In this issue of Blood, Isern and colleagues provide a comprehensive picture of the highly complex development of primitive erythroid cells from yolk sac over fetal liver to circulation at cellular and molecular resolution.1
Erythropoiesis is one of the best-studied processes in the whole field of cell biology. From the landmark determination of hemoglobin structure by Max F. Perutz,2 to work on globin locus control regions (LCRs),3 to the relatively recent discovery in 2005 of the Jak2 mutant V617F as a leading cause of erythrocytosis,4 the field has always remained active.
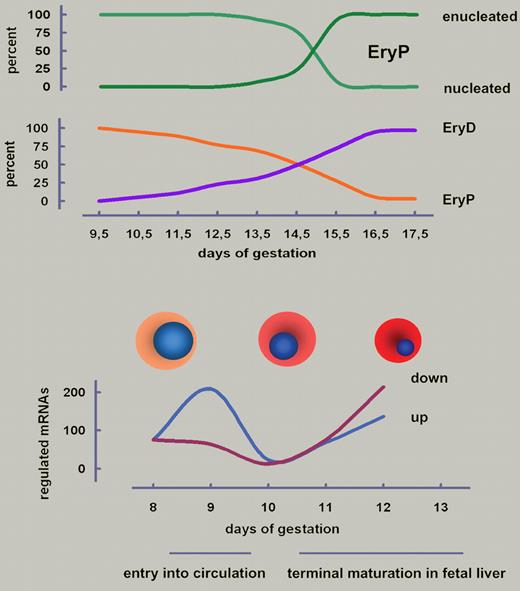
Development of primitive erythroid cells (EryPs) in the mouse. (Top) EryPs do enucleate but the whole process from onset of nuclear condensation at E10.5 to completed extrusion of nuclei at E15.5 is much slower than for definitive erythroid cells (EryDs; 2 days). At E14.5, approximately equal numbers of EryPs and EryDs are observed in embryonal blood. Data taken from Fraser et al.5 (Bottom) Entry of earliest EryPs into circulation is accompanied by a wave of gene activation. A second distinct wave of up- as well as down-regulation occurs during maturation in the fetal liver.1 Cell and nuclear diameters to scale.
Development of primitive erythroid cells (EryPs) in the mouse. (Top) EryPs do enucleate but the whole process from onset of nuclear condensation at E10.5 to completed extrusion of nuclei at E15.5 is much slower than for definitive erythroid cells (EryDs; 2 days). At E14.5, approximately equal numbers of EryPs and EryDs are observed in embryonal blood. Data taken from Fraser et al.5 (Bottom) Entry of earliest EryPs into circulation is accompanied by a wave of gene activation. A second distinct wave of up- as well as down-regulation occurs during maturation in the fetal liver.1 Cell and nuclear diameters to scale.
Despite all these successful efforts, there are still considerable gaps in our knowledge. One of these relates to primitive erythroid cells (EryP). Their emergence as nucleated progenitors in the yolk sac, developmental transitions toward coexistence with definitive erythrocytes (EryD) in the fetal liver and enucleated erythrocytes in the bloodstream, as well as their ultimate fate is incompletely understood.
The present study from Isern et al characterizes the life of EryPs in as-yet-unprecedented molecular detail, the culmination of years of intensive work.1 Their approach to open questions was facilitated by an elegant mouse model system, in which expression of green fluorescent protein (GFP) specifically in EryPs is driven by a human embryonic ϵ-globin gene promoter and micro-LCR.5,6 With this tool it could be confirmed that circulating EryPs (1) undergo synchronous maturation with nuclear condensation starting around embryonic day (E) 10.5, (2) exhibit a surprisingly slow enucleation lasting until ∼ E14.5, and (3) persist at least until after birth (see figure). Like EryDs, also EryPs require assistance of macrophages in erythroblastic islands within the fetal liver to get rid of their nuclei,6 sticking to central macrophages via various integrins and VCAM-1. (Note for completeness: adult late-stage erythroid maturation also involves such islands in bone marrow stroma.) EryPs develop from E7.5 on in the yolk sac and enter the earliest blood vessels as nucleated blasts at E9.5. EryDs emerge in fetal liver, but both, EryPs and EryDs, enucleate in this organ. Thus, the earliest circulating EryPs must have some homing capability that enables them to stay at least transiently in the fetal liver niche, a previously unrecognized feature.6
Isern and coworkers had already analyzed expression timelines and abundance levels for a number of factors characteristically associated with erythropoiesis, from cell surface markers like Ter119 or CD715 to cell adhesion molecules, to transcription factors like EKLF.7 Consequently, a full-blown genomics approach was the logical next step.
The current article by Isern et al describes the corresponding expression profiling and validation route taken and presents some unexpected results regarding necessary signaling and physiologic adaptations distinguishing EryP progenitors from EryPs from EryDs.1 Because there are no reliable surface markers for embryonic erythroid cells, isolation of the minute number of committed progenitors from an E7.5 embryo (∼ 200) was made possible through availability of the ϵ-globin-promoter-GFP transgenic mice and the fact that this promoter is already active in the yolk sac of E7.5 embryos, nicely shown in supplemental Video 1.1 After sorting for green cells, by clonogenic assays this cell population could be verified as true erythroid-committed progenitors that are already lost at E9.0 with onset of circulation and their movement to the embryo proper. To study the development of EryP progenitors to the ensuing EryPs, cell populations were isolated by FACS at daily intervals from E7.5 until E12.5, further sampling being precluded by nuclear condensation/enucleation.
The data from Illumina arrays showed that the number of significant changes in transcript abundance—up or down—peaked with entry of EryPs into circulation at E9 and the beginning of the fetal liver stage at E12. Hierarchical clustering demonstrated regulation of almost all usual suspects typical for progressive erythroid maturation, for example, induction of genes for heme synthesis or iron metabolism and down-regulation of mRNAs associated with DNA replication or ribosome biogenesis. Validation of array data by qPCR and flow cytometry was performed for an impressive battery of ∼ 20 mRNAs/proteins.
Contemporary profiling papers are made complete with functional assays, opening avenues for further studies. To pick out just 2 eventual future starting points, there were, for example, indications that the extremely rapid proliferation (4-hour doubling time) of EryP precursors in the yolk sac is linked to medium/high intensity of TGF-β1 signaling, maybe involving an autocrine loop. There are only a few reports on such an autocrine stimulation in hematopoiesis. The closest counterpart may be the development of very early chick embryo erythroid progenitors, whose proliferation is accelerated by TGF-β1 while withdrawal promotes differentiation.8 Given the pleiotropic effects of TGF-β on apoptosis, transformation, differentiation, inflammation, proliferation, etc, already this finding is worth a follow-up. A second observation can lead to an “ah, yes, of course” in hindsight. The yolk sac is an inherently hypoxic environment. Given the lack of circulation, oxygen supply depends on diffusion. Several genes from the profiling results, many involved in glucose metabolism, are known to be up-regulated by hypoxia. And indeed, in vitro cultivation of erythroid progenitors at low oxygen (2%-5%) kept these target genes highly active and proliferation rates high, whereas atmospheric oxygen levels resulted in down-regulation and smaller colonies. This may resemble what EryPs experience during entry into circulation, migration to fetal liver and adaptation to the new environment. To give the present picture on EryP development, further shading will, among other things, involve additional profiling or deep sequencing to further validate the genomics' side of the system. And in studying the most relevant novel targets, characterization of posttranscriptional mechanisms from mRNA stability/translational efficiency to protein modifications and degradation frequently is more demanding than their initial identification.
For obvious reasons, profiling studies in human hematopoiesis have so far mostly focused on disease phenotypes or individual developmental steps that were comparatively easy to follow. One recent study on global gene expression analysis of human erythroid progenitors from the CFU-E stage to late erythroblasts may serve as an example to this statement.9 In addition, other related studies were almost always concerned with definitive erythropoiesis. In this light, the extensive study of Isern et al will remain a highly valuable reference source for all groups interested in “normal” early erythropoiesis, whether human or mouse.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■