Abstract
During an analysis of T-cell responses against human renal cell carcinoma (RCC), we identified a CD4+ T-cell line that showed TCR-mediated recognition and lysis of nearly all RCC lines regardless of MHC type. We have now elucidated the nature of the ligand for this α/β TCR, and it contains no MHC-related moiety and does not involve classic peptide processing. First, matrix metalloproteinase 14 (MMP14) expressed on RCC cells releases membrane-bound TRAIL expressed by the T cell; then, soluble TRAIL binds to its receptor DR4 (TRAIL-R1), which is expressed on tumor cells, and this TRAIL-DR4 complex is recognized by the TCR through a complementarity-determining region 3α (CDR3α)–mediated interaction. Direct and specific antigen-TCR interaction was demonstrated when the immobilized recombinant TRAIL/DR4 complex stimulated the TCR. In addition, amino acid substitutions in the CDR3α of the TCR either obliterated or enhanced target-specific recognition. This description of the molecular nature of a non-MHC target structure recognized by a naturally occurring α/β TCR not only broadens our concept of what the TCR can recognize, but also raises the question of whether such a T cell could be of clinical utility against RCC.
Introduction
T cells expressing α/β TCRs classically bind to processed peptides presented by MHC molecules. This same interaction is thought to be central to the processes of thymic development and selection that shape the peripheral T-cell repertoire. Recent work has identified canonical amino acids in the TCR Vβ that participate in generic recognition of framework MHC domains that are critical to producing a normal T-cell repertoire, suggesting that such interactions may be hard-wired into the genome.1 Even when atypical TCRs recognizing nonclassic antigens have been found, there is an MHC-like presenting molecule involved. However, in murine systems, the germline deletion of β-2-microglobulin (B2M), MHC class II, CD4, and CD8 does not prevent thymic selection and does not obliterate all peripheral blood α/β T cells. In fact, many MHC-independent α/β T cells are present in the lymphoid periphery, and these cells respond in mixed-lymphocyte reactions to MHC-null stimulator cells, but the MHC-independent antigenic ligands they recognize are not known.2
In a previous study, we described the cloning of HC/2G-1, a CD4+ T cell from the blood of a patient with renal cancer that showed broad recognition of nearly all human renal cancer tumor lines despite those lines sharing no common MHC allele.3 Reactivity could not be blocked by anti–class I, class II MHC Ab, or anti-CD4 Ab, but was significantly reduced by an anti-TRAIL Ab.3 Anti-TRAIL Ab blocked not only killing of targets but also IFN-γ secretion by HC/2G-1 cells, suggesting that the involvement of TRAIL was not merely as an apoptosis-inducing ligand.3 Renal cell carcinoma (RCC) recognition by HC/2G-1 was mediated through its TCRs, as evidenced by the transfer of RCC reactivity to allogeneic peripheral blood T cells by the retroviral transduction of the HC/2G-1 TCR α and β genes.3 Our previous observation that this T-cell line showed equal recognition of a B2M-deficient RCC line and a B2M-transduced control line suggested that the recognition mechanism was B2M independent, precluding the participation of most MHC class Ib as well as class Ia molecules.3 Through a process of sequential expression screening of cDNA libraries from renal cancer lines, we have identified the specific antigenic complex recognized by HC/2G-1, and it appears to have no relationship to MHC molecules.
Methods
Reagents
Antibodies for flow cytometric analyses were DR4/PE (DJR1; eBioscience), DR5/PE (DJR2-4; eBioscience), DcR1/PE (DJR3; eBioscience), DcR2/PE (104918; R&D Systems), CD58/FITC (1C3; BD Biosciences), matrix metalloproteinase 14 (MMP14)/PE (IC9181P; R&D Systems), TRAIL NA/LE (RIK-2; BD Pharmingen), and the mouse IgG1 isotype control (16-4714; eBioscience). ELISA reagents were DuoSet DY375 (R&D Systems) for soluble TRAIL and matched pair Abs M700A and M701B (ThermoFisher Scientific) for IFN-γ. Recombinant human DR4-Fc, DR5-Fc, DcR1-Fc, DcR2-Fc, and IgG1-Fc were purchased from R&D Systems. Anti-CD3 Ab was purified from the culture supernatant of the hybridoma OKT3 (ATCC).
Cell lines
All RCC, EBV-B, and melanoma cell lines were established at the Surgery Branch, National Cancer Institute, National Institutes of Health (Bethesda, MD). RCC lines were as follows: RCC#1: 2245R, RCC#2: 2219R, RCC#3: 2390R, RCC#5: 2102R, RCC#6: 2246R, RCC#8: 2361R, RCC#9: 2362R, RCC#10: 2261R, and RCC#11: 1764R. Chinese hamster ovary (CHO)–K1 cells and 293T cells were purchased from ATCC. The 293gp cell line was a kind gift of Dr Paul F. Robbins (Surgery Branch, NCI, NIH).
Retroviral transductions
TCR transduction to PBLs was as described previously.3 Coding sequences of DR4, DR5, and CD58 cDNA were cloned into the pRx vector.4 For tumor cells, retrovirus was produced using the 293gp retrovirus producer cell line and the VSVG envelope. Virus in the culture supernatant was filtered by a syringe filter with 0.45-μm pore size, and was used for infection in the presence of 8 μg/mL of Polybrene for 6 hours at 37°C and 5% CO2.
Lentiviral transductions
MMP7 and MMP14 cDNA were cloned into the pCAG lentiviral vector.5 Each vector was transfected to 293T cells with pMDLg/pRRE, pRSV-Rev, and pMD.G (kind gifts of Dr Richard A. Morgan, Surgery Branch, NCI, NIH) using Lipofectamine 2000 (Invitrogen). One day after the transfection, the medium was replaced with fresh medium and harvested for gene transduction the next day. After filtration through 0.45-μm-pore filters, EBV-B cells were transduced in 6-well plates in the presence of 8 μg/mL Polybrene and centrifuged at 2200g for 4 hours at 32°C.
TCR mutagenesis
PCR-based site-directed mutagenesis of the retrovirus vector plasmid was done using the QuikChange kit (Stratagene).
cDNA library expression screening
Total RNA was purified from RCC#6 using RNeasy Maxi (QIAGEN), and was further purified using FastTrack MAG Maxi mRNA Isolation Kit (Invitrogen) to obtain poly(A)+ RNA. cDNA was synthesized with the SuperScript Plasmid System (Invitrogen), and cloned into pME18S vector with a modified linker that accommodates SalI-NotI fragments. ElectroMAX DH10B-competent cells (Invitrogen) were transformed by electroporation and, after the titration, E coli (∼ 150 clones/well) was inoculated into 96-well format culture blocks (10 blocks) and cultured overnight. Plasmids were purified by QIAprep 96 Turbo Miniprep Kit (QIAGEN), and transfected to human embryonic kidney epithelial cell line 293 (HEK-293), HEK-293/DR4, or human DR4–transduced CHO cells (CHO/hDR4) cells using Lipofectamine 2000 (Invitrogen) in flat-bottom, 96-well plates. After an overnight culture, HC/2G-1 cells (5000-20 000 cells) were added; after a 20- to 24-hour coculture, IFN-γ in the supernatant was measured by ELISA. Candidate wells were picked, and for the secondary and third screenings, subpool libraries (∼ 20 clones/well, 48 wells) and clone libraries (1 clone/well, 96 wells) were prepared and screened. Identified clones were sequenced to identify the cDNA that conferred HC/2G-1 recognition to recipient cells.
Stimulation with plastic-bound ligands
Flat-bottom, 96-well plates were coated overnight with a functional-grade purified anti-CD2 Ab (eBioscience), DR4-Fc carrier-free, DR5-Fc carrier-free, DcR1-Fc carrier-free, DcR2-Fc carrier-free, or IgG1-Fc carrier-free (R&D Systems), alone or in combination as indicated, at 10 μg/mL each in 100 μL of PBS. Wells were blocked with 200 μL of PBS/5% FBS for 1 hour, and soluble TRAIL (Biomol) in RPMI with 10% FBS (100 μL) was added at 10 μg/mL (Figures 3C and 4B) or at the concentrations indicated (Figure 4C) and incubated at 37°C for 3 hours. Wells were rinsed gently for 3 times with RPMI with 10% FBS, and HC/2G-1 cells (20 000-50 000 cells/well) in RPMI with 10% FBS were added. After a 24-hour culture, IFN-γ in the supernatant was measured by ELISA.
Quantitative RT-PCR
TaqMan Gene Expression Assays for DR4 (Hs00269492_m1) and β-actin (Hs03023880_g1) were purchased from Applied Biosystems. Plasmids encoding DR4 and β-actin were used as standards. Linear regression analysis was done with Prism software Version 5.0c (GraphPad).
Statistical analysis
One-way ANOVA and Tukey multiple comparison post hoc tests were done using Version 5.0c Prism software (GraphPad).
Informed consent
All human specimens were obtained from individuals from whom written informed consent was received before the study in accordance with the Declaration of Helsinki. All studies were approved by the National Institutes of Health.
Results
Expression cloning identifies TRAIL receptor-1 (DR4) as a component of target recognition by the HC/2G-1 T cell
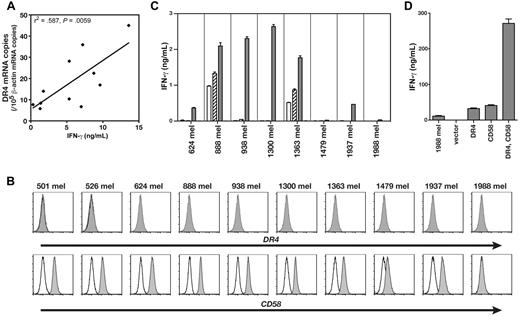
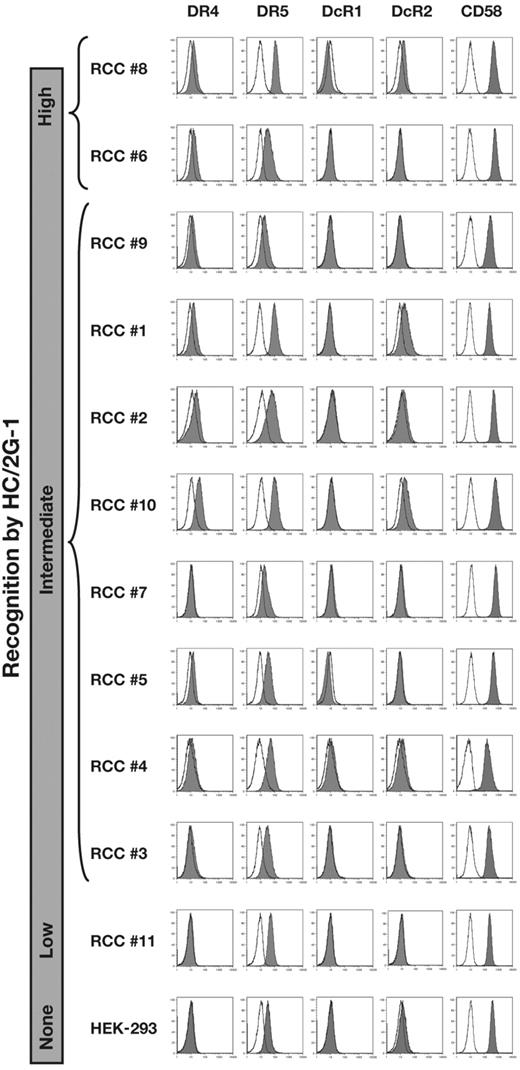
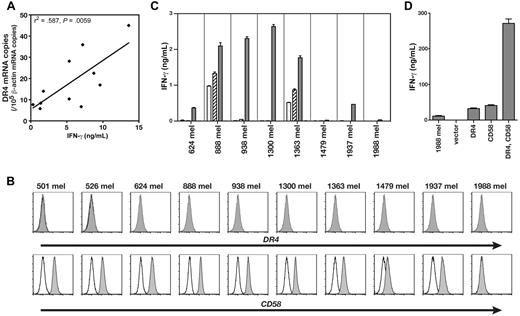
HC/2G-1 recognized multiple RCCs but not patient-matched EBV-B cells, precluding alloresponse as the mechanism of recognition (Figure 1A and Wang et al3 ). This observation implied that any putative restriction element had limited or no polymorphism. The transformed human embryonic kidney epithelial line HEK-293 was not recognized by HC/2G-13 and, on the assumption that HEK-293 cells might express any nonpolymorphic restriction element, we first conducted expression screening of an RCC cDNA library in HEK-293 to identify the antigen. This screening identified a cDNA clone that encoded one of the TRAIL receptors, DR4 (also known as CD261, TRAILR1, or TNFRSF10A), as the moiety that conferred recognition of HEK-293 cells by HC/2G-1 (Figure 1B). This was in agreement with our prior finding that HC/2G-1 reactivity was reduced by the anti-TRAIL Ab (Figure 1C and Wang et al3 ), and suggested that DR4 serves its functional role as a TRAIL receptor rather than as the source of a (processed) antigenic peptide. Four cell-surface receptors are known to bind to TRAIL with a similar affinity: DR4,6 DR5 (CD262, TRAILR2, or TNFRSF10B),7,8 DcR1 (CD263, TRAILR3, or TNFRSF10C),7,8 and DcR2 (CD264, TRAILR4, or TNFRSF10D).9 DR4 and DR5 are known as apoptosis-inducing receptors, whereas DcR1 and DcR2 lack a functional death domain, do not induce apoptosis, and represent decoy receptors. DR5 was naturally expressed on parental HEK-293 cells (Figure 2), and transduction of the DR5 gene into HEK-293 did not induce recognition by HC/2G-1 (Figure 1B). Flow cytometric analysis of TRAIL receptors on RCC cell lines showed no correlation between recognition by HC/2G-1 and expression of DR5, DcR1, or DcR2, whereas DR4 expression showed a weak correlation (Figure 2). DR4 expression in RCC lines and their recognition by HC/2G-1, as measured by quantitative RT-PCR and IFN-γ secretion, respectively, showed a statistically significant (R2 = .587 and P = .0059) correlation (Figure 3A). Melanoma cell lines are generally resistant to HC/2G-1 and express little or no DR4 (Figure 3B); however, when these melanoma lines were transduced with DR4, 6 of 8 lines were able to stimulate HC/2G-1 (Figure 3C). These findings showed that target cell expression of DR4 is a key element that determines reactivity to HC/2G-1. Further, inducing the expression of DR5 did not sensitize melanoma lines; in fact, 2 wild-type melanoma lines (888mel and 1363mel) that are weakly recognized by HC/2G-1 lost this reactivity when DR5 was overexpressed, implying that DR5 can function as a decoy receptor in HC/2G-1 recognition, presumably by competitive binding of TRAIL. This inhibitory decoy activity was also seen after overexpression of DcR1 or DcR2 on RCC lines (data not shown). Based on these observations, we concluded that DR4 binding to its natural ligand TRAIL was directly or indirectly involved in the triggering of HC/2G-1.
RCC recognition by HC/2G-1 is blocked by anti-TRAIL Ab and expression of DR4 (TRAIL-R1) and CD58 on target cells confers HC/2G-1 recognition. (A) RCC and EBV-B lines from 10 patients were cocultured with HC/2G-1 cells, and IFN-γ in the culture supernatant was detected by ELISA after 20 hours of coculture. Cell line numbers from the same patient are matched between EBV-B and RCC. HC/2G-1 was derived from the patient 1. Error bars indicate SEM. (B) HEK-293 and its mock-, DR4-, or DR5-transduced variants were cocultured with HC/2G-1 T cells, and T-cell activation was measured by IFN-γ ELISA. Error bars indicate SEM. (C) HC-2G-1 cells were preincubated with either an isotype-matched control Ab (gray bars) or an anti-TRAIL Ab (black bars), and cocultured with various RCC cell lines (left panel) or enzyme-digested fresh RCC cells (middle panel). In the right panel, MART127-35-reactive control CD8+ tumor-infiltrating lymphocytes (JKF6) were similarly preincubated with Abs and cocultured with 624mel, a human melanoma cell line. After a 20-hour coculture, IFN-γ in the supernatant was measured by ELISA. Error bars indicate SEM. (D-F) cDNA library expression screening using CHO cells stably transduced with DR4 as the transfection target identified CD58 as the second crucial element. The effect of DR4 and CD58 expression on recognition was specific to HC/2G-1 cells. CHO and CHO/hDR4 cells were transiently transfected with GFP, CD58, CD54, CD102, or CD106 cDNA expression plasmids. After 24 hours, HC/2G-1 T cells (D), MART127-35-reactive negative control CD8 T cells (E), or HLA-DRβ1*0401–restricted gp100 44-59-reactive negative control CD4 T cells (F) were added and IFN-γ in the supernatant was measured by ELISA at 20 hours of coculture. Error bars indicate SEM. Representative results from at least 3 similar experiments are presented.
RCC recognition by HC/2G-1 is blocked by anti-TRAIL Ab and expression of DR4 (TRAIL-R1) and CD58 on target cells confers HC/2G-1 recognition. (A) RCC and EBV-B lines from 10 patients were cocultured with HC/2G-1 cells, and IFN-γ in the culture supernatant was detected by ELISA after 20 hours of coculture. Cell line numbers from the same patient are matched between EBV-B and RCC. HC/2G-1 was derived from the patient 1. Error bars indicate SEM. (B) HEK-293 and its mock-, DR4-, or DR5-transduced variants were cocultured with HC/2G-1 T cells, and T-cell activation was measured by IFN-γ ELISA. Error bars indicate SEM. (C) HC-2G-1 cells were preincubated with either an isotype-matched control Ab (gray bars) or an anti-TRAIL Ab (black bars), and cocultured with various RCC cell lines (left panel) or enzyme-digested fresh RCC cells (middle panel). In the right panel, MART127-35-reactive control CD8+ tumor-infiltrating lymphocytes (JKF6) were similarly preincubated with Abs and cocultured with 624mel, a human melanoma cell line. After a 20-hour coculture, IFN-γ in the supernatant was measured by ELISA. Error bars indicate SEM. (D-F) cDNA library expression screening using CHO cells stably transduced with DR4 as the transfection target identified CD58 as the second crucial element. The effect of DR4 and CD58 expression on recognition was specific to HC/2G-1 cells. CHO and CHO/hDR4 cells were transiently transfected with GFP, CD58, CD54, CD102, or CD106 cDNA expression plasmids. After 24 hours, HC/2G-1 T cells (D), MART127-35-reactive negative control CD8 T cells (E), or HLA-DRβ1*0401–restricted gp100 44-59-reactive negative control CD4 T cells (F) were added and IFN-γ in the supernatant was measured by ELISA at 20 hours of coculture. Error bars indicate SEM. Representative results from at least 3 similar experiments are presented.
Expression of TRAIL receptors and CD58 on RCC lines. Expression of TRAIL receptors and CD58 on RCC lines and HEK-293 was analyzed by flow cytometry. Cell lines are presented in order of the recognition strength by HC/2G-1 from top to bottom.
Expression of TRAIL receptors and CD58 on RCC lines. Expression of TRAIL receptors and CD58 on RCC lines and HEK-293 was analyzed by flow cytometry. Cell lines are presented in order of the recognition strength by HC/2G-1 from top to bottom.
Expression of DR4 and CD58 on melanoma cells confers HC/2G-1 recognition. (A) Correlation between DR4 mRNA expression (as shown by quantitative RT-PCR) and strength of recognition by HC/2G-1 (as shown by IFN-γ secretion). Linear regression analysis yielded R2 = .587 and P = .0059. (B) Expression of DR4 and CD58 on melanoma cell lines was analyzed by flow cytometry. (C) Effect of retroviral transduction of DR4 or DR5 into human melanoma cell lines on their recognition by HC/2G-1. Parental (white bars), mock-transduced (hatched bars), DR4-transduced (gray bars), and DR5-transduced (black bars) were cocultured with HC/2G-1 cells, and the IFN-γ concentration was measured by ELISA. Error bars indicate SEM. (D) 1988mel, a human melanoma cell line that lacks expression of both DR4 and CD58, becomes sensitive to HC/2G-1 when transduced with both genes. 1988mel cells were retrovirally transduced with cDNA, as shown, and were tested for recognition by HC/2G-1 cells. Error bars indicate SEM. Representative data from 3 experiments are shown.
Expression of DR4 and CD58 on melanoma cells confers HC/2G-1 recognition. (A) Correlation between DR4 mRNA expression (as shown by quantitative RT-PCR) and strength of recognition by HC/2G-1 (as shown by IFN-γ secretion). Linear regression analysis yielded R2 = .587 and P = .0059. (B) Expression of DR4 and CD58 on melanoma cell lines was analyzed by flow cytometry. (C) Effect of retroviral transduction of DR4 or DR5 into human melanoma cell lines on their recognition by HC/2G-1. Parental (white bars), mock-transduced (hatched bars), DR4-transduced (gray bars), and DR5-transduced (black bars) were cocultured with HC/2G-1 cells, and the IFN-γ concentration was measured by ELISA. Error bars indicate SEM. (D) 1988mel, a human melanoma cell line that lacks expression of both DR4 and CD58, becomes sensitive to HC/2G-1 when transduced with both genes. 1988mel cells were retrovirally transduced with cDNA, as shown, and were tested for recognition by HC/2G-1 cells. Error bars indicate SEM. Representative data from 3 experiments are shown.
A CD2-CD58 interaction is a second component of tumor recognition by HC/2G-1 T cells
We found that CHO cells transduced with human DR4 were not recognized by HC/2G-1 (Figure 1D). Because hamster cells may not express functional homologs of human molecules necessary for recognition, we conducted additional cDNA library expression screening using CHO/hDR4 cells to look for such molecules. One cDNA was found to confer recognition to CHO/hDR4 cells but, unexpectedly, this cDNA turned out to encode human CD58 (LFA3), which is the counterpart for CD2 that is expressed on all T cells (Figure 1D). Other cell-adhesion molecules that are expressed on RCC, such as CD54 (ICAM1), CD102 (ICAM2), or CD106 (VCAM1), did not confer HC/2G-1 reactivity to CHO/hDR4 cells, suggesting a crucial role of the CD2-CD58 interaction in target recognition by HC/2G-1 cells (Figure 1D). CHO/hDR4/hCD58 cells were not recognized by CD2+ melanoma-reactive CD8 T cells (Figure 1E) or by CD4 T cells (Figure 1F), arguing against the possibility that the recognition of CHO/hDR4/hCD58 cells by HC/2G-1 was through nonspecific, CD2-mediated T-cell stimulation. Interestingly, transduction of both human DR4 and human CD58 conferred HC/2G-1 reactivity to the mouse melanoma cell line B16F10 (not shown) as well as CHO cells. 1988mel was one of the human melanoma cell lines that were not sensitized by the transduction of DR4 (Figure 3C). We examined the expression of CD58 on melanoma lines and found that 1988mel was deficient in CD58 expression (Figure 3B). Transducing CD58 cDNA into 1988mel/DR4 led to recognition by HC/2G-1 (Figure 3D), corroborating the importance of the CD2-CD58 interaction in tumor recognition by HC/2G-1 T cells. In another experiment, we immunized mice with RCC#6 to generate monoclonal Abs against the human RCC line. After screening approximately 2000 hybridoma culture supernatants, we found a monoclonal Ab that interfered with RCC recognition by HC/2G-1 cells. cDNA library expression screening revealed that the Ab binds to CD58—again confirming the importance of CD58 (data not shown).
Signal transduction by DR4 on target cells is not involved in recognition by HC/2G-1 T cells
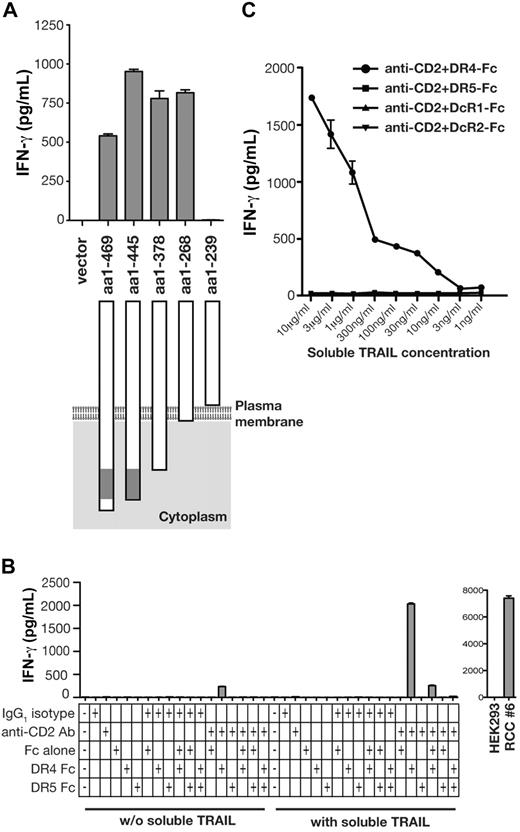
Recognition of CHO/hDR4/hCD58 was unexpected, because it implied that all additional components of the antigenic structure were conserved across species and did not clarify the role of DR4. To scrutinize the contribution of DR4, cDNA constructs encoding DR4 with variable intracellular truncations were prepared, transfected into CHO/CD58, and examined for recognition by HC/2G-1 (Figure 4A). This experiment showed that expression of the membrane-bound DR4 extracellular region was sufficient and that no intracellular component of DR4 was needed.
Activation of HC/2G-1 T cells by DR4 loaded with soluble TRAIL. (A) CHO/CD58 cells were transduced with retroviruses that encode various truncations of the carboxyl terminus of DR4, and their recognition by HC/2G-1 T cells was measured by IFN-γ ELISA. The gray box in DR4 denotes the death domain. (B) A 96-well plate was coated with anti-CD2 Ab, control Ab, recombinant DR4-Fc, DR5-Fc, and Fc, individually or in various combinations as indicated. After blocking with PBS/FBS for 1 hour, recombinant soluble TRAIL (10 μg/mL) solubilized in the culture medium was added where indicated. After 3 hours, wells were rinsed to remove unbound soluble TRAIL and HC/2G-1 T cells were added. After an overnight culture, IFN-γ in the supernatant was measured by ELISA. Specificity controls (HEK-293 as the negative control and RCC#6 as the positive control) are shown in the right panel. (C) Titration of soluble TRAIL was done using the same method as in panel B. After coating with anti-CD2 Ab and recombinant DR4-Fc, DR5-Fc, DcR1-Fc, or DcR2-Fc, wells were blocked with PBS with 5% FBS for 1 hour, and serially diluted soluble TRAIL in the culture medium was added as indicated. After incubating for 3 hours, unbound TRAIL was removed by rinsing and HC/2G-1 T cells were added. IFN-γ in the supernatant was measured by ELISA after an overnight culture. Error bars indicate SEM.
Activation of HC/2G-1 T cells by DR4 loaded with soluble TRAIL. (A) CHO/CD58 cells were transduced with retroviruses that encode various truncations of the carboxyl terminus of DR4, and their recognition by HC/2G-1 T cells was measured by IFN-γ ELISA. The gray box in DR4 denotes the death domain. (B) A 96-well plate was coated with anti-CD2 Ab, control Ab, recombinant DR4-Fc, DR5-Fc, and Fc, individually or in various combinations as indicated. After blocking with PBS/FBS for 1 hour, recombinant soluble TRAIL (10 μg/mL) solubilized in the culture medium was added where indicated. After 3 hours, wells were rinsed to remove unbound soluble TRAIL and HC/2G-1 T cells were added. After an overnight culture, IFN-γ in the supernatant was measured by ELISA. Specificity controls (HEK-293 as the negative control and RCC#6 as the positive control) are shown in the right panel. (C) Titration of soluble TRAIL was done using the same method as in panel B. After coating with anti-CD2 Ab and recombinant DR4-Fc, DR5-Fc, DcR1-Fc, or DcR2-Fc, wells were blocked with PBS with 5% FBS for 1 hour, and serially diluted soluble TRAIL in the culture medium was added as indicated. After incubating for 3 hours, unbound TRAIL was removed by rinsing and HC/2G-1 T cells were added. IFN-γ in the supernatant was measured by ELISA after an overnight culture. Error bars indicate SEM.
HC/2G-1 T cells are directly activated by soluble TRAIL bound to DR4
The findings from truncating DR4 led to the novel hypothesis that DR4 itself serves as the molecule that presents soluble TRAIL to HC/2G-1 cells. To test this hypothesis, a 96-well plate was coated with anti-CD2 Ab (as a surrogate for CD58) and recombinant DR4-Fc or DR5-Fc protein (extracellular domains fused to the Fc portion of Ig) with or without exogenous recombinant soluble TRAIL protein. HC/2G-1 cells were then added to each well and their activation was measured by 24-hour IFN-γ release. Supporting the new hypothesis, HC/2G-1 was activated only when the plate was coated with anti-CD2 Ab and DR4-Fc and soluble TRAIL was added (Figure 4B). Reminiscent of the findings after overexpressing DR5 in melanoma cells (Figure 3C), on plastic, adding DR5-Fc to anti-CD2 Ab and DR4-Fc was again inhibitory, presumably through a decoy receptor mechanism (Figure 4B). Titration of soluble TRAIL showed that as little as 10 ng/mL of soluble TRAIL could induce recognition when pulsed onto DR4-Fc, but not DR5-Fc, DcR1-Fc, or DcR2-Fc, demonstrating the specificity of HC/2G-1 for DR4 as the TRAIL-binding element (Figure 4C).
Recognition of the TRAIL/DR4 complex involves a specific interaction with the CDR3 of the HC/2G-1 TCR α chain
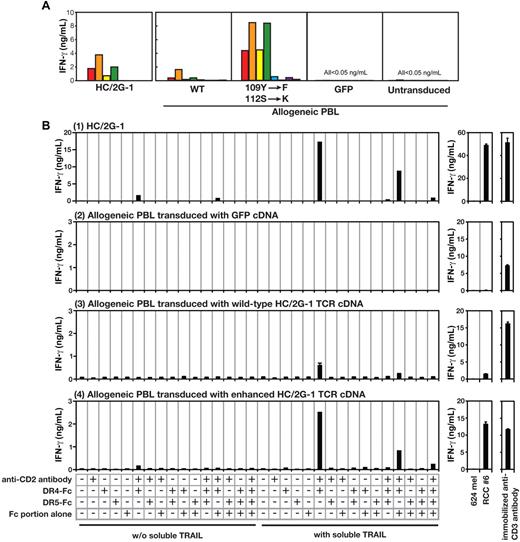
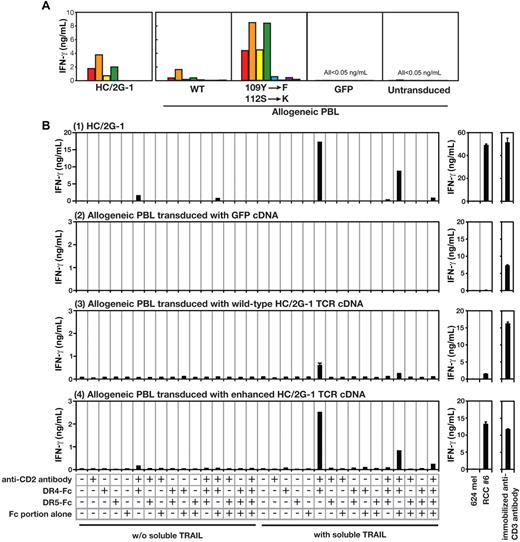
In our previous study, we showed that HC/2G-1 uses its TCR α and β receptors (GenBank accession numbers, EF101779 and EF101778, respectively) for target recognition.3 However, it was not known if this activation was through engagement of invariant domains of the TCR (as with superantigens) or if it was through specific binding via the complementarity-determining regions (CDRs). Subtle modifications of the CDRs of conventionally reactive TCRs have been shown to either destroy or improve TCR reactivity to specific MHC-peptide complexes.10,11 To determine whether this was true for HC/2G-1, we prepared 28 retroviral vectors containing alanine substitutions in the CDR2 and CDR3 regions of the TCR α (15 positions) and β (13 positions) chains. Allogeneic PBLs were retrovirally transduced with these mutated TCR genes and their reactivity to a panel of RCC tumor and control cell lines was examined. This experiment showed that replacing serine (position 112) in CDR3 of the TCR α chain increased reactivity to RCC lines without compromising specificity, whereas most substitutions at other positions reduced reactivity (compared with the wild-type HC/2G-1 receptor chains). We further focused on the CDR3 region of the TCR α chain and, after extensive trial-and-error testing, found that combining a tyrosine109 → phenylalanine with a serine112 → lysine substitution resulted in the strongest recognition of the RCC tumor panel without increasing reactivity to control target cells (Figure 5A and Wang et al12 ). We then investigated whether this enhanced TCR with a finely tuned CDR3α would also show enhanced reactivity and maintain specificity to plate-bound DR4 and TRAIL. This was indeed the case, and these mutations in the CDR3α not only recapitulated the pattern of recognition against plastic-adherent purified target molecules, but also showed the same augmented responses to RCC as wild-type TCR (Figure 5A-B).
HC/2G-1 uses the TCR α chain CDR3 for recognizing the DR4/TRAIL complex. (A) Allogeneic T cells transduced with the genes encoding wild-type HC/2G-1 TCR (WT), HC/2G-1 TCR with doubly mutated α chain and wild-type β chain (109Y-F, 112S-K), control GFP expressing retrovirus (GFP) or nothing (Untransduced) were cocultured with a panel of tumor lines. The parental HC/2G-1 T-cell clone is also shown (HC/2G-1). Red bars indicate RCC#1; orange bars, RCC#6; yellow bars, RCC#8; green bars, RCC#10; blue bars, RCC#11; indigo-blue bars, 624mel; purple bars, 938mel; and gray bars, medium alone. (B) Plate coating was done in the same way as in Figure 4B using anti-CD2 Ab, DR4-Fc, DR5-Fc, and IgG Fc in the combinations indicated. Activation of parental HC/2G-1 cells (1), allogeneic T cells transduced with GFP cDNA (2), allogeneic T cells transduced with wild-type HC/2G-1 TCR α and β chain cDNA (3), or allogeneic T cells transduced with mutated HC/2G-1 TCR α (109Y-F, 112S-K) and wild-type HC/2G-1 TCR β chain cDNA (4) were measured at 24 hours with IFN-γ ELISA. Stimulation by 624mel and RCC#6 is shown as a specificity control and by anti-CD3 Ab as a functional control. Error bars indicate SEM. Representative data from more than 3 experiments with similar results are shown.
HC/2G-1 uses the TCR α chain CDR3 for recognizing the DR4/TRAIL complex. (A) Allogeneic T cells transduced with the genes encoding wild-type HC/2G-1 TCR (WT), HC/2G-1 TCR with doubly mutated α chain and wild-type β chain (109Y-F, 112S-K), control GFP expressing retrovirus (GFP) or nothing (Untransduced) were cocultured with a panel of tumor lines. The parental HC/2G-1 T-cell clone is also shown (HC/2G-1). Red bars indicate RCC#1; orange bars, RCC#6; yellow bars, RCC#8; green bars, RCC#10; blue bars, RCC#11; indigo-blue bars, 624mel; purple bars, 938mel; and gray bars, medium alone. (B) Plate coating was done in the same way as in Figure 4B using anti-CD2 Ab, DR4-Fc, DR5-Fc, and IgG Fc in the combinations indicated. Activation of parental HC/2G-1 cells (1), allogeneic T cells transduced with GFP cDNA (2), allogeneic T cells transduced with wild-type HC/2G-1 TCR α and β chain cDNA (3), or allogeneic T cells transduced with mutated HC/2G-1 TCR α (109Y-F, 112S-K) and wild-type HC/2G-1 TCR β chain cDNA (4) were measured at 24 hours with IFN-γ ELISA. Stimulation by 624mel and RCC#6 is shown as a specificity control and by anti-CD3 Ab as a functional control. Error bars indicate SEM. Representative data from more than 3 experiments with similar results are shown.
MMP14 expressed on tumor cells releases soluble TRAIL for presentation by DR4 and may account for the specificity of HC/2G-1 for human RCC
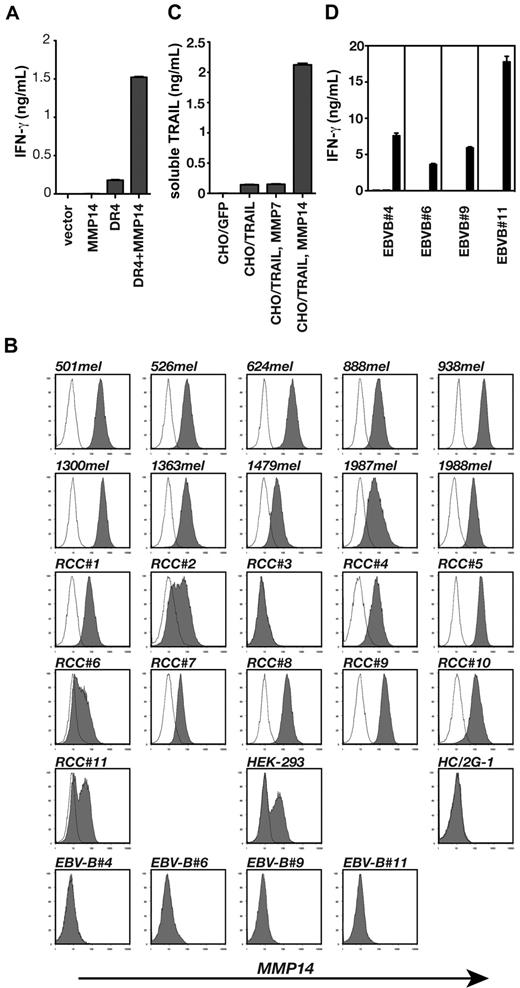
Despite identifying the multiple components that contribute to HC/2G-1 stimulation, it was still not clear why most RCC lines were recognized without the addition of exogenous TRAIL. In addition, EBV-B cells were not recognized by HC/2G-1 cells, but flow cytometric analyses showed that they express DR4 and CD58 at levels comparable with well-recognized RCC lines (data not shown). These observations indicated that there was still an unidentified component(s) that determines target cell sensitivity to HC/2G-1 T cells. As shown earlier, HEK-293 cells were recognized by HC/2G-1 when transduced with DR4, but the amount of IFN-γ secreted with HEK-293/DR4 in the assay (∼ 1 ng/mL) was not as high as against well-recognized RCC lines (∼ 5-10 ng/mL). We used HEK-293/DR4 as the transfection target for additional cDNA library expression screening, looking for missing components that would enhance IFN-γ release to maximal levels. This screening led to the identification of MMP14 or MT1-MMP as a gene that enhances HC/2G-1 reactivity when expressed in HEK-293 together with DR4 (Figure 6A). Flow cytometric analysis of MMP14 showed that most (10 of 11) RCC lines and melanoma lines (10 of 10) express MMP14, but EBV-B cells and HC/2G-1 cells do not (Figure 6B). Because MMP7 was known to cleave the Fas ligand (which, like TRAIL, belongs to the TNF family) to generate its soluble form,13 we investigated whether MMP14 could cleave membrane-bound TRAIL to release soluble TRAIL. CHO cells were transduced with both full-length, membrane-bound TRAIL and either MMP7 or MMP14. This resulted in the release of soluble TRAIL only with MMP14 expression (Figure 6C). More importantly, when EBV-B cells were transduced with MMP14 cDNA, they were recognized by HC/2G-1 (Figure 6D), indicating that the expression of a peptidase that can cleave TRAIL is needed for a cell to be recognized by HC/2G-1.
MMP14 on target cells releases soluble TRAIL and is the third component that determines tumor sensitivity to HC/2G-1 T cells. (A) HEK-293 cells were transiently transfected with GFP expression plasmid, MMP14 expression plasmid, or DR4-expressing plasmid as indicated, and their recognition by HC/2G-1 was examined by IFN-γ ELISA. (B) Flow cytometric analysis of MMP14 expression on various cell lines. (C) CHO cell lines expressing GFP, TRAIL, TRAIL with MMP7, or TRAIL with MMP14 were prepared by lentiviral transductions. Each cell line was plated into 96-well plates (100 000 cells/well) and after an overnight culture, soluble TRAIL in the supernatant was measured using ELISA. (D) EBV-B cell lines were lent, virally transduced with GFP (white bars), MMP7 (gray bars), or MMP14 (black bars). Their recognition by HC/2G-1 was examined by coculturing overnight and measuring IFN-γ in the supernatant using ELISA.
MMP14 on target cells releases soluble TRAIL and is the third component that determines tumor sensitivity to HC/2G-1 T cells. (A) HEK-293 cells were transiently transfected with GFP expression plasmid, MMP14 expression plasmid, or DR4-expressing plasmid as indicated, and their recognition by HC/2G-1 was examined by IFN-γ ELISA. (B) Flow cytometric analysis of MMP14 expression on various cell lines. (C) CHO cell lines expressing GFP, TRAIL, TRAIL with MMP7, or TRAIL with MMP14 were prepared by lentiviral transductions. Each cell line was plated into 96-well plates (100 000 cells/well) and after an overnight culture, soluble TRAIL in the supernatant was measured using ELISA. (D) EBV-B cell lines were lent, virally transduced with GFP (white bars), MMP7 (gray bars), or MMP14 (black bars). Their recognition by HC/2G-1 was examined by coculturing overnight and measuring IFN-γ in the supernatant using ELISA.
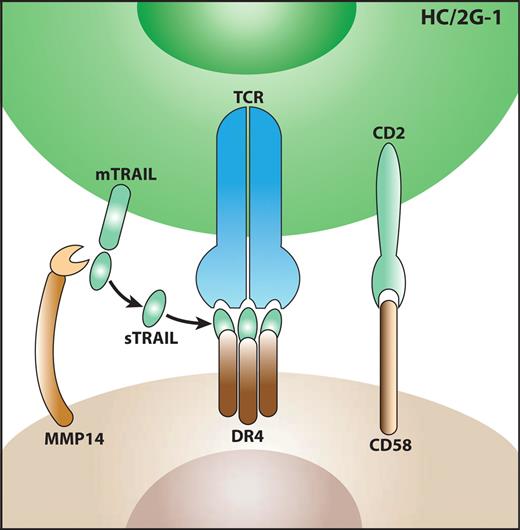
In summary, there are 3 crucial components that account for the target-recognition profile of HC/2G-1 cells: (1) CD58 needs to be expressed to participate in a cell-cell interaction, (2) a local source of soluble TRAIL is needed (such as MMP14-mediated cleavage from the T-cell membrane), and (3) DR4 is needed to form the DR4/TRAIL antigen complex (Figure 7).
Model of the target-recognition mechanism by HC/2G-1 T cells. First, HC/2G-1 and tumor cells form a cell-cell interaction through CD2 and CD58. Next, peptidases such as MMP14 expressed on tumor cells cleave membrane-bound TRAIL (mTRAIL) and release soluble TRAIL (sTRAIL). Then, sTRAIL binds to DR4 and is recognized by HC/2G-1 TCR.
Model of the target-recognition mechanism by HC/2G-1 T cells. First, HC/2G-1 and tumor cells form a cell-cell interaction through CD2 and CD58. Next, peptidases such as MMP14 expressed on tumor cells cleave membrane-bound TRAIL (mTRAIL) and release soluble TRAIL (sTRAIL). Then, sTRAIL binds to DR4 and is recognized by HC/2G-1 TCR.
Discussion
The elucidation of antigen processing and presentation and the role of MHC molecules was a seminal advance in our understanding of T-cell function. This paradigm of small peptides being processed from whole proteins, transported to compartments where they are loaded onto MHC molecules, and then exported to the cell surface where these complexes engage the receptors of T cells remains true for the majority of T cells in the peripheral repertoire. Variations on this mechanism have been described, with native molecules complexed with semi-invariant MHC-like presenting molecules, as is the case for some natural killer T (NKT) cells recognizing glycolipids bound to CD1d using a limited repertoire of TCR α or β chains.14 Other variations include α/β T cells interacting directly with Hfe, an MHC class Ib molecule without peptide-presenting ability,15 and MUC1, a tumor-specific cell-surface antigen composed of tandemly repeated peptide sequences.16 However, the crystal structures of these 2 cases have not been reported, and the detailed mechanism of their interaction is not known. In γ/δ T cells, crystal-structure analysis of the direct interaction between the γ/δ T-cell clone G8 TCR and the MHC class Ib molecule T22 has been resolved.17 In this case, TCR uses the recombined long δ chain CDR3 that is composed of 12 amino acids. In terms of the CDR3 length, TCR δ is generally longer than TCR β and is more similar to Igs, which is deduced to be the structural basis of the MHC-independent γ/δ T-cell receptor interaction with its ligands.18 In our case, the length of HC/2G-1 CDR3α/β is typical of TCRα/β (CDR3α = 9 residues; CDR3β = 8 residues), and the HC/2G-1 TCR is less likely to form an Ig-like structure. The crystallized structure of TRAIL bound to DR4 has not been described, but the structure of TRAIL bound to DR5 has been.19,20 Homotrimeric DR5 engages trimerized TRAIL with a tulip-like structure, which has no obvious similarities to the structures of minimal determinant peptides complexed with either MHC class I or MHC class II. A recent study by Scott-Browne et al proposed that 3 amino acids in the germline-encoded CDR2 regions of TCR β chains (βY46, βY48, and βE54) often contact MHC and control MHC-restricted thymic selection.21 These amino acids are also found frequently in the same positions in human TCR β chains. Of 48 functional TCR Vβ chains, βY46 is found in 28 chains, βY48 in 12 chains, and βD/E54 in 33 chains. There are 8 TCR Vβ chains that do not have any of these amino acids, and the TCR Vβ2 (TRBV20-1) used by HC/2G-1 is one of them. Extensive future structural studies will be needed to determine the nature of the binding of this TCR, but those await crystallization of the complete ligand.
The possibilities remain that this TCR may have been conventionally selected in the thymus by an unidentified cross-reactive antigen presented by MHC class I or class II or generated by a random mutation. However, the previously demonstrated lack of coreceptor dependence of the HC/2G-1 TCR3 and the broad correlation of function and target structure expression shown here suggest that other explanations of how this T cell found its way into the periphery may be required. Van Laethem et al proposed that the sequestration of the kinase Lck by CD4/8 coreceptors prevents the selection of non–MHC-restricted TCRs.2 This may not be relevant to HC/2G-1 cells, which are CD4+ restricted but not MHC restricted. Alternative possible mechanisms of the HC/2G-1 selection could be unrecognized interactions between CD4 and the TRAIL/DR4 complex or a mutation in Lck lowering its affinity to CD4 and resulting in the increase of free Lck available to TCR and the positive selection of HC/2G-1 cells. Our preliminary analysis on the HC/2G-1 Lck sequence by RT-PCR has shown that there are no mutations (data not shown), precluding the second possibility. Further study is needed to elucidate the selection mechanism of HC/2G-1–like cells.
The triggering complex that binds to the CDR of the HC/2G-1 TCR appears to be soluble TRAIL specifically bound to its receptor, DR4. Although there was a significant correlation between DR4 mRNA expression and recognition by HC/2G-1, the correlation between DR4 expression by FACS and tumor recognition was weak. This weak correlation could be explained as follows. First, it is not DR4 but the complex of DR4 and TRAIL that is recognized by the TCR. The abundance of this complex can be influenced by the expression of cell-surface MMP-14 blunting the high DR4 expression. Second, the sensitivity of ligand detection by TCR (1-10 molecules per cell22 ) surpasses the sensitivity of Ab-based FACS detection. In fact, by FACS, 888mel looks virtually negative for DR4 expression; however, when we constructed a cDNA library from 888mel and screened for a cDNA that conferred HC/2G-1 recognition to HEK-293, we rediscovered DR4 cDNA (data not shown). This strongly supports the role of DR4 in 888mel recognition by HC/2G-1. Third, the presence of positive and negative modulators such as DR5, DcR1, DcR2, and CD58 may further obscure this correlation.
TRAIL/Apo2L is better known as an effector molecule that is expressed on cytotoxic CD4+ T cells,23,24 NK cells,25-27 and NKT cells.28 It belongs to the TNF family, and its ability to induce apoptosis through DR4 and DR5 in tumor cells but not in normal cells has provoked interest as a cancer-therapeutic agent, using both recombinant TRAIL and agonist Abs to its receptors.29 The release of TRAIL is mediated by proteolysis, here demonstrated to be from RCC-bound MMP14. The native mechanisms for release of membrane-bound TRAIL were not previously known, but the similarities of this system with those of other TNF family members such as Fas and TNF suggest that this may be an important endogenous mechanism. However, MMP14 is probably not the only enzyme that releases soluble TRAIL, as exemplified by HC/2G-1 recognition of RCC#3, which does not express MMP14. Whether TRAIL can be released from other sources (such as coexisting NK and NKT cells) or by other mechanisms remains a subject of future study.
A CD58-CD2 interaction appears to play a crucial role in target recognition by HC/2G-1. Bachmann et al investigated the role of CD2 in T-cell activation and showed that CD2 signaling lowers the activation threshold of LCMV-reactive T cells.30 Because HC/2G-1 does not benefit from CD4 or CD8 interaction with MHC, CD2-CD58 may play this role.
One major question left unanswered is whether other T cells with HC/2G-1–like activity exist as a component of the normal T-cell repertoire. If they do, it would be unlikely that the primary function of these T cells is to eradicate RCC. In preliminary experiments, we found that transfecting cDNA of mitochondrial antiviral signaling protein (MAVS, also known as IPS-131 and VISA32 ) into HEK-293/DR4 cells strongly enhanced the recognition of these cells by HC/2G-1. We also found that the amount of mRNA for DR4 and MMP14 are up-regulated and the recognition by HC/2G-1 was enhanced when an RCC line was transfected with poly I:C to mimic a virus infection (data not shown). This observation may suggest that the activation of an innate virus-detection pathway results in changes in the expression of molecules such as DR4 and MMP14. This merger of innate and adaptive responses to viral infection is one possible hypothesis for the normal role of HC/2G-1–like cells.
In summary, a conventional-appearing α/β TCR has been shown to engage a radically different antigenic determinant, soluble TRAIL bound to its receptor DR4. A postulated immunologic synapse also uses CD2 and CD58 to engage T cells and target cells. With the T cell delivering its own ligand (TRAIL) in an inactive, membrane-bound form, a specific release mechanism is also required, here attributed to proteolysis by RCC-bound MMP14, although the involvement of other peptidases remains a possibility. Elucidation of an unprecedented target-recognition mechanism by one T-cell clone would not warrant a change in paradigm. However, this work broadens the realm of possibilities of moieties that specifically engage the CDRs of the α/β TCR and also suggests the need for additional, perhaps novel, mechanisms of thymic selection.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Steven A. Rosenberg for support and for critically reviewing the manuscript and Nicholas P. Restifo, Jack A. Bennink, Paul F. Robbins, Kannan Natarajan, Anastasia Tikhonova, Naomi Taylor, and Alfred Singer for review and discussion.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, Bethesda, MD.
National Institutes of Health
Authorship
Contribution: K.H. designed and executed the experiments and prepared the manuscript; Q.J.W. performed TCR modifications; T.I. provided discussions and experimental ideas; and J.C.Y. supervised the project and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of T.I. is Department of Dermatology, University of Yamanashi, Yamanashi, Japan.
Correspondence: Ken-ichi Hanada, 10 Center Dr, Bldg 10 CRC, Rm 3W-3840, Bethesda, MD 20892; e-mail: hanadak@mail.nih.gov.