Abstract
Specific interactions of transcription factors (TFs) with their targets are crucial for specifying gene expression programs during cell differentiation. How specificity is maintained despite limited selectivity of individual TF-DNA interactions is not fully understood. RUNX1 TF is among the most frequently mutated genes in human leukemia and an important regulator of megakaryopoiesis. We used megakaryocytic cell lines to characterize the network of RUNX1 targets and cooperating TFs in differentiating megakaryocytes and demonstrated how dynamic partnerships between RUNX1 and cooperating TFs facilitated regulatory plasticity and specificity during this process. After differentiation onset, RUNX1 directly activated a large number of genes through interaction with preexisting and de novo binding sites. Recruitment of RUNX1 to de novo occupied sites occurred at H3K4me1-marked preprogrammed enhancers. A significant number of these de novo bound sites lacked RUNX motif but were occupied by AP-1 TFs. Reciprocally, AP-1 TFs were up-regulated by RUNX1 after 12-O-tetradecanoylphorbol-13-acetate induction and recruited to RUNX1-occupied sites lacking AP-1 motifs. At other differentiation stages, additional combinatorial interactions occurred between RUNX1 and its coregulators, GATA1 and ETS. The findings suggest that in differentiating megakaryocytic cell lines, RUNX1 cooperates with GATA1, AP-1, and ETS to orchestrate cell-specific transcription programs through dynamic TF partnerships.
Introduction
The RUNX transcription factors (TFs) are key regulators of cell lineage and differentiation in several important developmental pathways. They regulate transcription in a context-dependent manner through binding to the consensus core DNA sequence PyGPyGGT.1 RUNX1 functions as key regulator in embryonic and adult hematopoiesis.2 Consistent with its important roles, haploinsufficiency, resulting from heterozygous loss-of-function mutations, is associated with familial platelet disorder and predisposition to acute myeloid leukemia (FPD-AML).3,4 Sporadic heterozygous mutations in RUNX1 are also leukemogenic.5,6 RUNX1 resides on human chromosome 21, and chromosomal translocations involving RUNX1, including 8;21, 3;21, and 12;21, are among the most frequent leukemia-associated translocations.7 In addition, patients with Down syndrome (DS), the phenotypic manifestation of trisomy 21, have 500 fold-increased risk of developing acute megakaryoblastic leukemia (DS-AMKL/AML-M7) relative to normal persons.8
RUNX1 plays an important role in megakaryopoiesis, the process leading to production of megakaryocytes, the polyploid precursors of platelets.9,10 Megakaryocytes share a common precursor with erythrocytes known as the megakaryocyte erythroid progenitor, which gives rise to both megakaryocytic and erythroid lineages.9,10 Overexpression of RUNX1 in myeloid cell lines induces megakaryocytic differentiation,11,12 whereas induced Runx1 deficiency in bone marrow results in impaired megakaryocytic maturation and reduced blood platelet number (thrombocytopenia).13 Although the cellular differentiation stages of megakaryopoiesis are well characterized, the regulatory programs responsible for the implementation of this process are largely unknown, as are the global RUNX1-regulatory mechanisms and direct target genes that drive this differentiation process.
RUNX1, in conjunction with additional sequence-specific TFs, regulates hematopoietic cell differentiation programs through specific interaction with its target genes after developmental signals.14 In complex metazoan genomes, sequence recognition of binding site motifs by TFs is by itself not sufficient to discriminate bona fide binding sites from background genomic sequences. Hence, additional parameters, such as chromatin structure and interactions with cooperating TFs, determined the functionality of potential binding sites. In a typical scenario, only a fraction of the numerous potential TF-binding site motifs in the genome is occupied at a given state, and even smaller subsets directly regulate transcription. This flexible selectivity creates a dense network of TF-genome interactions, which is currently difficult to predict and/or understand. Most importantly, it is unclear how to discern functionally important TF-genome interactions from transient or spurious ones and hence define the interactions that play active role in transcriptional regulation.15 Protein-protein interactions between TFs that simultaneously engage DNA16 add another layer of complexity challenging our current understanding of transcriptional control.
Here we used 12-O-tetradecanoylphorbol-13-acetate (TPA)–treated K56217 and CMK cells to model megakaryocytic differentiation and to explore cell immediate response to a differentiation signal. We found that RUNX1 acts as an essential regulator of immediate gene expression and characterized its genome-wide occupancy profile before and after induction of differentiation. A combination of genome-wide chromatin immunoprecipitation (ChIP)–sequencing (ChIP-seq) occupancy and gene expression profiles was used to identify a subset of RUNX1 sites directly involved in regulatory response. Additional ChIP-seq and sequence analysis delineated the epigenomic landscape of H3K4me1/H3K27me3 and cooperating TFs that participate in RUNX1-mediated cell response to TPA. The data provide the first genome-wide profile of RUNX1-occupancy before and during megakaryocytic differentiation and revealed a set of functional target genes downstream to a complex landscape of numerous RUNX1-binding sites. The analysis elucidated how the limited sequence specificity of RUNX1 is diversified by the epigenomic makeup (H3K4me1 vs H3K27me3) and the binding landscape of RUNX1 cooperating TFs. It shows that RUNX1 and its partners act in a coordinated manner affecting gene expression outcome. The data suggest that stage-specific combinatorial interactions, in addition to epigenomic makeup, dynamically shape the transcriptional program during megakaryocytic differentiation.
Methods
Cells
K562 and CMK cells were maintained in RPMI medium supplemented with 10% fetal bovine serum (Invitrogen), 2mM l-glutamine, and penicillin/streptomycin at 37°C and 5% CO2. K562 cells were treated with 40nM TPA (Sigma-Aldrich) to induce megakaryocytic differentiation. For the generation of stable knockdown of RUNX1 in K562 cells (K562RUNX1-KD), RUNX1 pGIPZ lentiviral shRNAmir vector V2LHS_150257 (Open Biosystems; RHS4531-NM_001754) was transfected into K562 using the Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. For selection of RUNX1 knockdown cells, culture was supplemented with puromycin (2 μg/mL, Sigma-Aldrich) and medium replaced every 72 hours. Nonsilencing lentiviral shRNAmir vector (Open Biosystems) was used for negative controls.
Further information on immunoprecipitation and Western blot analysis of RUNX1 in cell lysates of K562, K562-TPA, and CMK cells as well as generation of primary fetal liver-derived megakaryocytes is included in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
ChIP-seq
ChIP was performed essentially as described.18 Briefly, cross-linked chromatin from approximately 108 K562 cells, before or 24 hours after treatment with TPA (40nM) or from approximately 108 cells CMK or 107 mature fetal liver-derived megakaryocytes, was prepared and fragmented to an average size of approximately 200 bp by 40 cycles of sonication (30 seconds each) in 15-mL tubes using the Bioruprtor UCD-200 sonicator (Diagenode). For immunoprecipitation, the following antibodies were added to 12 mL of diluted, fragmented chromatin: 30 μL of homemade anti-RUNX119 raised against the protein C-terminal fragment; antimonomethyl-histone H3 (Lys4) and anti-trimethyl-histone H3 (Lys27) (Millipore), anti-C-FOS (Santa Cruz Biotechnology), anti-FOS-B (Cell Signaling Technology), and anti-GATA1 (Abcam). Rabbit preimmune serum was used as control. DNA was purified using QIAquick spin columns (QIAGEN) and sequencing performed using Illumina genome analyzer IIx, according to the manufacturer's instructions. Two biologic repeats were conducted and separately sequenced with each cell line and/or physiologic condition. For ChIP-seq analysis, Illumina sequencing short reads (36 bp) were aligned to the human genome (hg18) using the Eland program (Illumina). Multiple reads were discriminated and coverage profile generated by elongating reads to 200 bp according to mapped strand. Coverage profile was analyzed in bins of 50 bp unless otherwise noted. Nonimmune serum ChIP was used to discard regions with higher than expected background coverage (> 6 mapped elongated reads).
Further information on ChIP quantitative polymerase chain reaction (PCR) and ChIP-seq data, validation by reporter construct transfection assays is included in supplemental data.
Microarray processing and analysis
RNA was isolated by EZ-RNA (Biologic Industries), according to the manufacturer's instructions. Purified RNA was reverse-transcribed, amplified, and labeled with Affymetrix GeneChip whole transcript sense target labeling kit. Labeled cDNA from TPA-treated or untreated K562 cells was analyzed using Affymetrix human exon ST 1.0 microarrays, according to the manufacturer's instructions. Microarrays were scanned using GeneChip scanner 3000 7G. Microarray data were normalized using dChip model-based expression. All microarray data are available in the GEO public database under accession number GSE24779.
Further information on gene expression assay by quantitative reverse-transcribed (RT)–PCR is included in supplemental data.
Analysis of genomic regions encompassing promoters and enhancers
Annotated transcription start sites (TSSs) were downloaded from the UCSC site (January 2010 version). Each genomic locus was associated with the nearest TSS (see Figures 2,Figure 3,Figure 4–5 for analysis). Loci at a distance of up to 3K were categorized as “promoter regions,” whereas loci at a distance between 3K and 200K were categorized as “enhancer regions.”
Distribution of RUNX1 genomic occupancy indicated numerous strong binding peaks under all tested conditions as well as a significant number of weaker ones (supplemental Figure 10). This behavior was even more pronounced when H3K4me1 profiles were considered. These observations and the notion that a flexible wide range of interaction specificities exist for certain transcription factors suggested that it would be impossible (or indeed undesirable) to apply a single universal definition of a RUNX-binding site. As the main goal of the analysis was to obtain data on the global behavior of RUNX1 and its potential cooperating TFs, we applied a simple coverage threshold to detect RUNX1 sites and candidate enhancers. Genomic regions with high binding coverage in the nonimmune serum control ChIP-seq experiments (n > 6) were discarded. Contiguous regions with high binding coverage were grouped together to form distinct binding sites or enhancer regions. The cut-off for RUNX1 was based on the top 0.05% of data in the K562 profile (weighted coverage > 13). The cutoffs for all other tracks were scaled proportionally to the number of reads in the track. The ChIP-seq analysis coverage statistics and derived cutoff values are shown in supplemental Table 3. Further information about analysis of ChIP-seq data are described in the supplemental data.
Results
RUNX1 expression in megakaryocytic cell lines
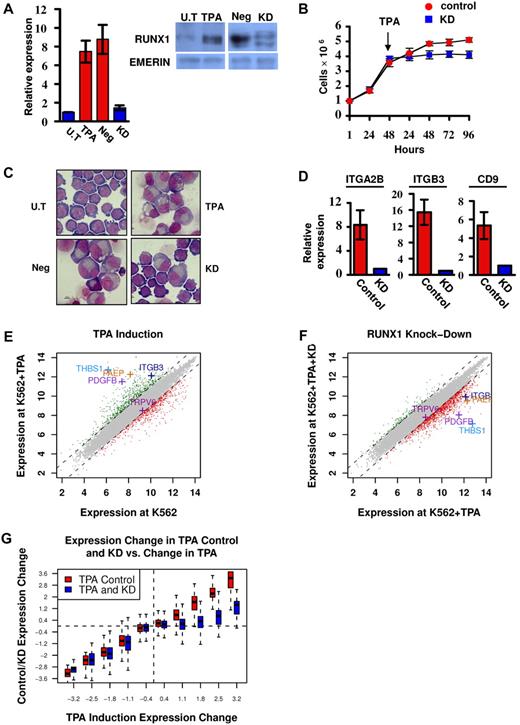
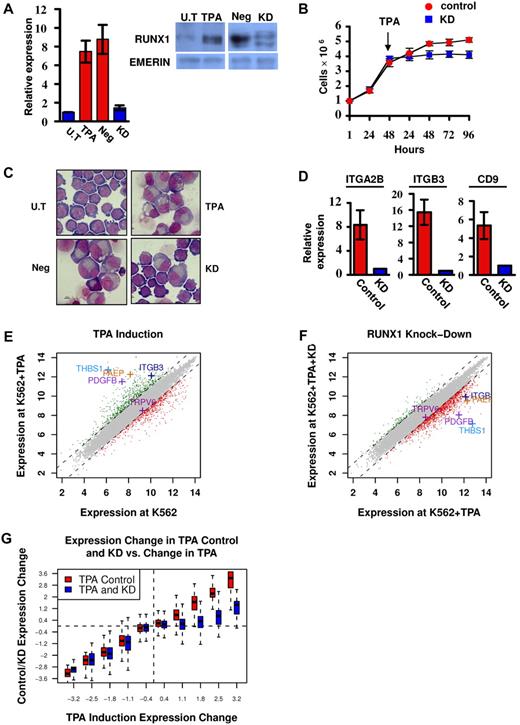
RUNX1 is highly expressed in megakaryocytic cell lines, including CMK and Meg01 (supplemental Figure 1A-B). In the multipotent cell line K562, RUNX1 expression was up-regulated on induction of megakaryocytic differentiation by TPA (Figure 1A) as was also observed by Elagib et al.12 These findings indicated that analysis of these cell lines under attenuated RUNX1 expression (Figure 1A) would furnish important information on the transcriptional program regulated by RUNX1 during megakaryopoiesis.
Phenotypic and gene expression response of K562 and K562Runx1KD cells to TPA treatment. (A) Quantitative RT-PCR and Western blot analysis of RUNX1 expression in untreated (U.T) and TPA-treated (TPA) cells as well as in TPA-treated cells stably expressing shRNA-miR (K562RUNX1KD), which knocked down RUNX1 expression (KD) compared with nonsilencing shRNA-miR control (Neg). Quantitative RT-PCR data are mean ± SD of 2 independent experiments performed in triplicates. Western blot of nuclear extract using anti-RUNX1 antibodies. Emerin was used as control of protein loading (supplemental data). (B) Proliferation assay using K562-TPA Neg (red) and KD (blue) cells. A total of 1 × 106 cells were grown in culture and counted every 24 hours. TPA was added 48 hours after seeding of cells. Data are mean ± SD of 2 independent experiments performed in triplicates. (C) Cell morphology changes. Representative microscopic images of May-Grunwald-Giemsa stained untreated (U.T), TPA, TPA Neg, and TPA KD K562 cells. Images were viewed with a Nikon Eclipse 800 microscope with a 100×/1.25 numeric aperture oil objective lens, captured using a Nikon DXM1200 digital camera, and processed with Nikon ACT-1 2.63 software. (D) Expression of megakaryocytic markers in K562 (Control) and K562RUNX1KD (KD) in response to TPA. Quantitative RT-PCR analysis of ITGA2B (CD41), ITGB3 (CD61), and CD9 expression in K562 cells. Data are mean ± SD of 2 independent experiments performed in quadruplicates. The decreased expression in TPA-treated K562RUNX1KD relative to K562 control was significant (P < .01) for all 3 markers. Primers used for quantitative RT-PCR assays are listed in supplemental Table 2. (E) TPA-induced transcriptional changes. Genes are plotted based on their expression level (log scale) in K562-TPA cells versus their basal level in untreated cells. Genes showing 2-fold (1 in log scale) increase or decrease in expression levels are indicated in green or red, respectively. Indicated are examples of up-regulated genes known to play an important role in megakaryocytic differentiation along with the reference TRPV6 gene. (F) RUNX1 knockdown impaired TPA-induced transcriptional activation. Gene expression levels in K562-TPA versus K562Runx1KD-TPA cells, stably expressing a TPA-responsive RUNX1 shRNA-miR, are shown in supplemental Table 1. Note the change in RUNX1-dependent expression of genes indicated in panel E. (G) Change in RUNX1-dependent transcriptional regulation in TPA-treated cells. Genes were divided into groups according to differential expression after TPA treatment. Shown are boxplots representing the expression change distribution in K562Runx1KD cells (blue) and K562CONTROL (red) for each group of genes. In RUNX1 expression KD, transcriptional repression (left section) did not change, whereas transcriptional activation (right section) was almost completely abolished.
Phenotypic and gene expression response of K562 and K562Runx1KD cells to TPA treatment. (A) Quantitative RT-PCR and Western blot analysis of RUNX1 expression in untreated (U.T) and TPA-treated (TPA) cells as well as in TPA-treated cells stably expressing shRNA-miR (K562RUNX1KD), which knocked down RUNX1 expression (KD) compared with nonsilencing shRNA-miR control (Neg). Quantitative RT-PCR data are mean ± SD of 2 independent experiments performed in triplicates. Western blot of nuclear extract using anti-RUNX1 antibodies. Emerin was used as control of protein loading (supplemental data). (B) Proliferation assay using K562-TPA Neg (red) and KD (blue) cells. A total of 1 × 106 cells were grown in culture and counted every 24 hours. TPA was added 48 hours after seeding of cells. Data are mean ± SD of 2 independent experiments performed in triplicates. (C) Cell morphology changes. Representative microscopic images of May-Grunwald-Giemsa stained untreated (U.T), TPA, TPA Neg, and TPA KD K562 cells. Images were viewed with a Nikon Eclipse 800 microscope with a 100×/1.25 numeric aperture oil objective lens, captured using a Nikon DXM1200 digital camera, and processed with Nikon ACT-1 2.63 software. (D) Expression of megakaryocytic markers in K562 (Control) and K562RUNX1KD (KD) in response to TPA. Quantitative RT-PCR analysis of ITGA2B (CD41), ITGB3 (CD61), and CD9 expression in K562 cells. Data are mean ± SD of 2 independent experiments performed in quadruplicates. The decreased expression in TPA-treated K562RUNX1KD relative to K562 control was significant (P < .01) for all 3 markers. Primers used for quantitative RT-PCR assays are listed in supplemental Table 2. (E) TPA-induced transcriptional changes. Genes are plotted based on their expression level (log scale) in K562-TPA cells versus their basal level in untreated cells. Genes showing 2-fold (1 in log scale) increase or decrease in expression levels are indicated in green or red, respectively. Indicated are examples of up-regulated genes known to play an important role in megakaryocytic differentiation along with the reference TRPV6 gene. (F) RUNX1 knockdown impaired TPA-induced transcriptional activation. Gene expression levels in K562-TPA versus K562Runx1KD-TPA cells, stably expressing a TPA-responsive RUNX1 shRNA-miR, are shown in supplemental Table 1. Note the change in RUNX1-dependent expression of genes indicated in panel E. (G) Change in RUNX1-dependent transcriptional regulation in TPA-treated cells. Genes were divided into groups according to differential expression after TPA treatment. Shown are boxplots representing the expression change distribution in K562Runx1KD cells (blue) and K562CONTROL (red) for each group of genes. In RUNX1 expression KD, transcriptional repression (left section) did not change, whereas transcriptional activation (right section) was almost completely abolished.
RUNX1 is a key gene expression regulator during megakaryocytic differentiation of K562 cells
K562 cells readily differentiate along the megakaryocytic lineage after TPA treatment,17 providing a well-characterized system for studying megakaryopoiesis.12 Treatment with TPA induced a profound decrease in cell proliferation (Figure 1B) and changes in cell morphology (Figure 1C), as was previously reported.17 These changes that were characteristic of megakaryocytic differentiation include increased cell size and cytoplasm-to-nucleus ratio, reduced basophilic staining of cytoplasm, appearance of lobulated nuclei (Figure 1C), and increase in the expression of megakaryocytic markers (Figure 1D). Of note, knockdown (KD) of RUNX1 in K562 cells (K562Runx1KD; Figure 1A) resulted in marked diminution of the TPA effect on proliferation, cell morphology, and expression of megakaryocytic markers (Figure 1B-D).
Gene expression analysis of K562 cells before and after TPA treatment (Figure 1E) revealed an extensive transcriptional response in the first 48 hours of treatment. Changes included repression of genes involved in growth-related pathways, such as ribosomal proteins and DNA synthesis, and induction of numerous genes in pathways involved in megakaryocytic differentiation (supplemental Table 1). Significantly, comparable analysis in K562Runx1KD cells (Figure 1F) showed that approximately 80% of these induced megakaryopoietic genes displayed low response to TPA in the absence of RUNX1 (Figure 1F-G). In contrast, KD of RUNX1 did not systematically compromise the repression of immediate TPA-responding genes (Figure 1G). These results identified a large set of TPA-responsive genes whose transcriptional regulation was RUNX1-dependent (supplemental Table 1) and established TPA-treated K562 versus K562Runx1KD cells as a unique system for analyzing the molecular events underlying RUNX1-mediated regulation during megakaryocytic differentiation in this cell line.
Induction of differentiation in K562 involves de novo recruitment of RUNX1 to a large number of genomic sites
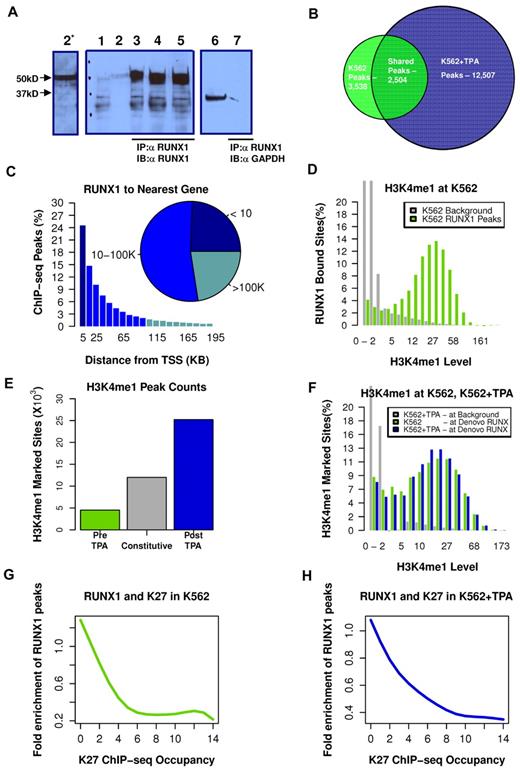
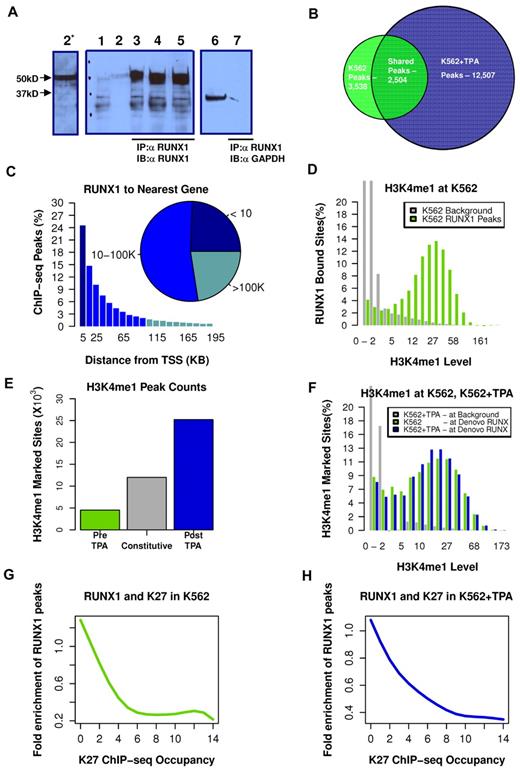
We used our highly specific anti-RUNX1 antibodies (Figure 2A; and supplemental data) in ChIP-seq experiments to map RUNX1 binding in K562 before and after TPA treatment. The genome-wide RUNX1-binding profiles were then combined with genome-wide mapping of enhancer/promoter regions by H3K4me1/H3K27me3 ChIP-seq analysis. Before induction of megakaryocytic differentiation, RUNX1 occupied several thousand loci (3532 permissive threshold sites). After induction, the original RUNX1-binding sites were largely preserved (Figure 2B; supplemental Figure 2). But in addition, a large number of de novo RUNX1 regions became occupied; increasing the number of RUNX1 bound sites by more than 3-fold, to a total of 12 507 bound sites (Figure 2B). These data support the finding that RUNX1 plays a pivotal role in regulating the TPA-induced transcriptional program in K562 cells.
TPA-dependent recruitment of RUNX1 to preprogrammed remote enhancer regions. (A) Validation of anti-RUNX1 antibody efficacy and specificity in immunoprecipitation (IP). IP of RUNX1 from CMK, K562, or K562-TPA whole cell lysates by the anti-RUNX1 antibodies used in the ChIP-seq experiments. Lane 1 indicates protein-A agarose beads (supplemental data) were incubated without cell lysate. Lane 2, Western blot using 50 μg of protein of cell lysate and anti-RUNX1 antibodies (α RUNX1). Lane 2*, Longer exposure of lane 2. Lanes 3 to 5, Western blot analysis of proteins immunoprecipitated from CMK (2), K562 (3), and K562-TPA cell lysates. Lane 6, Western blot analysis of CMK cell lysate using anti-GAPDH antibodies (α GAPDH). Lane 7, Western blot analysis, using anti-GAPDH antibodies, of proteins immunoprecipitated from CMK by anti-RUNX1 antibodies. (B) Venn diagram showing genome-wide RUNX1 occupancy in K562 and K562-TPA cells. RUNX1-binding peaks from ChIP-seq analysis (covered beyond a threshold and lacking significant coverage in control experiment) before and 24 hours after TPA treatment were compared. (C) Distribution of RUNX1 ChIP-seq peaks relative to the TSS. Shown are RUNX1 peak frequencies relative to the distance from the nearest annotated TSS. Although regions proximal to TSSs show increased RUNX1 binding, the majority of RUNX1 peaks (inset) were localized more than 10 kb from the nearest TSS. (D) Enrichment of RUNX1 binding in regions with open chromatin. Distributions of H3K4me1 ChIP-seq readouts at RUNX1 bound sites (light green) and at background regions (gray) demonstrate that RUNX1 binding was largely restricted to H3K4me1-marked remote enhancers. (E) Change in H3K4me1-marked regions after TPA induction. Histograms represent the numbers of ChIP-seq H3K4me1-marked genomic regions that were identified in both pre– and post–TPA-treated cells (constitutive, gray), in pre–TPA-treated cells (green), and in TPA-induced cells alone (blue). (F) De novo occupied RUNX1 sites are marked by H3K4me1 before TPA induction. Histograms represent the distributions of H3K4me1 ChIP-seq readouts before (light green) and after (blue) TPA in de novo occupied RUNX1 sites. The distribution of H3K4me1 levels at background regions (gray) served as a control. The data demonstrate that de novo RUNX1-occupied regions were marked open by H3K4me1 before TPA induction and before RUNX1 binding. (G-H) RUNX1 binding is negatively correlated with H3K27me3 ChIP-seq occupancy. A RUNX1 enrichment value is computed by dividing the frequency of RUNX1 peaks over the defined part of the genome by the RUNX1 peak frequency over the entire genome. Plotted are RUNX1 enrichment values as a function of H3K27me3 occupancy, before (G) and after (H) TPA induction.
TPA-dependent recruitment of RUNX1 to preprogrammed remote enhancer regions. (A) Validation of anti-RUNX1 antibody efficacy and specificity in immunoprecipitation (IP). IP of RUNX1 from CMK, K562, or K562-TPA whole cell lysates by the anti-RUNX1 antibodies used in the ChIP-seq experiments. Lane 1 indicates protein-A agarose beads (supplemental data) were incubated without cell lysate. Lane 2, Western blot using 50 μg of protein of cell lysate and anti-RUNX1 antibodies (α RUNX1). Lane 2*, Longer exposure of lane 2. Lanes 3 to 5, Western blot analysis of proteins immunoprecipitated from CMK (2), K562 (3), and K562-TPA cell lysates. Lane 6, Western blot analysis of CMK cell lysate using anti-GAPDH antibodies (α GAPDH). Lane 7, Western blot analysis, using anti-GAPDH antibodies, of proteins immunoprecipitated from CMK by anti-RUNX1 antibodies. (B) Venn diagram showing genome-wide RUNX1 occupancy in K562 and K562-TPA cells. RUNX1-binding peaks from ChIP-seq analysis (covered beyond a threshold and lacking significant coverage in control experiment) before and 24 hours after TPA treatment were compared. (C) Distribution of RUNX1 ChIP-seq peaks relative to the TSS. Shown are RUNX1 peak frequencies relative to the distance from the nearest annotated TSS. Although regions proximal to TSSs show increased RUNX1 binding, the majority of RUNX1 peaks (inset) were localized more than 10 kb from the nearest TSS. (D) Enrichment of RUNX1 binding in regions with open chromatin. Distributions of H3K4me1 ChIP-seq readouts at RUNX1 bound sites (light green) and at background regions (gray) demonstrate that RUNX1 binding was largely restricted to H3K4me1-marked remote enhancers. (E) Change in H3K4me1-marked regions after TPA induction. Histograms represent the numbers of ChIP-seq H3K4me1-marked genomic regions that were identified in both pre– and post–TPA-treated cells (constitutive, gray), in pre–TPA-treated cells (green), and in TPA-induced cells alone (blue). (F) De novo occupied RUNX1 sites are marked by H3K4me1 before TPA induction. Histograms represent the distributions of H3K4me1 ChIP-seq readouts before (light green) and after (blue) TPA in de novo occupied RUNX1 sites. The distribution of H3K4me1 levels at background regions (gray) served as a control. The data demonstrate that de novo RUNX1-occupied regions were marked open by H3K4me1 before TPA induction and before RUNX1 binding. (G-H) RUNX1 binding is negatively correlated with H3K27me3 ChIP-seq occupancy. A RUNX1 enrichment value is computed by dividing the frequency of RUNX1 peaks over the defined part of the genome by the RUNX1 peak frequency over the entire genome. Plotted are RUNX1 enrichment values as a function of H3K27me3 occupancy, before (G) and after (H) TPA induction.
Analysis of RUNX1 occupancy site locations, relative to the nearest TSSs of annotated genes, revealed that approximately 80% of RUNX1 bound sites were situated more than 5 kb away from any TSS (Figure 2C), and approximately 25% were in “gene deserts” (> 100 kb from the nearest TSS). The majority of de novo RUNX1-occupied sites are therefore either not functional or affect transcription through long-range promoter-enhancer interactions. The apparent plasticity and wide distribution of RUNX1 occupancy landscape suggested that RUNX1 regulates gene expression via multiple interactions with genomic chromatin and other transcriptional regulators.
RUNX1 is preferentially recruited to sites of preprogrammed open chromatin
H3K4me1 marks chromatin of genomic regions associated with enhancer activity.20 Using H3K4me1 ChIP-seq, we analyzed the chromatin landscape, before and after the massive recruitment of RUNX1 to de novo TPA-induced sites on switch-on of the differentiation program. In K562 cells, RUNX1 binding is largely confined to regions displaying H3K4me1 occupancy (Figure 2D). After induction of megakaryocytic differentiation, the genomic landscape of H3K4me1 regions expanded and changed, as a large group of loci (∼ 25 000) acquired de novo monomethylation at H3K4 (Figure 2E blue), but fewer lost their existing marks (Figure 2E green). A third group, designated “constitutive” was marked with H3K4me1 in both pre– and post–TPA-treated cells (Figure 2E gray). Importantly, the numerous de novo (postinduction) RUNX1-occupied sites belonged to this constitutively marked H3K4me1 group, sites that were already marked with H3K4me1 before induction (Figure 2F).
Analysis of H3K27me3 ChIP-seq readout in RUNX1 bound peaks indicated a general lack of overlap between RUNX1-occupied enhancers and the polycomb repressive histone marker (Figure 2G-H). Together, these results are consistent with the conclusion that RUNX1 recruitment, during the first 24 hours after induction, did not require extensive chromatin remodeling and that the newly engaged enhancers were actually accessible before induction but became occupied by RUNX1 only after the onset of the differentiation program.
Transcriptional activation of target genes is tightly correlated with RUNX1 recruitment to remote binding sites
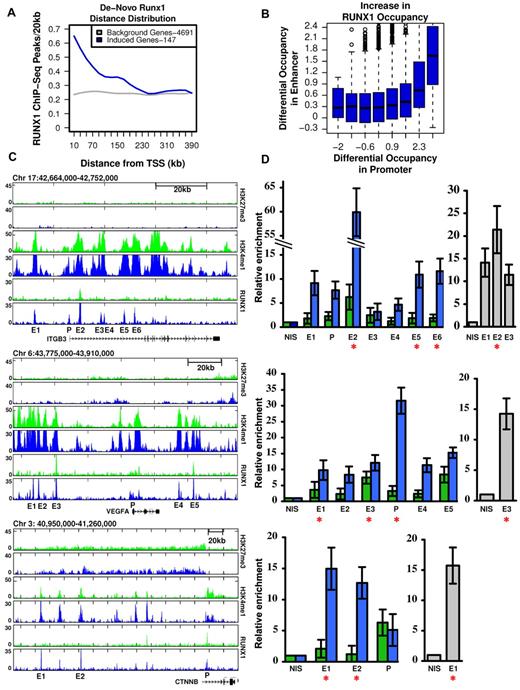
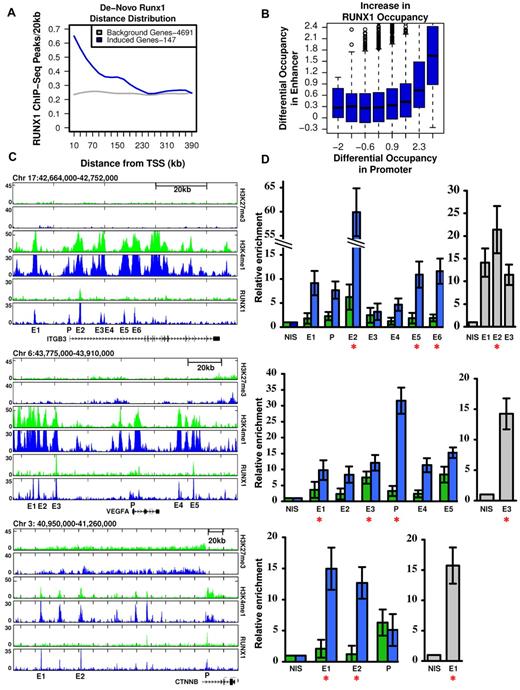
As shown in Figures 1 and 2, megakaryocytic differentiation of K562 cells was largely dependent on RUNX1-mediated gene expression (Figure 1) and was associated with a vast increase in de novo occupied RUNX1 sites (Figure 2B-C). This would argue that de novo RUNX1 bound genomic elements directly regulate TPA-induced genes. Consistent with this possibility, the stringently selected group of 147 genes (supplemental Table 1), which were TPA-induced and RUNX1-regulated, displayed a significant enrichment for de novo RUNX1 occupancy within 250 kb around their activated TSSs (Figure 3A). Interestingly, this defined subset of apparently direct RUNX1 targets (marked in supplemental Table 1) contained a preponderance of genes important for megakaryopoiesis. Together, the data establish a causal link between de novo RUNX1 occupancy and TPA induction of differentiation. Importantly, the finding that most (∼ 80%) of these de novo RUNX1 bound sites, in proximity to activated genes, were localized far away from the TSSs (Figures 2C, 3A), indicates that RUNX1 regulates its target genes primarily through long-range enhancer promoter interactions. Of note, a significant statistical dependency (Spearman = 0.07, P < 1.46e57) was observed between increased RUNX1 occupancy at gene promoters versus their surrounding enhancers (Figure 3B), underscoring the importance of remote enhancer-promoter interaction in RUNX1-mediated response to TPA.
RUNX1-dependent transcriptional activation is linked to remote de novo RUNX1 occupancy sites. (A) Enrichment of de novo RUNX1 binding in proximity of activated genes. The density (peaks/20 kb, y-axis) of de novo RUNX1 bound sites is plotted relative to TSSs for 147 bona fide RUNX1-regulated genes after TPA induction (blue) and for genes not activated by TPA (gray). (B) De novo binding of RUNX1 at long-range enhancer regions significantly correlates with its de novo binding at gene promoters. Log of maximal difference in RUNX1 ChIP-seq coverage before and after TPA within 3 kb (promoter region) and 200 kb (enhancer region) around the TSS was computed. Boxplots represent the distribution of differential RUNX1 occupancy at enhancers (y-axis) for groups of promoters with similar differential RUNX1 occupancy (x-axis). The outliers represent values more than 90th percentile. (C) Remote constitutive and de novo RUNX1-occupied sites and H3K4me1/H3K27me3 profile at key TPA-responsive RUNX1-regulated megakaryocytic gene loci. RUNX1, H3K4me1, and H3K27me3 ChIP-seq readouts, before (green) and after (blue) TPA treatment, in several genomic loci encompassing megakaryocytic important genes that were activated after TPA in a RUNX1-dependent manner. (D) Quantitative evaluation of RUNX1 ChIP-seq results in K562 and K562-TPA cells and murine primary megakaryocytes. Quantitative PCR analysis of RUNX1 binding to regions spanning ITGB3, VEGFA, and CTNNB indicated in panel C. Data are mean ± SD of 2 independent ChIP-quantitative PCR experiments performed in quadruplicates using K562 cells (green), K562-TPA cells (blue), and murine primary megakaryocytes (gray). Red asterisks represent regions containing RUNX motifs. Primers used for quantitative PCR assays are listed in supplemental Table 2, and details are described in supplemental data.
RUNX1-dependent transcriptional activation is linked to remote de novo RUNX1 occupancy sites. (A) Enrichment of de novo RUNX1 binding in proximity of activated genes. The density (peaks/20 kb, y-axis) of de novo RUNX1 bound sites is plotted relative to TSSs for 147 bona fide RUNX1-regulated genes after TPA induction (blue) and for genes not activated by TPA (gray). (B) De novo binding of RUNX1 at long-range enhancer regions significantly correlates with its de novo binding at gene promoters. Log of maximal difference in RUNX1 ChIP-seq coverage before and after TPA within 3 kb (promoter region) and 200 kb (enhancer region) around the TSS was computed. Boxplots represent the distribution of differential RUNX1 occupancy at enhancers (y-axis) for groups of promoters with similar differential RUNX1 occupancy (x-axis). The outliers represent values more than 90th percentile. (C) Remote constitutive and de novo RUNX1-occupied sites and H3K4me1/H3K27me3 profile at key TPA-responsive RUNX1-regulated megakaryocytic gene loci. RUNX1, H3K4me1, and H3K27me3 ChIP-seq readouts, before (green) and after (blue) TPA treatment, in several genomic loci encompassing megakaryocytic important genes that were activated after TPA in a RUNX1-dependent manner. (D) Quantitative evaluation of RUNX1 ChIP-seq results in K562 and K562-TPA cells and murine primary megakaryocytes. Quantitative PCR analysis of RUNX1 binding to regions spanning ITGB3, VEGFA, and CTNNB indicated in panel C. Data are mean ± SD of 2 independent ChIP-quantitative PCR experiments performed in quadruplicates using K562 cells (green), K562-TPA cells (blue), and murine primary megakaryocytes (gray). Red asterisks represent regions containing RUNX motifs. Primers used for quantitative PCR assays are listed in supplemental Table 2, and details are described in supplemental data.
This finding raised the possibility that some of the identified RUNX1 promoter-occupancy sites resulted from initial binding at remote enhancers followed by chromosomal looping.21,22 This interpretation is illustrated by ChIP-seq readouts of several TPA-induced RUNX1-regulated megakaryocytic genes encompassing remote newly occupied RUNX1-binding sites spanning H3K4me1-rich H3K27me3-poor regions (Figure 3C). RUNX1 occupancy along the regions shown in Figure 3C was further evaluated using quantitative ChIP-PCR. RUNX1 binding was detected in all ChIP-seq peaks tested (Figure 3D), although it was higher at sites containing RUNX motifs. Moreover, Runx1 binding to several homologous mouse regions was detected by quantitative ChIP-PCR using fetal liver-derived murine megakaryocytes (Figure 3D).
Sequence specificity of RUNX1 occupancy sites
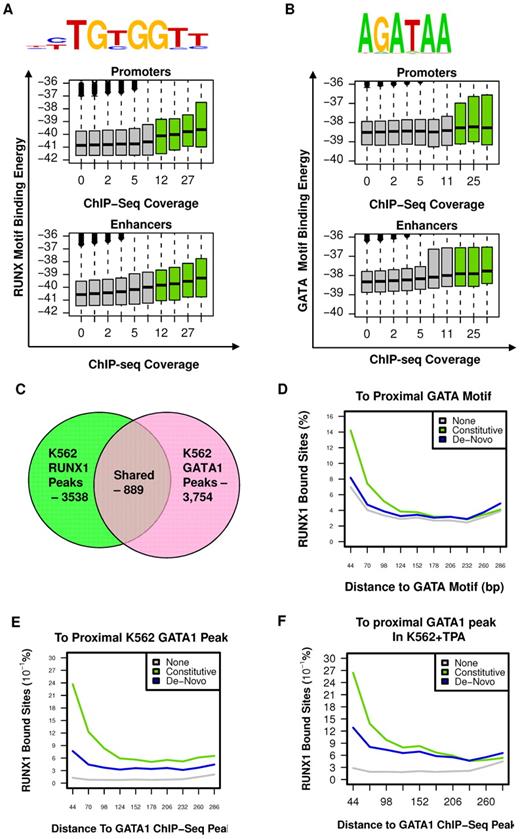
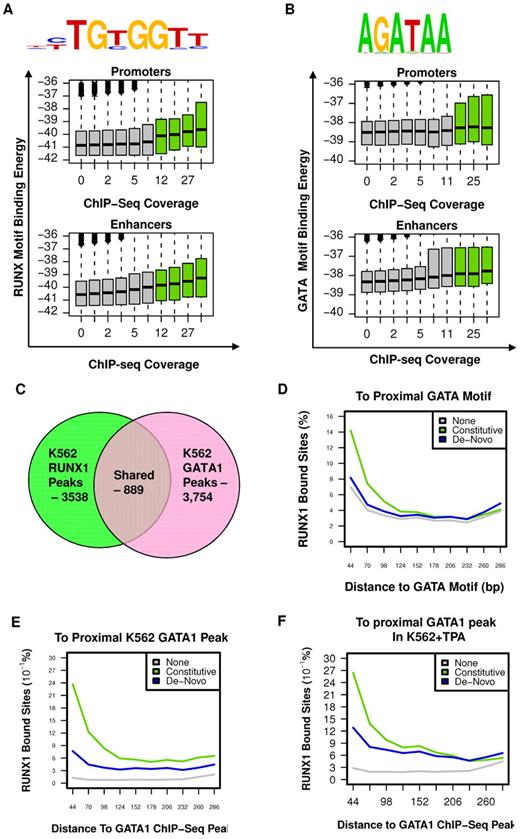
To characterize the sequence specificity of constitutive RUNX1 occupancy sites, we searched for DNA sequence motifs, within RUNX1 bound regions, before TPA treatment, compared with a background set consisting of H3K4me1-enriched regions lacking RUNX1 occupancy (for details, see supplemental data). This analysis confirmed the existence of a RUNX motif, which specified RUNX1 binding to a unique subset of enhancer and promoter elements distinguished from background enhancers (Figure 4A). Interestingly, although this RUNX1-specific motif was highly significant (P < 10−53), it was detected in only approximately 40% of occupied sites, when setting the background motif percentage at 5%. This occurred even when RUNX1 sites were selected from the H3K4me1-marked enhancers rather than considering the entire genome. Such loose specificity, previously found for other mammalian TFs,23-25 suggested that additional sequences and/or cofactors are required to specify RUNX1 binding. On the other hand, analysis of the correspondence between predicted RUNX1-binding potential (binding energy, see supplemental Methods) and the actual level of RUNX1 ChIP-seq in vivo revealed a weak but statistically significant correlation (Pearson = 0.08, P < 10−300, Figure 4A), even for RUNX motifs of less than optimal sequence. This wide pattern of correlation suggested that weaker, suboptimal RUNX motifs were still playing a role in specifying RUNX-binding sites, probably in cooperation with additional TF motifs. Interestingly, the correlation between RUNX sequence motif-binding energy and actual RUNX1 binding was weaker at promoter regions than at enhancer regions (in promoters: Pearson = 0.06, P < 10−137; in enhancers: Pearson = 0.09, P < 10−300), supporting the thesis that some of the reported RUNX1 promoter occupancy resulted from chromatin looping.
RUNX1 and GATA1 are over-represented at constitutive RUNX1-occupied regions. (A) RUNX motif-binding energy is correlated with RUNX1 ChIP-seq occupancy. Indicated RUNX motif was inferred directly from RUNX1 occupancy peaks (see supplemental Methods). Boxplots represent the distributions of motif-binding energies (y-axis) in groups of regions with increasing RUNX1 ChIP-seq coverage (x-axis). Only regions marked with H3K4me1 were considered. The regions that passed the threshold and were thus defined as RUNX1 bound peaks are shown in green boxplots. Data are plotted separately for promoters (< 3 kb from TSS, top panel) and enhancers (> 3 kb from TSS, bottom). (B) The GATA binding motif is correlated with RUNX1 ChIP-seq occupancy. A GATA motif was inferred directly from RUNX1 constitutively bound peaks. The correspondence between RUNX1 occupancy and GATA motif-binding energies is presented as in panel A (but using a GATA motif model instead of a RUNX motif model). Data reveal a correlation between RUNX1 ChIP-seq occupancy and the intensity of GATA motifs. Boxplots represent the distributions of motif-binding energies (y-axis) in groups of H3K4me1-marked regions with increasing RUNX1 ChIP-seq readout coverage (x-axis). (C) Venn diagram summarizing the overlap between RUNX1 and GATA1 bound sites in K562. (D-F) Co-occurrence analysis of RUNX1 and GATA1. Shown are distributions of distances from RUNX1 bound sites to the nearest GATA motif (D), to the nearest GATA1 ChIP-seq peak in K562 cells (E), and to the nearest GATA1 ChIP-seq peak in K562-TPA cells (F). Distance distributions were computed separately for constitutively occupied RUNX1 sites (light green), de novo occupied RUNX1 sites (blue), or RUNX1 motifs in H3K4me1-marked regions without significant RUNX1 occupancy (gray).
RUNX1 and GATA1 are over-represented at constitutive RUNX1-occupied regions. (A) RUNX motif-binding energy is correlated with RUNX1 ChIP-seq occupancy. Indicated RUNX motif was inferred directly from RUNX1 occupancy peaks (see supplemental Methods). Boxplots represent the distributions of motif-binding energies (y-axis) in groups of regions with increasing RUNX1 ChIP-seq coverage (x-axis). Only regions marked with H3K4me1 were considered. The regions that passed the threshold and were thus defined as RUNX1 bound peaks are shown in green boxplots. Data are plotted separately for promoters (< 3 kb from TSS, top panel) and enhancers (> 3 kb from TSS, bottom). (B) The GATA binding motif is correlated with RUNX1 ChIP-seq occupancy. A GATA motif was inferred directly from RUNX1 constitutively bound peaks. The correspondence between RUNX1 occupancy and GATA motif-binding energies is presented as in panel A (but using a GATA motif model instead of a RUNX motif model). Data reveal a correlation between RUNX1 ChIP-seq occupancy and the intensity of GATA motifs. Boxplots represent the distributions of motif-binding energies (y-axis) in groups of H3K4me1-marked regions with increasing RUNX1 ChIP-seq readout coverage (x-axis). (C) Venn diagram summarizing the overlap between RUNX1 and GATA1 bound sites in K562. (D-F) Co-occurrence analysis of RUNX1 and GATA1. Shown are distributions of distances from RUNX1 bound sites to the nearest GATA motif (D), to the nearest GATA1 ChIP-seq peak in K562 cells (E), and to the nearest GATA1 ChIP-seq peak in K562-TPA cells (F). Distance distributions were computed separately for constitutively occupied RUNX1 sites (light green), de novo occupied RUNX1 sites (blue), or RUNX1 motifs in H3K4me1-marked regions without significant RUNX1 occupancy (gray).
GATA motif enrichment and GATA1/RUNX1 co-occupancy at constitutive RUNX1 bound sites
Before induction of megakaryocytic gene expression program by TPA (Figure 1), RUNX1 was bound at 3538 genomic sites of which at least 2504 were also occupied after TPA treatment (Figure 2B) and were stringently defined as constitutively occupied regions. Sequence analysis of these regions revealed significant enrichment for GATA box motifs (Figure 4B, P < 1e−24). Genome-wide analysis of distance distribution of RUNX-GATA motifs revealed that at constitutively occupied regions the 2 motifs were coupled, whereas in de novo RUNX1 bound regions this coupling was weak (Figure 4C). This significant association between RUNX1-GATA at constitutively occupied RUNX1 regions was confirmed by analysis of previously published26,27 GATA-1 ChIP-seq data in K562 cells. It showed that 25% of RUNX1 bound regions were co-occupied by GATA1 (Figure 4D-E; supplemental Figure 3A).
This latter finding was in clear contrast to the limited co-occurrence of RUNX1 and GATA1 bound sites revealed by GATA1 ChIP-seq analysis of TPA-treated K562 cells (Figure 4F). The ChIP-seq data were further confirmed by independent ChIP-PCR analysis of RUNX1 and GATA1 on several RUNX1 target genes (supplemental Figure 3B), indicating lack of GATA1 binding at de novo occupied regions. The co-occurrence of RUNX and GATA motifs at constitutively bound RUNX1 sites and the ChIP-seq co-occupancy of GATA1 and RUNX1 before TPA treatment strongly indicated that combination of the 2 TFs plays a role in early stages of the differentiating program. Supporting this idea are the findings that RUNX1 and GATA-1 have an essential role in megakaryopoiesis6,13,28-31 and functionally cooperate in this process.12,14 It suggests that RUNX1 modulates the broad regulatory activity spectrum of GATA-1, known to regulate development of other hematopoietic lineages.27,32-34
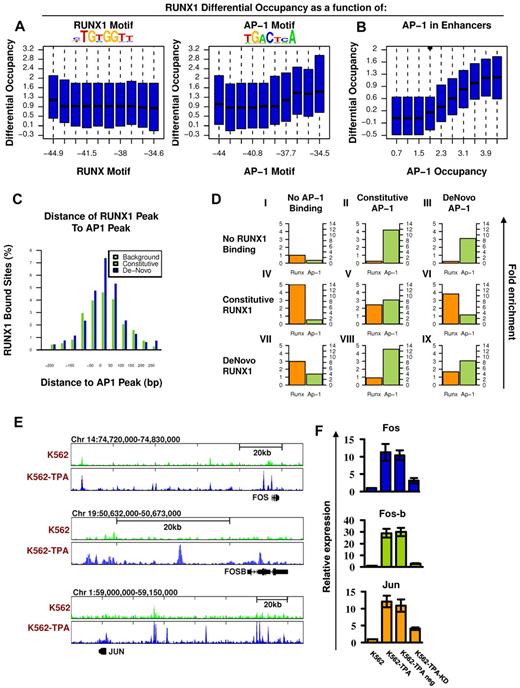
AP-1 motifs enrichment and AP-1/RUNX1 co-occupancy at de novo RUNX1 bound sites
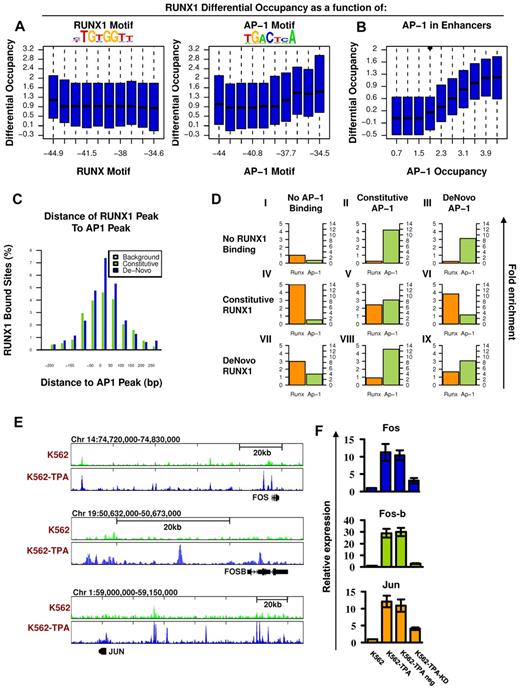
To further analyze the genomic characteristics underlying RUNX1 recruitment after TPA treatment, we examined the sequence compositions composed of de novo RUNX1 occupancy regions. Motif analysis revealed that, although RUNX motif was enriched at de novo occupied regions, the motif alone was not sufficient to distinguish them from the similarly enriched constitutive sites. On the other hand, the analysis revealed a highly specific enrichment of the AP-1 motif (TGACTCA) at the de novo RUNX1 sites (Figure 5A). Moreover, the estimated binding energy at the AP-1 motifs was positively correlated with differential ChIP-seq occupancy of RUNX1 in TPA-treated versus nontreated cells (Figure 5A right), in contrast to the lack of such correlation to the binding energy of the RUNX motif itself (Figure 5A left).
Co-occurrence of RUNX1 and AP-1 at de novo RUNX1 bound sites and RUNX1-mediated increase of AP-1 in response to TPA. (A) RUNX1 and AP1 motifs were inferred directly from de novo RUNX1 bound sites. Distributions of differential (after vs before TPA treatment) RUNX1 ChIP-seq readouts are plotted for groups of regions with increasing binding energy of RUNX (left) or AP-1 (right) motifs. Unlike RUNX motifs, AP-1 motifs are significantly correlated with differential (± TPA) RUNX1 occupancy. (B) Correlation between differential occupancy of RUNX1 and AP-1 occupancy. Shown are the distributions of increased RUNX-1 occupancy (y-axis; after vs before TPA treatment) in groups of loci with range of AP-1 occupancy level after TPA treatment (x-axis). AP-1 occupancy is a good predictor for TPA-induced RUNX1 recruitment. (C) Co-occurrence of AP-1 and RUNX1 ChIP-seq peaks at de novo RUNX1-occupied sites. Shown are the distributions of distances between RUNX1 peaks and most proximal AP-1 peaks at constitutively (light green) or de novo (+TPA blue) RUNX1 sites. For a reference, the distribution of distances for the nearest AP-1 sites from random H3K4me1 enhancers lacking RUNX1 binding is presented (gray). (D) Combinatorial analysis of RUNX1 and AP-1 sites and motifs. The relative fold enrichments of RUNX (orange) and AP-1 (green) motifs relative to the genomic background are shown for groups I to IX of loci with various combinatorial ChIP-seq readouts of the 2 factors. Constitutive sites are those observed in K562 cells both before and after TPA treatment. De novo sites are those observed only after TPA treatment. The pattern represented in groups VI and VIII suggest that RUNX1 occupancy enables AP-1 recruitment to regions lacking AP-1 motif; and vice versa, AP-1 binding enables recruitment of RUNX1 to sites lacking RUNX motifs. (E) TPA-dependent binding of RUNX1 to 3 AP-1 genes. RUNX1 ChIP-seq tracks at loci encompassing FOS, FOSB, and JUN before (light green) and after (blue) TPA, in K562 cells. De novo RUNX1 bound sites in remote enhancers are noted. (F) Quantitative RT-PCR analysis of FOS, FOS-B, and JUN expression in K562 cells. Data are mean ± SD of 3 independent experiments performed in triplicates. The increased expression of FOS, FOS-B, and JUN in K562-TPA relative to K562 and in K562-TPA with RUNX1 knockdown control (TPA neg) relative to K562 KD was significant (K562 ± TPA, P = .01, .005, and .005; and K562 ± KD, P = .01, .001, and .01).
Co-occurrence of RUNX1 and AP-1 at de novo RUNX1 bound sites and RUNX1-mediated increase of AP-1 in response to TPA. (A) RUNX1 and AP1 motifs were inferred directly from de novo RUNX1 bound sites. Distributions of differential (after vs before TPA treatment) RUNX1 ChIP-seq readouts are plotted for groups of regions with increasing binding energy of RUNX (left) or AP-1 (right) motifs. Unlike RUNX motifs, AP-1 motifs are significantly correlated with differential (± TPA) RUNX1 occupancy. (B) Correlation between differential occupancy of RUNX1 and AP-1 occupancy. Shown are the distributions of increased RUNX-1 occupancy (y-axis; after vs before TPA treatment) in groups of loci with range of AP-1 occupancy level after TPA treatment (x-axis). AP-1 occupancy is a good predictor for TPA-induced RUNX1 recruitment. (C) Co-occurrence of AP-1 and RUNX1 ChIP-seq peaks at de novo RUNX1-occupied sites. Shown are the distributions of distances between RUNX1 peaks and most proximal AP-1 peaks at constitutively (light green) or de novo (+TPA blue) RUNX1 sites. For a reference, the distribution of distances for the nearest AP-1 sites from random H3K4me1 enhancers lacking RUNX1 binding is presented (gray). (D) Combinatorial analysis of RUNX1 and AP-1 sites and motifs. The relative fold enrichments of RUNX (orange) and AP-1 (green) motifs relative to the genomic background are shown for groups I to IX of loci with various combinatorial ChIP-seq readouts of the 2 factors. Constitutive sites are those observed in K562 cells both before and after TPA treatment. De novo sites are those observed only after TPA treatment. The pattern represented in groups VI and VIII suggest that RUNX1 occupancy enables AP-1 recruitment to regions lacking AP-1 motif; and vice versa, AP-1 binding enables recruitment of RUNX1 to sites lacking RUNX motifs. (E) TPA-dependent binding of RUNX1 to 3 AP-1 genes. RUNX1 ChIP-seq tracks at loci encompassing FOS, FOSB, and JUN before (light green) and after (blue) TPA, in K562 cells. De novo RUNX1 bound sites in remote enhancers are noted. (F) Quantitative RT-PCR analysis of FOS, FOS-B, and JUN expression in K562 cells. Data are mean ± SD of 3 independent experiments performed in triplicates. The increased expression of FOS, FOS-B, and JUN in K562-TPA relative to K562 and in K562-TPA with RUNX1 knockdown control (TPA neg) relative to K562 KD was significant (K562 ± TPA, P = .01, .005, and .005; and K562 ± KD, P = .01, .001, and .01).
In addition, co-occurrence analysis revealed a significant coupling between AP-1 and RUNX motifs at de novo RUNX1 occupancy regions (supplemental Figure 4) and between AP-1 motif and RUNX1 binding to regions lacking proximal RUNX motif (supplemental Figure 5). Importantly, using ChIP-seq, we also demonstrated RUNX1/AP-1 co-occupancy of the FOS AP-1 component in K562-TPA cells (supplemental Figure 4), found that FOS ChIP-seq peaks were highly enriched with an AP-1 motif (supplemental Figure 6), and confirmed their significant co-occurrence with RUNX1 sites (Figure 5B). All in all, after induction, FOS occupancy was found to be in high correlation with RUNX1 recruitment (Figure 5B-C).
Next, we explored the nature of RUNX1/AP-1 co-occupancy by analyzing the relations between RUNX1 and AP-1 binding and their DNA motifs. For this purpose, we used a dataset combining the ENCODE-derived cFOS ChIP-seq occupancy in untreated K56226 and our FOS and FOSB ChIP-seq data in K562-TPA cells (Figure 5D). RUNX1 and AP-1 bound sites were highly enriched for their respective motifs. However, de novo RUNX1 bound sites lack RUNX motifs when recruited to constitutive AP-1 sites (group VIII in Figure 5D, only 5% have the motif compared with 25% of the stand-alone RUNX1 sites). Conversely, de novo AP-1 bound sites have a marked reduction in AP-1 motifs when recruited to constitutive RUNX1 sites (group VI in Figure 5D, 20% have the motif compared with 50% in stand-alone sites). In joint AP-1/RUNX1-binding sites (either constitutive or de novo), both motifs are enriched but to a lesser degree. According to this analysis, RUNX1 and AP-1 are capable of recruiting each other to target sites. This conclusion is supported by finding that the 2 TFs physically interact.16 After TPA induction, levels of both TFs increased and facilitated de novo recruitment of AP-1/RUNX1 complexes to H3K4me1-marked sites, either new or previously occupied by only one of them (supplemental Figure 11).
Interestingly, the finding that after TPA treatment RUNX1 bound to H3K4me1-marked regions upstream of FOS, FOS-B, and JUN (Figure 5E) and up-regulated their expression (Figure 5F; supplemental Table 1) raised the possibility that TPA induction triggered a regulatory cascade in which RUNX1 up-regulated AP-1 expression, thereby facilitating recruitment of RUNX1-AP-1 modules to a new set of target genes.
Enrichment of ETS TF motif proximal to RUNX1 bound sites in CMK cells
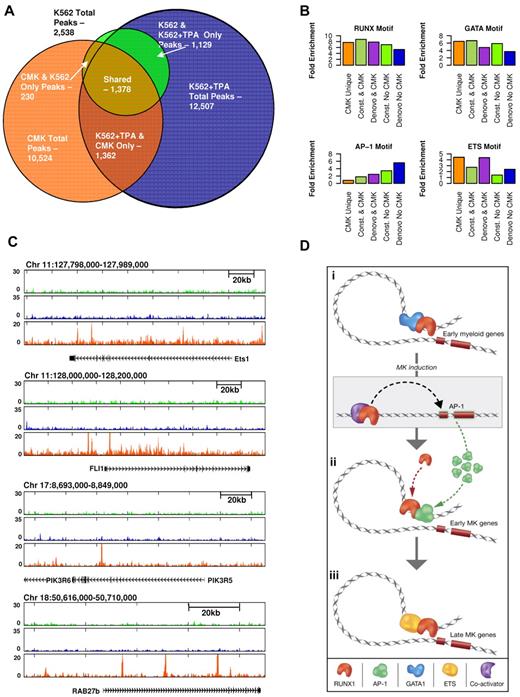
The commonly used megakaryoblastic cell line CMK35,36 is considered more differentiated than K562 as it expresses late markers of megakaryocytes and platelet differentiation.35,37 Using this cell line, we used RUNX1 ChIP-seq to further address the plasticity of RUNX1 occupancy during megakaryopoiesis. Analysis revealed a substantial overlap between sites bound by RUNX1 in CMK and K562 cells but also identified a significant number (∼ 7000) of CMK-specific RUNX1-occupied sites (Figure 6A; supplemental Figure 7). Sequence analysis revealed ETS TF motifs in close proximity to CMK-specific RUNX1 bound sites, in clear distinction from the K562 sites (Figure 6B). ETS family members were previously shown to cooperate with RUNX1.38-41 Interestingly, analysis of RUNX1 occupancy patterns in loci of several genes expressed in CMK, revealed differential binding of RUNX1 to 2 ETS TFs (ETS1 and FLI1) in CMK compared with K562 cells (Figure 6C). Differential binding of RUNX1 in proximity to PIK3R5/6 and RAB27b genes was also noted (Figure 6C). These genes are known to play role in late stages of megakaryopoiesis and platelet formation,42,43
Proximity of RUNX1-occupied sites to ETS motifs in CMK cells and schematic illustration of regulatory interplay between RUNX1 and cooperating TFs. (A) Venn diagram showing the relationships btween RUNX1 ChIP-seq occupancy profiles in K562, K562-TPA, and CMK cells. A substantial number (∼ 7000) of RUNX1 bound sites are CMK specific. (B) RUNX1 occupancy profile at several CMK specific loci. Shown are RUNX1 occupancy profiles in K562 cells before (light green) and after (blue) TPA, and in CMK cells (orange), at loci spanning the ETS1, FLI1, PIK3R6, and RAB27b genes. (C) A multimodal RUNX1 occupancy landscape of distinct TF motif combinations characterizing the megakaryopoietic gene expression program. RUNX1 bound sites in K562, K562-TPA, and CMK were grouped according to their co-occurrence with motifs of RUNX, GATA, AP-1, and ETS. Histograms show percentage of occurrence of motifs with binding energies in the top 5% of background. ETS motifs are highly prevalent in CMK cells, whereas the AP-1 motif is clearly biased to de novo occupied TPA-induced sites. (D) A schematic model summarizing our hypothesis about stage-specific RUNX1-mediated regulation. RUNX1 (orange crescent) is preferentially bound to remote enhancers and cooperates with GATA1 (blue cluster) to regulate early myeloid genes (orange rectangles). On induction, RUNX1 recruits coactivators (purple crescent) to activate the AP1 genes. Thereafter, AP1 TFs (green clusters) facilitate the binding of RUNX1 to early megakaryocytic genes, thereby launching and driving the differentiation program. At subsequent differentiation stage (CMK), RUNX1 cooperates with ETS family TFs (yellow ellipses) to activate a different set of megakaryocytic genes. This scenario underscores the notion that RUNX1 functions in a context-dependent manner to regulate the transcriptional program in differentiating megakaryocytic cell lines.
Proximity of RUNX1-occupied sites to ETS motifs in CMK cells and schematic illustration of regulatory interplay between RUNX1 and cooperating TFs. (A) Venn diagram showing the relationships btween RUNX1 ChIP-seq occupancy profiles in K562, K562-TPA, and CMK cells. A substantial number (∼ 7000) of RUNX1 bound sites are CMK specific. (B) RUNX1 occupancy profile at several CMK specific loci. Shown are RUNX1 occupancy profiles in K562 cells before (light green) and after (blue) TPA, and in CMK cells (orange), at loci spanning the ETS1, FLI1, PIK3R6, and RAB27b genes. (C) A multimodal RUNX1 occupancy landscape of distinct TF motif combinations characterizing the megakaryopoietic gene expression program. RUNX1 bound sites in K562, K562-TPA, and CMK were grouped according to their co-occurrence with motifs of RUNX, GATA, AP-1, and ETS. Histograms show percentage of occurrence of motifs with binding energies in the top 5% of background. ETS motifs are highly prevalent in CMK cells, whereas the AP-1 motif is clearly biased to de novo occupied TPA-induced sites. (D) A schematic model summarizing our hypothesis about stage-specific RUNX1-mediated regulation. RUNX1 (orange crescent) is preferentially bound to remote enhancers and cooperates with GATA1 (blue cluster) to regulate early myeloid genes (orange rectangles). On induction, RUNX1 recruits coactivators (purple crescent) to activate the AP1 genes. Thereafter, AP1 TFs (green clusters) facilitate the binding of RUNX1 to early megakaryocytic genes, thereby launching and driving the differentiation program. At subsequent differentiation stage (CMK), RUNX1 cooperates with ETS family TFs (yellow ellipses) to activate a different set of megakaryocytic genes. This scenario underscores the notion that RUNX1 functions in a context-dependent manner to regulate the transcriptional program in differentiating megakaryocytic cell lines.
To derive unbiased information regarding the relationship between sequence motifs and different modules of RUNX1 binding, we systematically calculated the fold enrichment of each motif associated with RUNX1 occupancy in the different binding modules (Figure 6C). The results correspond well to the experimental data indicating a common prevalence of RUNX motif in all classes and additional motifs, of RUNX1-cooperating TFs, including GATA, AP-1, and ETS, which were biased toward class specificity. Importantly, their enrichment varied according to megakaryocytic differentiation stages: GATA at K562 constitutive sites, AP-1 at TPA-induced sites, and ETS at CMK-specific sites.
Interestingly, when RUNX1 ChIP-seq data for Jurkat T cells40 were included in the co-occurrence analysis, it was found that AP-1 motif was significantly under-represented, whereas a pronounced enrichment for the motif of TF PBX1B (GATGTG) was noted (supplemental Figure 8),44,45 raising the possibility that, in T cells, RUNX1 also cooperates with PBX1B. Comparison of the overall RUNX1-binding profile showed that the highest overlap with T cells was found among CMK ChIP-seq data (supplemental Figure 9).
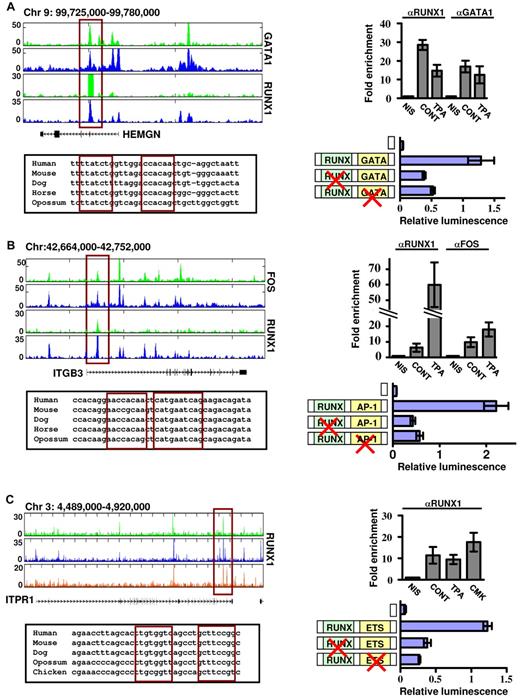
We next assessed the functional cooperation between RUNX1 and its collaborating TFs using reporter assays in megakaryocytic cell lines. Regulatory regions of several biologically relevant RUNX1 target genes, identified as co-occupied by our Chip-seq experiments, were cloned into reporter constructs and tested (Figure 7). HEMGN promoter was coactivated by RUNX1 and GATA1 in noninduced K562 cells (Figure 7A), whereas the intronic regulatory region of ITGB3 conferred RUNX1-AP-1–dependent reporter expression in K562-TPA cells (Figure 7B) and ITPR1 regulatory region, which was bound by RUNX1 at various differentiation stages, was cooperatively activated by RUNX1 and ETS TFs in Meg01 cells (Figure 7C). Collectively, the complementary outcome of these functional assays and the ChIP-seq occupancy data show that at, different stages of megakaryocytic cell line differentiation, RUNX1 sequentially cooperates with GATA1, AP-1, and ETS TFs to drive the transcription program (Figure 6D).
Functional cooperation between RUNX1 and its partner TFs GATA1, AP-1, and ETS. Reporter assays in megakaryocytic cell lines demonstrate cooperation between RUNX1-GATA (A), RUNX1-AP-1 (B), and RUNX1-ETS (C) in regulation of gene expression of HEMGN, ITGB3, and ITPR1 genes. (A) Top left: RUNX1 and GATA1 ChIP-seq tracks in K562 and K562-TPA cells proximal to the HEMGN locus. The regulatory region cloned in vectors and used in transfection assays is indicated by the red rectangle, as are the evolutionary conserved RUNX and GATA binding sites (bottom left) located within this region. Top right: ChIP-quantitative PCR validation of RUNX1 and GATA1 binding to the indicated ChIP-seq region. Shown are independent ChIP assays followed by quantitative PCR using K562 and K562-TPA cells. Bottom right: Dual-luciferase reporter assays in transfected K562 cells using PGL4.73 vector alone or with intact/mutated HEMGN regulatory region (supplemental data). (B) Top left: FOS and RUNX1 ChIP-seq tracks proximal to the ITGB3 locus in K562 and K562-TPA cells. The regulatory region cloned in vectors and used in transfection assays is indicated by the red rectangle, as are the evolutionary conserved RUNX and AP-1 binding sites (bottom left) located within this region. Top right: ChIP-quantitative PCR validation of RUNX1 and AP-1 binding to the indicated ChIP-seq region. Shown are independent ChIP assays followed by quantitative PCR using K562 and K562-TPA cells. Bottom right: Dual-luciferase reporter assays in transfected K562 cells using PGL4.73 vector alone or with intact/mutated ITGB3 regulatory region (see supplemental Methods). (C) Top left: RUNX1 ChIP-seq tracks proximal to the ITRP1 locus in K562, K562-TPA, and CMK cells. The regulatory region cloned in vectors and used in transfection assays is indicated by the red rectangle, as are the evolutionary conserved RUNX and ETS binding sites (bottom left) located within this region. Top right: ChIP-quantitative PCR validation of RUNX1 binding to the indicated ChIP-seq region. Shown are independent ChIP assays followed by quantitative PCR using K562, K562-TPA, and CMK cells. Bottom right: Dual-luciferase reporter assays in transfected K562 cells using PGL4.73 vector alone or with intact/mutated ITPR1 regulatory region (see supplemental Methods). (A-C) Data of quantitative PCR and dual-reporter assays represent mean ± SE of at least 2 biologic repeats performed in triplicates.
Functional cooperation between RUNX1 and its partner TFs GATA1, AP-1, and ETS. Reporter assays in megakaryocytic cell lines demonstrate cooperation between RUNX1-GATA (A), RUNX1-AP-1 (B), and RUNX1-ETS (C) in regulation of gene expression of HEMGN, ITGB3, and ITPR1 genes. (A) Top left: RUNX1 and GATA1 ChIP-seq tracks in K562 and K562-TPA cells proximal to the HEMGN locus. The regulatory region cloned in vectors and used in transfection assays is indicated by the red rectangle, as are the evolutionary conserved RUNX and GATA binding sites (bottom left) located within this region. Top right: ChIP-quantitative PCR validation of RUNX1 and GATA1 binding to the indicated ChIP-seq region. Shown are independent ChIP assays followed by quantitative PCR using K562 and K562-TPA cells. Bottom right: Dual-luciferase reporter assays in transfected K562 cells using PGL4.73 vector alone or with intact/mutated HEMGN regulatory region (supplemental data). (B) Top left: FOS and RUNX1 ChIP-seq tracks proximal to the ITGB3 locus in K562 and K562-TPA cells. The regulatory region cloned in vectors and used in transfection assays is indicated by the red rectangle, as are the evolutionary conserved RUNX and AP-1 binding sites (bottom left) located within this region. Top right: ChIP-quantitative PCR validation of RUNX1 and AP-1 binding to the indicated ChIP-seq region. Shown are independent ChIP assays followed by quantitative PCR using K562 and K562-TPA cells. Bottom right: Dual-luciferase reporter assays in transfected K562 cells using PGL4.73 vector alone or with intact/mutated ITGB3 regulatory region (see supplemental Methods). (C) Top left: RUNX1 ChIP-seq tracks proximal to the ITRP1 locus in K562, K562-TPA, and CMK cells. The regulatory region cloned in vectors and used in transfection assays is indicated by the red rectangle, as are the evolutionary conserved RUNX and ETS binding sites (bottom left) located within this region. Top right: ChIP-quantitative PCR validation of RUNX1 binding to the indicated ChIP-seq region. Shown are independent ChIP assays followed by quantitative PCR using K562, K562-TPA, and CMK cells. Bottom right: Dual-luciferase reporter assays in transfected K562 cells using PGL4.73 vector alone or with intact/mutated ITPR1 regulatory region (see supplemental Methods). (A-C) Data of quantitative PCR and dual-reporter assays represent mean ± SE of at least 2 biologic repeats performed in triplicates.
Discussion
Cellular differentiation progresses through a cascade of coordinated transcriptional events involving dynamic interplay between TFs and epigenetic changes. This interplay affects chromatin structure and regulates gene expression by permitting or restricting transcription. Here we studied the mechanisms by which RUNX1 interacts with cellular genomic DNA sequences and the epigenomic makeup to regulate the megakaryocytic transcriptional program. The data indicate that RUNX1 functions as a key regulator mediating the differentiation process through stage-dependent cooperation with other TFs.
RUNX1 plays a pivotal role in megakaryopoiesis
Using differentiating megakaryocytic cell-line models, we provided, for the first time, a systematic genome-wide flowchart of the RUNX1 occupancy patterns and regulatory targets in differentiating megakaryocytic cell lines. Our findings identified hundreds of previously unknown RUNX1 target genes based on their RUNX1-dependent response and its recruitment to sites proximal to their TSSs. The results delineated the molecular events underlying RUNX1 site selection specificity and its cooperation with coregulators and underscored the pivotal role of RUNX1 in executing the megakaryopoietic gene expression program.
RUNX1-mediated gene expression is regulated by interactions of epigenetically preprogrammed enhancers and target promoters
TPA-induced gene expression in K562 cells was largely dependent on RUNX1, attesting to its crucial role in megakaryocytic differentiation. Interestingly, 24 hours after treatment, RUNX1 binds to numerous new occupancy sites. Analysis of the H3K4me1 pattern before and 24 hours after TPA treatment demonstrated that the de novo RUNX1-binding regions were preprogrammed with open chromatin before activation. This finding placed RUNX1 at the center of an ordered cell differentiation process in which the epigenomic landscape was preorganized to meet subsequent regulatory requirements. The detailed RUNX1 genome-wide occupancy and associated gene expression patterns provided new important insights on the interaction of RUNX1 with the epigenome. RUNX1 binds preferentially to regions remote from its target genes. This was demonstrated by the correlation between the differential RUNX1 binding at target gene promoters and nearby enhancers and the better correspondence between RUNX1 binding and RUNX motifs at remote enhancers compared with promoters.
GATA1, AP-1, and ETS emerge as key RUNX1 cooperators in megakaryopoiesis
Sequence analysis of regions occupied by RUNX1 before induction of the megakaryocytic differentiation program indicated enrichment for the RUNX-GATA motif pair, suggesting that RUNX1 and GATA1 might cooperate during early stages of megakaryopoiesis. Experimental evidence in favor of this conclusion was granted by analysis of the recently reported data of GATA1 occupancy in K562 cells,26 GATA1 ChIP-seq analysis in K562-TPA cells, and ChIP-PCR of RUNX1 target genes. These findings pertain to the possibility that impaired RUNX1-GATA1 cooperation in early fetal liver hematopoiesis resulted in increased proliferation of megakaryocytic progenitors and contributes to the subsequent development of DS-AMKL. The data presented here underscore the involvement of GATA1 in regulating gene expression of several hematopoietic lineages, as was recently highlighted by the comprehensive analysis of its genome-wide occupancy in differentiating erythroid cells.27,32,33,46
TPA-induced, de novo occupied RUNX1 sites were highly enriched in AP-1 sequence motifs, whereas constitutive RUNX1 sites were not. ChIP-seq and sequence analysis demonstrated coupling between RUNX1 and AP-1 binding and have indicated that constitutively bound RUNX1 recruited AP-1 to regions lacking AP-1 motifs, whereas constitutively bound AP-1 recruited RUNX1 to sites lacking RUNX motifs. Interestingly, in the K562-TPA system, RUNX1 up-regulated AP-1 genes, raising the intriguing possibility of a regulatory cascade in which RUNX1-mediated expression of AP-1 acts to facilitate its binding to a new set of target genes required for megakaryocytic differentiation (Figure 6D).
The CMK cell line expresses late megakaryocytic markers, including platelet peroxidase and glycoprotein IIb/IIa,35,37 and is therefore considered more differentiated than 48-hour TPA-treated K562 cells. Interestingly, analysis of RUNX1 occupancy regions in CMK cells revealed a pronounced enrichment of the RUNX-ETS motif pair. This finding suggested that in CMK cells RUNX1 cooperated with members of the ETS TF family to drive megakaryocytic gene expression. Of note, both GATA1 and ETS family members were previously shown to cooperate with RUNX1 in various in vitro and cell transfection assays.9,12,39-41 On the other hand, to the best of our knowledge, this is the first time that AP-1 is implicated as a major transcriptional collaborator of RUNX1, although its involvement in megakaryocyte differentiation was previously reported.47,48
The methodologies introduced here may facilitate evaluation of RUNX1 function in other differentiation programs underlying the etiology of 8;21 leukemia as well as provide insights into the mechanisms underlying TF-DNA interaction specificity. The data suggest that genomic interactions of RUNX1 are highly dynamic and specified by a combination of genomic inputs, which include RUNX-binding sites and cell type–specific epigenetic makeup, such as H3K4me1-marked enhancers, and protein-protein interactions with other sequence specific TFs. Importantly, the data show that the interaction of RUNX1 with its cooperating TFs is critically important for determining its occupancy profiles at different developmental stages. Protein-protein interactions of TFs generate an additional layer of complexity superimposed on the genomic sequence and epigenetic makeup, thereby enhancing the diversity of RUNX1 binding landscapes, and the repertoire of its regulated genes. The generality of this phenomenon will be clarified as more genomic occupancy data become available. A model delineating the plasticity and spatial dynamics of RUNX1 occupancy and interactions during the 3-stage differentiation process is shown in Figure 6D.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Daniela Amann-Zalcernstein and Dr Shirley Horn-Saban for help in Ilumina sequencing, Dr Ester Feldmesser for help in bioinformatics analysis, Dalia Goldenberg for technical assistance, and Dr Joseph Lotem and Dr Ditsa Levanon for helpful comments and discussions throughout the work.
This work was supported by the European Union AnEUploidy project, the Jerome Lejeune Foundation, and the Israel Science Foundation (Y.G.) and the Alon Fellowship and the Israel Science Foundation (A.T.).
Authorship
Contribution: N.P., A.T., and Y.G. conceived and designed the experiments; N.P. performed the experiments; and N.P., R.J., A.T., and Y.G. analyzed the data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yoram Groner, Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, 76100 Israel; e-mail: yoram.groner@weizmann.ac.il.
References
Author notes
N.P. and R.J. contributed equally to this study.