YS through successive stages of erythroblast differentiation in the circulation. Many of these probes target less well-annotated transcripts or transcript isoforms of known genes. There are 21 174 unique genes in the University of California-Santa Cruz mouse mm9 refseq protein coding gene database, and the Illumina mouse-6 v1.1 microarray used in this study contains probes for 18 970 or 89.6% of the well-annotated refseq mouse genes. Analyses were performed using all probes with Entrez ID annotations found with the lumiMouseAll.db version 1.6.1 annotation package. (A) Changes (increased or decreased) in transcript numbers during consecutive stages of EryP development. The graph represents the total numbers of transcripts showing a change of greater than 2-fold (P < .01). Dotted red line indicates increasing expression; dotted gray line indicates decreasing expression. Peaks in transcription variation were identified during the windows from E8.5-E9.5 (transition from the YS to the circulation stage, 273 transcripts) and from E11.5-E12.5 (fetal liver stage, 351 transcripts). (B) Plot representations of 7 specific clusters of transcripts with similar temporal expression patterns. Clusters were subclassified into 3 groups, representing genes that are progressively up-regulated (clusters 1, 2, and 3, red lines); down-regulated (clusters 4, 5, and 6, green lines); or up-regulated through E11.5 and then down-regulated rapidly over the next 24 hours of development (gray line, cluster 7). The peaks in transcription variation indicated in panel A are especially evident in clusters 1, 4, and 7 (relatively sharp increases or decreases in expression, E8.5-E9.5, corresponding to the transition from the YS to the circulation stage) and in clusters 3, 6, and 7 (abrupt increases or decreases in expression, E11.5-E12.5, corresponding to fetal liver stage, when EryP complete their maturation and enucleate). Each individual point (○) represents the mean gene expression of the cluster genes from one microarray experiment. Each line connects mean values for all replicates. (C) Overrepresented gene ontologies for the clusters shown in panel A. (D) Expression of a representative gene that is up-regulated (Gata1) and one that is down-regulated (Igf2) in the EryP microarray dataset, analyzed using qRT-PCR. Expression levels were normalized relative to ubiquitin b (Ubb).
Global gene expression profiling of the primitive erythroid lineage. Labeled cRNA samples were hybridized to Illumina Mouse WG-6 v1.1 Expression BeadChip genome-wide arrays. Quality control of array data was performed using the Bioconductor lumi R package. The filtered genes were clustered into 7 major patterns using the maSigPro algorithm. We were able to survey a genome-wide probe set representing 46 630 murine transcripts, encompassing the emergence of EryP progenitors in the YS through successive stages of erythroblast differentiation in the circulation. Many of these probes target less well-annotated transcripts or transcript isoforms of known genes. There are 21 174 unique genes in the University of California-Santa Cruz mouse mm9 refseq protein coding gene database, and the Illumina mouse-6 v1.1 microarray used in this study contains probes for 18 970 or 89.6% of the well-annotated refseq mouse genes. Analyses were performed using all probes with Entrez ID annotations found with the lumiMouseAll.db version 1.6.1 annotation package. (A) Changes (increased or decreased) in transcript numbers during consecutive stages of EryP development. The graph represents the total numbers of transcripts showing a change of greater than 2-fold (P < .01). Dotted red line indicates increasing expression; dotted gray line indicates decreasing expression. Peaks in transcription variation were identified during the windows from E8.5-E9.5 (transition from the YS to the circulation stage, 273 transcripts) and from E11.5-E12.5 (fetal liver stage, 351 transcripts). (B) Plot representations of 7 specific clusters of transcripts with similar temporal expression patterns. Clusters were subclassified into 3 groups, representing genes that are progressively up-regulated (clusters 1, 2, and 3, red lines); down-regulated (clusters 4, 5, and 6, green lines); or up-regulated through E11.5 and then down-regulated rapidly over the next 24 hours of development (gray line, cluster 7). The peaks in transcription variation indicated in panel A are especially evident in clusters 1, 4, and 7 (relatively sharp increases or decreases in expression, E8.5-E9.5, corresponding to the transition from the YS to the circulation stage) and in clusters 3, 6, and 7 (abrupt increases or decreases in expression, E11.5-E12.5, corresponding to fetal liver stage, when EryP complete their maturation and enucleate). Each individual point (○) represents the mean gene expression of the cluster genes from one microarray experiment. Each line connects mean values for all replicates. (C) Overrepresented gene ontologies for the clusters shown in panel A. (D) Expression of a representative gene that is up-regulated (Gata1) and one that is down-regulated (Igf2) in the EryP microarray dataset, analyzed using qRT-PCR. Expression levels were normalized relative to ubiquitin b (Ubb).
Hierarchical clustering of the gene expression data were performed to identify functionally related groups of genes that cooperate in biologic processes involved in erythroid cell maturation. Genes clustered into 7 major patterns (Figure 2B). We analyzed Gene Ontology (GO) terms for biologic processes and found that each cluster was enriched for distinct functional classes of genes (Figure 2C). The GO terms that are overrepresented in clusters of genes that are up-regulated during EryP maturation include heme biosynthesis, iron transport, macromolecule catabolism, and autophagy (Figure 2C), all processes associated with erythroid maturation. Conversely, the GO terms overrepresented in clusters containing genes that are down-regulated include DNA replication, ribosome and nucleolus biogenesis, and glucose metabolism, suggesting that early EryPs are heavily engaged in these processes and that these processes decrease in significance as the cells mature. Cluster 7, in which genes are up-regulated through E11.5 and are then rapidly down-regulated, includes genes involved in DNA packaging and chromatin assembly (Figure 2C). The peak at E11.5 may represent a “transition state” through which EryPs pass as they enter the terminal stages of differentiation.
To validate the microarray results, we confirmed the temporal expression pattern of a battery of transcripts detected in the EryP dataset using RT-PCR (supplemental Figure 3C) and/or FACS (supplemental Figure 3D). Representative qRT-PCR analyses of genes (Gata1 and Igf2) whose expression was found to increase or decrease, respectively, during development are shown in Figure 2D. The transcription patterns identified by microarray (not shown) and qRT-PCR analyses (supplemental Figure 3C) were very similar. Additional validation of the microarray data were obtained using FACS analysis of cell surface proteins such as CD31, β1-integrin, endothelial cell-selective adhesion molecule (ESAM), and c-kit data (Figures 4B-C, 5C and supplemental Figure 3D); these were down-regulated on EryPs, as predicted from the transcriptional profiling. Other genes and their translated proteins, such as Transferrin Receptor (TfR; CD71) and Glycophorin A (Gypa; Ter119), were up-regulated (supplemental Figure 3C-D).
Molecular signatures of the earliest EryP progenitors
The earliest hematopoietic progenitors in the developing mouse embryo are thought to arise from a population of mesoderm that transiently expresses the receptor tyrosine kinase Flk-1 (vascular endothelial growth factor [VEGF] receptor-2 or Kdr). Therefore, we investigated whether mesodermal or endothelial genes are expressed in EryP progenitors. Whereas Kdr mRNA was not detected in E7.5 EryPs (supplemental Figure 3A), the protein was detected on the surface of a subset (∼ 50%) of EryP/GFP(+) cells at this stage (Figure 3A), suggesting transcription of the Kdr gene in an earlier precursor. Cdh5, which encodes the endothelial adhesion protein vascular endothelial cell–cadherin (VE-cadherin), was expressed by EryPs at E7.5 and was down-regulated by E8.5. VE-cadherin protein was expressed on a subset of GFP(+) cells at E7.5 and, like Flk-1, was rapidly lost around the time of erythroid commitment (Figure 3A).
Molecular signatures of EryP progenitors. (A) Hemangioblastic marker expression by early EryPs. FACS plots showing expression of Flk-1 and VE-cadherin surface protein in GFP(+) cells from ϵ-globin::H2B-EGFP embryos at E7.5 and E8.5. (B) Selected genes that are activated during EryP progenitor expansion from E7.5-E8.5. Transcripts listed showed a low adjusted P value and median expression level < 9.5 (log2 scale) for triplicates at E7.5 and were up-regulated ≥ 3.5-fold by E8.5. Samples were amplified before hybridization to the microarray. Expression cutoff, 7.2. (C) Expression of activated β-catenin (*β-cat) in EryP at E8.0. Cells from dispersed E8.0 ϵ-globin::H2B-EGFP embryos were cytocentrifuged onto slides and then immunostained. (D-E) Expression of a TCF/Lef::H2B-GFP transgenic reporter for the canonical Wnt-signaling pathway13 in an ∼ E7.75 embryo. (D) Anterior view of late headfold (LHF) stage TCF/Lef::H2B-GFP embryo counterstained with Hoechst to highlight nuclei. The image is a 3D reconstruction of a z-stack and was acquired using a Zeiss LSM 510 microscope outfitted with a plan-apochromat 20×/0.75 NA lens. White box indicates the blood islands of the YS. Scale bar, 100 μm. (E) High-magnification view of the boxed region in panel D. Scale bar, 50 μm.
Molecular signatures of EryP progenitors. (A) Hemangioblastic marker expression by early EryPs. FACS plots showing expression of Flk-1 and VE-cadherin surface protein in GFP(+) cells from ϵ-globin::H2B-EGFP embryos at E7.5 and E8.5. (B) Selected genes that are activated during EryP progenitor expansion from E7.5-E8.5. Transcripts listed showed a low adjusted P value and median expression level < 9.5 (log2 scale) for triplicates at E7.5 and were up-regulated ≥ 3.5-fold by E8.5. Samples were amplified before hybridization to the microarray. Expression cutoff, 7.2. (C) Expression of activated β-catenin (*β-cat) in EryP at E8.0. Cells from dispersed E8.0 ϵ-globin::H2B-EGFP embryos were cytocentrifuged onto slides and then immunostained. (D-E) Expression of a TCF/Lef::H2B-GFP transgenic reporter for the canonical Wnt-signaling pathway13 in an ∼ E7.75 embryo. (D) Anterior view of late headfold (LHF) stage TCF/Lef::H2B-GFP embryo counterstained with Hoechst to highlight nuclei. The image is a 3D reconstruction of a z-stack and was acquired using a Zeiss LSM 510 microscope outfitted with a plan-apochromat 20×/0.75 NA lens. White box indicates the blood islands of the YS. Scale bar, 100 μm. (E) High-magnification view of the boxed region in panel D. Scale bar, 50 μm.
We identified a large group of genes that are expressed at high levels at E7.5 but are down-regulated within the next 24 hours of development (supplemental Figure 3B). These transcriptional decreases are likely required for progenitor expansion and perhaps also for the earliest steps in EryP differentiation. The sharpest decrease was observed for Gata2, which is required for the proliferation and survival of hematopoietic progenitors.21 Down-regulation of other transcription factor genes (Runx1, its coregulator Cbfb, Lmo2, and Etv2) was also detected.
Several Wnt/β-catenin pathway genes (encoding the transcription factor β-catenin, the Wnt receptor Fzd7, and the secreted activator Rspo3) were also expressed in EryPs at E7.5 and were then down-regulated (supplemental Figure 3B and data not shown). We investigated the potential involvement of canonical Wnt activity in EryP progenitors using an antibody against activated (stabilized) β-catenin. As shown in Figure 3C, EryPs (green nuclei) stained with the antibody. That the Wnt pathway is active in these cells is further suggested by the bright fluorescence of an H2B-GFP–transgenic Wnt/β-catenin reporter (TCF/Lef::H2B-GFP)13 in the blood islands of the YS (Figure 3D-E).
Several collagen genes were expressed in E7.5 and E8.5 EryPs (supplemental Figure 3B), suggesting that EryP progenitors and their early descendants secrete components of the extracellular matrix of the YS. Genes encoding cytoskeleton, receptor, and cell-communication proteins were also down-regulated between E7.5 and E8.5, perhaps reflecting the imminent transition of EryPs from the YS into the bloodstream.
The developmental window from E7.5-E8.5 was also associated with substantial increases in gene expression. Most prominent among these were the embryonically expressed ϵY-, ζ-, and α-globin genes (Hbb-y, Hba-x, and Hba-a1, respectively) and the anion transporter band 3/Slc4a1 (Figure 3B). Additional erythroid lineage–specific genes, including Gypa and Alas synthase 2, were already expressed at significant levels at E7.5 and were further induced by E8.5 (Figure 3B). Others, including Eklf/Klf1, Tal1/Scl, Erythrocyte band 4.1, and Fog-1, were expressed at high levels at both E7.5 and E8.5 (supplemental Figure 3B and supplemental Table 3).
Gene expression changes during the transition of EryPs from the YS to the circulation stage of their development
During the transition from progenitors to differentiating erythroblasts (E8.5-E9.5), expression of the transcriptional regulators Gata1 and Cited2 increased, whereas expression of Lmo2, Hmga1, and Gatad2a decreased (Figure 4A). Among other classes of genes whose expression decreased from E8.5-E9.5, most notable were those involved in cell signaling, glucose metabolism, and nitric oxide metabolism (dimethylarginine dimethylaminohydrolase1 [Ddah1]; Figure 4A and supplemental Table 4B).
Changes in gene and protein expression during the transition of EryPs from the YS to the embryonic circulation. (A) Gene identifiers of EryP progenitors. Transcripts expressed by E8.5 EryPs are grouped into functional categories. The fold change from E8.5 and E9.5 is shown. Positive value, up-regulated; negative value, down-regulated. (B) FACS profiles of CD41 protein expression on ϵ-globin::H2B-EGFP embryos from E7.5-E9.5. Expression increases from E7.5-E8.5 and then declines by E9.5. Expression is undetectable at later stages (not shown). (C) FACS histograms showing down-regulation of adhesion molecules on ϵ-globin::H2B-EGFP EryPs during the transition from the YS stage to the circulation.
Changes in gene and protein expression during the transition of EryPs from the YS to the embryonic circulation. (A) Gene identifiers of EryP progenitors. Transcripts expressed by E8.5 EryPs are grouped into functional categories. The fold change from E8.5 and E9.5 is shown. Positive value, up-regulated; negative value, down-regulated. (B) FACS profiles of CD41 protein expression on ϵ-globin::H2B-EGFP embryos from E7.5-E9.5. Expression increases from E7.5-E8.5 and then declines by E9.5. Expression is undetectable at later stages (not shown). (C) FACS histograms showing down-regulation of adhesion molecules on ϵ-globin::H2B-EGFP EryPs during the transition from the YS stage to the circulation.
At E7.5, EryP progenitors express Pecam1 and integrins α5, α6, β1, and CD41 (Figure 4B-C). By E8.5, αV-, β2-, and β3-integrins and CD44 were also detected (Figure 4C). From E8.5-E9.5, expression of 4 adhesion molecule genes (Figure 4A) and their encoded proteins (Figure 4B-C and supplemental Figure 3D) decreased. Most integrin protein and RNA expression was lost by E10.5 (not shown).
Growth factor and cytokine pathways in EryP progenitors
The growth factor requirements of EryP-progenitor cells have not been well defined. Erythropoietin (Epo) is the sole cytokine known to stimulate the formation of EryP progenitors in vitro3 ; EpoR expression was detected as early as E7.5 (Figure 5A). Profiling of E7.5 and E8.5 EryPs allowed us to identify 3 growth factor–signaling pathways that might regulate this lineage at the progenitor stage: c-kit/stem cell factor (SCF), Tie-2/Angiopoietin, and TGFβ1 receptor 1/TGFβ. At E7.5, EryPs express mRNAs for receptors (c-kit, Tie-2, Acvr2b, Tgfbr1, and Tgfbr3) and downstream signaling components (eg, Smad3 and Smad4) of these pathways (Figure 5A). With the exception of EpoR and Vegfa, their transcription began to decrease by E8.5 (Figure 5A).
Growth factor and cytokine pathways in primitive erythroid progenitors. (A) Changes in expression of genes encoding growth factor or cytokine receptors and downstream signaling components in E7.5 and E8.5 EryPs. Shown in this table are median expression levels (log2) from the microarray and linear fold change in expression. (B) Effect of TGF-β1 on formation of EryP progenitors. EryPs were FACS sorted from whole E8.5 embryos and plated in the methylcellulose colony assays in the presence of the indicated concentrations of TGF-β1. Data represent the average of triplicate samples from 4 experiments; error bars represent SEM. (C) Expression of c-kit and Tie-2 protein on EryP at E7.5, E8.5, and E9.5. (D) c-kit marks EryP progenitors within the GFP(+) cell population from E8.5 embryos. Cells were FACS sorted and plated in triplicate in methylcellulose progenitor assays. Colonies were scored at day 5. One representative experiment of 3 is shown; error bars represent SEM (E) Tie-2 marks EryP progenitors within the GFP(+) cell population from E8.5 embryos. Cells were FACS sorted and plated in triplicate in methylcellulose progenitor assays. Colonies were scored at day 5. One representative experiment of 3 is shown; error bars represent SEM (F) Real-time RT-PCR analysis of mRNA expression of Tgf-β1, Ang-1, Scf, and Epo in FACS-sorted EryPs, visceral endoderm, and endothelial cells from YS or in cells from whole embryos at E8.5. Expression levels are shown relative to Ubb.
Growth factor and cytokine pathways in primitive erythroid progenitors. (A) Changes in expression of genes encoding growth factor or cytokine receptors and downstream signaling components in E7.5 and E8.5 EryPs. Shown in this table are median expression levels (log2) from the microarray and linear fold change in expression. (B) Effect of TGF-β1 on formation of EryP progenitors. EryPs were FACS sorted from whole E8.5 embryos and plated in the methylcellulose colony assays in the presence of the indicated concentrations of TGF-β1. Data represent the average of triplicate samples from 4 experiments; error bars represent SEM. (C) Expression of c-kit and Tie-2 protein on EryP at E7.5, E8.5, and E9.5. (D) c-kit marks EryP progenitors within the GFP(+) cell population from E8.5 embryos. Cells were FACS sorted and plated in triplicate in methylcellulose progenitor assays. Colonies were scored at day 5. One representative experiment of 3 is shown; error bars represent SEM (E) Tie-2 marks EryP progenitors within the GFP(+) cell population from E8.5 embryos. Cells were FACS sorted and plated in triplicate in methylcellulose progenitor assays. Colonies were scored at day 5. One representative experiment of 3 is shown; error bars represent SEM (F) Real-time RT-PCR analysis of mRNA expression of Tgf-β1, Ang-1, Scf, and Epo in FACS-sorted EryPs, visceral endoderm, and endothelial cells from YS or in cells from whole embryos at E8.5. Expression levels are shown relative to Ubb.
To determine whether the TGF-β-signaling pathway can modulate the expansion of EryP progenitors, we tested recombinant forms of the 3 Tgfbr1 ligands TGFβ1, TGFβ2, and TGFβ3 in clonogenic assays. TGF-β1 showed a dose-dependent effect on progenitor activity: growth of EryP progenitors was inhibited at a low concentration (0.02 ng/mL) and stimulated at high concentrations (0.2 or 2 ng/mL) of this cytokine (Figure 5B). Neither TGFβ2 nor TGFβ3 had any effect on the numbers of EryP progenitors (data not shown).
c-kit and Tie-2 proteins were detected on the surface of a significant subset of EryPs at E7.5 and E8.5, but were nearly absent by E9.5 (Figure 5C). To determine whether c-kit marks EryP progenitors within the GFP(+) population (Figure 1D), GFP(+)c-kit(+) and GFP(+)c-kit(−) cells were sorted from E8.5 embryos and plated in clonogenic assays in methylcellulose (Figure 5D). The GFP(+) fraction that expressed c-kit gave rise to EryP colonies, whereas those cells lacking c-kit failed to do so (Figure 5D right panel). Therefore, EryP progenitors are highly enriched within the c-kit(+) population.
Whereas it has been reported that EryP progenitors are present within Tie-2(+) cell populations,14 these are heterogeneous. To determine the extent to which EryP progenitors are enriched in the Tie-2(+) population of GFP(+) cells, EryPs were FACS sorted from E8.5 embryos and plated in methylcellulose. The Tie-2(+) fraction of GFP(+) cells showed a 4-fold enrichment of EryP-progenitor activity (Figure 5E). The numbers of EryP progenitors were not consistently stimulated by the addition of recombinant angiopoietin-1 (Ang-1), a Tie-2 ligand, or the c-kit ligand SCF, to the cultures (data not shown). It may be necessary to develop serum-free conditions for the growth of EryP progenitors in vitro to unmask possible Tie-2 and/or c-kit activity.
We next investigated which cell types of the YS express genes encoding the cognate ligands for Tgfbr1 (Tgfβ1), c-kit (SCF), and Tie-2 (Ang-1). Cells of the 3 major YS lineages were FACS sorted from E8.5 embryos on the basis of expression of Flk-1 (endothelial), GFP from a Afp-GFP transgene22 (visceral endoderM) and GFP from the ϵ-globin::H2B-EGFP transgene (EryP). Gene expression was analyzed using qRT-PCR (Figure 5F). YS endothelial cells expressed transcripts for all 3 ligands, Tgf-β1, SCF, and Ang-1, whereas only Ang-1 was expressed in visceral endoderm. Therefore, EryP-progenitor activity may be regulated by growth factors produced by the endothelial and endodermal cells of the YS. Interestingly, Tgfβ1 was also transcribed in EryP (Figure 5F), suggesting the possibility of autocrine and/or paracrine signaling.
Changes in glycolytic gene expression during EryP maturation
Glucose metabolism was an overrepresented functional category in cluster 5, in which genes are progressively down-regulated (Figure 2B). To determine whether high glycolytic activity is a hallmark of EryP progenitors, we performed a qRT-PCR analysis (supplemental Figure 3) of genes predicted from the microarray (Figure 6A) to be transcribed in EryP and known to play pivotal roles in glucose metabolism. Hexokinase 2 (Hk2), Pyruvate kinase M2 (Pkm2), Lactate dehydrogenase a (Ldha), and Phosphoglycerate kinase 1 (Pgk1) were all transcribed at high levels in E8.5 YS EryPs, a profile characteristic of aerobic glycolysis (the “Warburg effect”23,24 ), but their expression decreased dramatically in cells that had entered the circulation (E9.5 and on; Figure 6A). In contrast, Pyruvate kinase, liver/red blood cell (Pklr) was not expressed in progenitor-stage EryPs but was rapidly up-regulated in circulating EryPs (Figure 6A).
Hypoxia regulates EryP-progenitor activity. (A) Expression of genes involved in glucose metabolism during EryP maturation. Relative mRNA levels from the microarrays expressed on a log2 scale. Note the isoform switching from Pgk1 to Pklr. (B) Transcripts known to be induced by hypoxia are down-regulated during EryP maturation. Absolute expression (log2 scale) and fold change in expression are shown for the period from E8.5-E11.5. Expression cutoff, 6.0. (C) Hypoxia increases EryP-progenitor numbers in culture. E8.5 EryPs were FACS sorted, plated in methylcellulose, and incubated under atmospheric or low-oxygen (5%) conditions. Total EryP colony numbers were scored at day 5. (D) Increase in EryP colony size under low-oxygen conditions. Photographs of representative EryP colonies grown at atmospheric or low oxygen conditions are shown. Scale bar, 50 μm. The graph displays the mean radius of EryP colonies (day 4) grown at atmospheric or low oxygen. The EryP colonies that formed in low oxygen were significantly larger than those formed at atmospheric oxygen (33 vs 23.3 μm mean colony radius, respectively). (E) Giemsa staining of cytocentrifuged cells from EryP colonies grown at atmospheric or low oxygen and harvested at day 4. Scale bar, 50 μm. (F) Expression of hypoxia-regulated genes in EryP colonies grown at atmospheric or low oxygen. Colonies were harvested at day 4 and RNA was prepared for qRT-PCR analysis. Expression levels are shown relative to Ubb. Cells grown under hypoxic conditions maintain higher-level expression of these genes than cells grown at atmospheric oxygen. These genes are normally down-regulated as EryP progenitors mature (panel A).
Hypoxia regulates EryP-progenitor activity. (A) Expression of genes involved in glucose metabolism during EryP maturation. Relative mRNA levels from the microarrays expressed on a log2 scale. Note the isoform switching from Pgk1 to Pklr. (B) Transcripts known to be induced by hypoxia are down-regulated during EryP maturation. Absolute expression (log2 scale) and fold change in expression are shown for the period from E8.5-E11.5. Expression cutoff, 6.0. (C) Hypoxia increases EryP-progenitor numbers in culture. E8.5 EryPs were FACS sorted, plated in methylcellulose, and incubated under atmospheric or low-oxygen (5%) conditions. Total EryP colony numbers were scored at day 5. (D) Increase in EryP colony size under low-oxygen conditions. Photographs of representative EryP colonies grown at atmospheric or low oxygen conditions are shown. Scale bar, 50 μm. The graph displays the mean radius of EryP colonies (day 4) grown at atmospheric or low oxygen. The EryP colonies that formed in low oxygen were significantly larger than those formed at atmospheric oxygen (33 vs 23.3 μm mean colony radius, respectively). (E) Giemsa staining of cytocentrifuged cells from EryP colonies grown at atmospheric or low oxygen and harvested at day 4. Scale bar, 50 μm. (F) Expression of hypoxia-regulated genes in EryP colonies grown at atmospheric or low oxygen. Colonies were harvested at day 4 and RNA was prepared for qRT-PCR analysis. Expression levels are shown relative to Ubb. Cells grown under hypoxic conditions maintain higher-level expression of these genes than cells grown at atmospheric oxygen. These genes are normally down-regulated as EryP progenitors mature (panel A).
Oxygen tension is a regulator of EryP-progenitor activity and gene expression
The glucose metabolism–related genes (Figure 6A), as well as others identified in the transcriptome analysis (aldolase a [Aldoa], Vegfb, and the glucose transporter genes Slc2a3 and Slc2a1; Figure 6B) are known to be regulated by hypoxia.24 Therefore, we next investigated whether EryP development is sensitive to changes in oxygen tension, and in particular whether hypoxia can regulate progenitor activity and gene expression.
EryP progenitors arise in the hypoxic environment of the precirculation YS, where oxygen availability is limited by diffusion. To determine whether the maintenance and/or expansion of EryP progenitors is regulated by oxygen levels, GFP(+) cells were sorted from E8.5-transgenic embryos, plated in methylcellulose, and cultured under atmospheric (∼ 21%) or low (2%-5%) oxygen conditions. EryP-progenitor numbers were increased by 2.5-fold in low oxygen (Figure 6C). The EryP colonies that formed in low oxygen were significantly larger than those formed in atmospheric oxygen (Figure 6D), suggesting that hypoxia is a growth signal for EryP progenitors. The increase in colony size was not due to cellular hypertrophy, because cells from colonies cultured under either low or atmospheric oxygen were similar in size (Figure 6E).
The glucose metabolism genes Ldha1, Pkm2, Pgk1, and Hk2 and the endothelial growth factor gene Vegfb are expressed in EryPs (Figure 6A-B) and are known to be responsive to oxygen tension.24 A qRT-PCR analysis of cells from EryP colonies revealed that expression of these genes was higher in cells cultured under low than under atmospheric oxygen (Figure 6F). Thus, oxygen levels regulate gene expression in maturing EryPs and/or EryP progenitors.
Discussion
Emergence of primitive hematopoietic progenitors in the late-gastrulation-stage embryo
During gastrulation, the embryo develops 3 germ layers, the descendant lineages of which must then be established. We have prospectively identified and isolated progenitors and maturing progeny for EryPs, the earliest embryonic cell type to be specified from mesoderm1 at the end of mammalian gastrulation, and have systematically profiled their genome-wide transcriptomes throughout their development. In this study, we focused primarily on the progenitor and early-circulation stages of the EryP lineage. This resource adds to the existing whole-embryo transcriptomes25 by providing information for the single most abundant lineage at several overlapping stages (E7.5-E9.5) of development.
A transcriptional roadmap of EryPs throughout their development
EryPs arise as a cohort that matures in a stepwise, essentially synchronous manner.8 This feature, combined with the ϵ-globin::H2B-EGFP transgenic mouse line, facilitated the isolation of these cells at distinct stages of their development and allowed us to generate a microarray database that provides a timeline of gene expression over 6 days of embryogenesis. In contrast to EryPs, the production of EryDs within the fetal liver or bone marrow occurs continuously, so isolation of these cells at discrete stages is not straightforward. To date, the transcriptional profiles reported for erythroid cells represented only a single stage or heterogeneous populations or were derived from cells differentiated in vitro.26-30
Transcription in maturing EryPs is characterized by 2 discrete waves that correlate with key developmental hallmarks. The first wave (E8.5 ∼ E9.5) coincides with the transition from YS erythropoiesis to the entry of EryPs into the bloodstream, with concomitant loss of progenitor activity. Once EryPs are in circulation (E9.5-E11.5), gene expression changes are more limited, until the second wave of transcriptional variation (E11.5 ∼ E12.5) that corresponds to a period of extensive morphologic changes, decreased cell division rates, cytoskeletal remodeling, and nuclear condensation and extrusion. These changes are reflected in the GO functions that are enriched in developing EryPs, including nuclear organization, DNA packaging, and chromatin assembly.
As would be predicted from our previous validation of the ϵ-globin::histone H2B-GFP transgenic mouse line as a model for primitive erythropoiesis,11,12 a large number of erythroid genes is expressed in the GFP(+) but not the GFP(−) population at high levels from E7.5 on. Many of the genes identified in this study are known to or likely to serve important functions in the development of the definitive erythroid lineage. For example, genes encoding the well-studied transcription factors Gata1, Gata2, Runx1, Eklf/Klf1, Lmo2, Ldb1, and Stat5 are expressed in both lineages. Less well-known transcripts shared by the 2 lineages include the tetraspanin gene Penumbra,31 Homeodomain interacting protein kinase 2 (Hipk2),32 a putative mitochondrial transporter gene involved in the heme biosynthesis pathway (Slc25a39),33 and the antioxidant peroxiredoxin gene Prdx2.34
The EryP transcriptome includes some interesting surprises. For example, Ddah1 encodes an enzyme involved in the metabolism of methylarginines, molecules that inhibit nitric oxide synthase35 and regulate VEGF-mediated angiogenesis.36 Other examples include the fragilis gene family members Ifitm2 (fragilis3) and Ifim3 (fragilis1), which regulate the migration of primordial germ cells37 ; Sox5, which encodes an HMG domain transcription factor related to Sry38 ; and Muscleblind-1, a regulator of alternative splicing in muscle development.39 Future studies should help to clarify the functions of these genes in the EryP lineage.
Regulation of hematopoietic progenitor activity
An interesting feature of primitive erythropoiesis is the transient appearance of progenitors in the YS and the abrupt loss of these cells around the onset of circulation. The molecular events underlying this transition include loss of membrane receptors for growth factors, orchestrated down-regulation of a cohort of cell-adhesion molecules, and massive alterations in metabolism. The growth factor receptors TGFβR1, c-kit, and Tie-2 are expressed on YS EryPs from E7.5-E8.5 and are rapidly down-regulated by E9.5. Genes encoding their ligands for TGFβ1, SCF, and Ang1 are expressed by one or more cell types of the YS, suggesting that local sources of these proteins are available in this microenvironment. Whereas TGFβ1 has been reported to inhibit the growth of erythroid progenitors from adult bone marrow,40 its activity has not been evaluated for EryPs. We observed a dose-dependent response of EryP progenitors to TGFβ1, with suppression of growth at a low concentration and enhancement at high concentrations, raising the possibility that TGFβ1 functions as a morphogen for these cells.
Wnt/β-catenin signaling is required for the formation of the primitive streak41 and has been shown to regulate hematopoietic specification of mesoderm in differentiating embryonic stem cells in vitro.42,43 However, whether EryP cell fate is regulated by Wnt signaling in the embryo, and whether that pathway functions in EryP progenitors themselves or in a mesodermal precursor is unknown. Several Wnt/β-catenin pathway genes are expressed in EryPs from E7.5 embryos and are rapidly down-regulated as the cells mature. In this study, we provide evidence that the Wnt pathway is active and functions autonomously in EryP progenitors in the blood islands of the YS.
A variety of adhesion proteins are expressed on EryP progenitors but not on circulating EryPs. These observations suggest that, within the YS, EryPs may form tight associations with each other and/or with surrounding endothelial cells, as suggested by electron microscopy.2 Loss of adhesion proteins from the surface of EryPs in the YS might facilitate their entry into the bloodstream. In the zebrafish, release of EryPs into the circulation requires the activity of a metalloprotease.44 Interestingly, 2 metalloprotease genes, Adam17 and mmp2, are expressed in YS-stage EryPs (supplemental Figure 3B).
EryP progenitors, hypoxia, and the Warburg effect
During their maturation in successive microenvironments within the embryo,11 EryPs must adapt to changes in oxygen and nutrient supplies. We found that the number of EryP progenitors was enhanced by low oxygen concentrations in culture. It is intriguing that the transcriptome of EryP progenitors revealed a signature generally associated with cancer and other rapidly proliferating cells, which catabolize glucose aerobically and produce high levels of lactate in the cytosol, rather than using the more energy-efficient mitochondrial pathway known as the “Warburg effect.”23,24 EryP progenitors express high levels of Pkm2 and Hk2, genes that play critical roles in the Warburg effect.23,24 The reciprocal expression of the pyruvate kinase genes Pkm2 and Pklr in progenitors and maturing EryPs, respectively, indicates that isoform switching is a feature of EryP differentiation, and presumably reflects progressive adaptation of the rapidly dividing progenitors to new metabolic requirements. There may be a functional association between the development of the placenta and the onset of circulation and the abrupt loss of progenitor activity as the cells enter the bloodstream. Whether the aerobic glycolytic profile of EryP progenitors simply reflects the particular energy demands of these rapidly dividing cells or is a more unique feature of primitive erythropoiesis is not yet clear.
The rapidly dividing EryP-progenitor cell population effectively constitutes a transient amplifying pool from which large numbers of primitive erythroblasts are generated. Using the ϵ-globin::H2B-EGFP transgenic mouse system to measure the numbers of EryPs at E7.5 and E8.5, we found that these cells represent a huge fraction of the embryo at these stages. Indeed, by E8.5, EryPs comprise nearly half of all cells and are by far the most abundant cell type in the embryo. Thus, the embryo has set aside enormous resources specifically for the development of this single lineage.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Saghi Ghaffari for providing access to the hypoxia chamber incubator; the Mount Sinai Flow Cytometry Shared Resource Facility for assistance with cell sorting; and Drs Julie Baker, Jane Little, Philippe Soriano, and Daniel Weinstein for insightful comments on the manuscript.
Transgenic mice were produced by the Mount Sinai Mouse Genetics Shared Research Facility, which is supported by National Institutes of Health/National Cancer Institute grant R24 CA88302. This work was supported by grants from the National Institutes of Health to M.H.B. (RO1 HL62248, DK52191, and EB02209), to P.G.G. (RO1 HL65448, DK62039, and P30 DK072442), and to A.-K.H. (RO1 HD052115 and DK084391); from the Roche Foundation for Anemia Research (grant 9699367999, cycle X to M.H.B.); and from the New York State Department of Health (NYSTEM grant N08G-024 to M.H.B.).
National Institutes of Health
Authorship
Contribution: J.I., Z.H., S.T.F., and M.H.B. designed the experiments, analyzed the data, and prepared the figures; J.I., Z.H., and S.T.F., performed the experiments; S.N. performed the 3D time-lapse imaging; A.F.-V. generated and imaged the TCF/Lef::H2B-GFP strain; R.M. performed the β-catenin immunostaining; A.-K.H. provided the TCF/Lef::H2B-GFP mouse line and live imaging expertise; V.S., D.T., and P.G.G. helped design the microarray experiments and analyze and prepare data for publication; and J.I., S.T.F., and M.H.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of J.I. is Department of Cardiovascular Developmental Biology, Fundación Centro Nacional de Investigaciones Cardiovasculares (CNIC) Carlos III, Madrid, Spain. The current affiliation of S.T.F. is Discipline of Physiology, School of Medical Sciences, University of Sydney, Camperdown, Australia.
Correspondence: Margaret H. Baron, MD, PhD, Mount Sinai School of Medicine, Box 1079, 1468 Madison Ave, Annenberg 24-04E, New York, NY 10029-6574; e-mail: margaret.baron@mssm.edu.