Abstract
Mantle cell lymphoma (MCL) is a mostly incurable malignancy arising from naive B cells (NBCs) in the mantle zone of lymph nodes. We analyzed genomewide methylation in MCL patients with the HELP (HpaII tiny fragment Enrichment by Ligation–mediated PCR) assay and found significant aberrancy in promoter methylation patterns compared with normal NBCs. Using biologic and statistical criteria, we further identified 4 hypermethylated genes CDKN2B, MLF-1, PCDH8, and HOXD8 and 4 hypomethylated genes CD37, HDAC1, NOTCH1, and CDK5 when aberrant methylation was associated with inverse changes in mRNA levels. Immunohistochemical analysis of an independent cohort of MCL patient samples confirmed CD37 surface expression in 93% of patients, validating its selection as a target for MCL therapy. Treatment of MCL cell lines with a small modular immunopharmaceutical (CD37-SMIP) resulted in significant loss of viability in cell lines with intense surface CD37 expression. Treatment of MCL cell lines with the DNA methyltransferase inhibitor decitabine resulted in reversal of aberrant hypermethylation and synergized with the histone deacetylase inhibitor suberoylanilide hydroxamic acid in induction of the hypermethylated genes and anti-MCL cytotoxicity. Our data show prominent and aberrant promoter methylation in MCL and suggest that differentially methylated genes can be targeted for therapeutic benefit in MCL.
Introduction
Mantle cell lymphoma (MCL) is an aggressive and mostly incurable B-cell malignancy accounting for 5% of non-Hodgkin lymphomas (NHLs). MCL arises from naive B cells (NBCs) in the mantle zone of lymph node follicles and is characterized by the t(11,14) chromosomal translocation leading to overexpression of cyclin D1 (CCND1).1 However, murine models overexpressing CCND1 in the absence of other oncogenes, such as MYC, do not develop lymphoma,2 implying that additional pathogenic mechanisms are involved in MCL. Cip/Kip proteins have an important role in the formation of active CDK4/cyclin D complexes. In NHLs other than MCL, p27(Kip) protein expression is inversely related to the proliferation activity of the tumors.3 Apoptosis-related genes such as BCL2 have also been found to be altered in MCL with the use of different approaches. Homozygous deletions of BIM, a member of the BCL2 family, have also been found in MCL.4
Epigenetic changes, such as methylation of gene promoters, have been shown to contribute to the pathogenesis of both solid and hematologic malignancies.5 Single-locus studies analyzing methylation in MCL patient samples have shown hypermethylation of key genes, such as cell-cycle regulators p14ARF and CDKN2A,6,7 protein phosphatase SHP-18 and Rho–adenosine triphosphatase PARG-1.9 However these studies did not compare the methylation to NBCs, the normal counterparts of these malignant cells.10 Several lines of evidence conclusively show NBCs to be the cell of origin of MCL, including immunoglobulin heavy chain mutation status, t(11,14) chromosomal breakpoint analysis,11 and gene expression microarrays.12,13 To develop a more comprehensive understanding of aberrant DNA methylation in MCL, we performed a genomewide analysis of MCL DNA methylation and gene expression with the use of purified normal NBCs as controls. We report that promoter DNA methylation is aberrantly distributed in the MCL genome compared with normal NBCs, and we identified aberrantly hypermethylated and hypomethylated genes that provide a basis for rational targeted therapy in this disease.
Methods
Patient samples
Tissues and blood samples were obtained from patients newly diagnosed with MCL before any treatment after informed consent in accordance with the Declaration of Helsinki. Sample collection and laboratory studies were in compliance with institutional review board and Helsinki protocols. CD19+ cells from 22 patients treated at the National Institutes of Health were purified by magnetic bead sorting from peripheral blood or pheresis products before freezing to ensure greater than 90% purity for HpaII tiny fragment Enrichment by Ligation–mediated polymerase chain reaction (HELP PCR) analysis. Fourteen patients treated at the Hackensack University Medical Center with leukemic-phase MCL had peripheral blood or pheresis products frozen without any selection but with sufficiently high peripheral blast differential counts or concurrent flow cytometric analysis to ensure greater than 85% purity of CD19+ B cells. For controls, purified normal immunoglobulin D positive (IgD+) NBCs were obtained with the use of magnetic bead sorting from specimens from 10 healthy donors undergoing routine tonsillectomy (for nonneoplastic indications) at the Children's Hospital at Montefiore (Bronx, NY).
DNA methylation analysis by HELP
Genomic DNA was isolated from samples and cell lines with the use of a standard high-salt procedure, and the HELP assay was carried out as previously described.14,15 The assay uses comparative isoschizomer profiling, interrogating cytosine methylation status on a genomic scale. Briefly, genomic DNA from the samples was digested by a methylcytosine-sensitive enzyme HpaII in parallel with MspI, which is resistant to DNA methylation, and then the HpaII and MspI products were amplified by ligation-mediated PCR. PCR conditions have been optimized to amplify fragments between 200 and 2000 base pair (bp), thus ensuring the preferential amplification of cytosine-phosphate-guanosine (CpG) dinucleotide-dense regions. Each fraction is then labeled with a specific dye and cohybridized onto a microarray designed to cover HpaII amplifiable fragments (HAFs) across the genome.15 The differential digestion of DNA by the 2 restriction enzymes HpaII (methylation sensitive) and MspI (methylation insensitive) assays the methylation of genomic DNA covered in the microarray probes. Detailed descriptions of HELP methods and conditions have been previously published.14,15 DNA methylation was measured as the log (HpaII/MspI) ratio, ranging from −5.91 to 5.73, where HpaII reflects the hypomethylated fraction of the genome and MspI represents the whole genome reference. Fractions were labeled with the use of cyanine–labeled random primers (9-mers) and then hybridized onto a human HG17 custom-designed oligonucleotide array (50-mers) covering 25 626 HAFs located at gene promoters and imprinted regions. HAFs are defined as genomic sequences contained between 2 flanking HpaII sites found within 200 to 2000 bp from each other. Each HAF on the array is represented by 15 individual probes randomly distributed across the microarray slide. All samples for microarray hybridization were processed at the Roche-NimbleGen Service Laboratory. Scanning was performed with the use of a GenePix 4000B scanner (Axon Instruments). PCR fragment length bias was corrected by quantile normalization. Further quality control and data analysis of HELP microarrays were performed as described in Thompson et al.16 All microarray data have been submitted to the Gene Expression Omnibus repository (accession no. GSE19243).17
Gene expression microarrays
Gene expression data were obtained using Affymetrix Human Genome U133A 2.0 or Plus2 GeneChips; mRNA isolation, labeling, hybridization, and quality control were carried out as described before.18 Raw data were processed using the Robust Multi-Averaging (RMA) algorithm and Affymetrix Expression Console software. Data are available in the NCBI Gene Expression Omnibus database (accession number GSE19243).17
Microarray data analysis
Unsupervised clustering of HELP and gene expression data by principal component analysis was performed with the use of R 2.8.2 statistical software and GenePattern software (http://www.broad.mit.edu/cancer/software/genepattern/). Supervised analysis of the methylation data were carried out with a moderated t test with Benjamini-Hochberg correction with a significance level of P less than .001 and an absolute difference in methylation greater than 1.5 between the means of the 2 populations (eg, NBCs vs MCL) to increase the likelihood of detecting biologically significant changes in methylation levels.
Gene network and gene ontology analysis
Ingenuity Pathway Analysis software and the Database for Annotation, Visualization and Integrated Discovery19 were used to carry out network composition analyses. HpaII-amplifiable fragments on the HELP microarray were annotated to the nearest gene up to a maximum distance of 5 kilobases from the transcription start site.
Quantitative DNA methylation analysis by MassARRAY EpiTyper
Validation of HELP findings was performed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry by MassARRAY (Sequenom) as previously described.20 Briefly, PCR primers specific for bisulfite-converted genomic DNA were designed with the use of Sequenom EpiDesigner to cover the flanking HpaII sites for a given HAF, as well as any other HpaII sites found up to 2000 bp upstream of the downstream site and up to 2000 bp downstream of the upstream site (primer sequences indicated in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The PCR amplification products from bisulfite-treated genomic DNA were then transcribed in vitro into a single-stranded RNA followed by base-specific cleavage reactions (T-specific or C-specific cleavage) with endoribonuclease. Methylation influenced base-specific cleavage mass signal pattern through C to U sequence changes introduced into the bisulfite-treated genomic DNA and appeared as G/A generated from the reverse strand by base-specific cleavage. The cleaved products were detected by matrix-assisted laser desorption ionization time-of-flight mass spectrometry and analyzed by EpiTYPER software v1.0 (Sequenom). These G/A variations in cleaved products resulted in a mass shift of 16 Da per CpG, representing a signal pair pattern of unmethylated and methylated DNA. The presence/absence of mass signals was indicative of the methylation status of CpGs in the interrogated sequence, and the ratio of the peak areas of paired mass signals can be used to estimate the relative amounts of methylation. MassARRAY EpiTyper enables quantitative screening of large sequences with high resolution in a large cohort of samples, without the disadvantage of being easily biased by the limitation in the number of clones analyzed in clonal bisulfate sequence and the low sequencing distance in pyrosequence. Direct comparison between MassARRAY and the conventional clonal bisulfite sequence or bisulfite pyrosequence has shown to be highly correlated and comparable.21,22
Cell culture and drug treatment
The cell lines Granta 519, HBL-2, JeKo-1, JVM-2, Mino, SP-49, SP-53, UPN1, and Z138 were grown in a humidified incubator at 37°C and 5% CO2 with RPMI 1640 medium (Cellgro) supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products), 2mM l-glutamine, 100 U/mL penicillin G, and 100 μg/mL streptomycin (Cellgro). Decitabine (DAC; Sigma-Aldrich) was dissolved in sterile water at a concentration of 1mM and stored at −80°C. Cells were counted and seeded at a concentration of 1 × 106 cells/mL into 6- or 12-well plates. For methylation array experiments, DAC was added to achieve concentrations of 0.1, 0.5, or 1μM and exposure continued for 2 days. For viability and gene induction studies, DAC was added to the cells to achieve a concentration of 0.1 or 0.5μM on day 0, day 1, and day 2. Viability was estimated on day 3. In case of combined DAC and suberoylanilide hydroxamic acid (SAHA) treatment, cells were exposed to 0.5 or 1μM SAHA on day 0. For in vitro treatment of cells with antibodies, cells were suspended in media at a density of 1 × 106 cells/mL immediately after isolation. CD37-SMIP (Trubion Pharmaceuticals) treatment used a 5 μg/mL concentration except for the dose-response studies. Trastuzumab (Genentech) and rituximab (Genentech) were used at 10 μg/mL. The cross-linker, (Fc specific) goat anti–human IgG (Jackson ImmunoResearch Laboratories Inc) was added to the cell suspension 5 minutes after adding the primary antibodies, at a concentration 5 times that of the primary antibodies (ie, 25 μg/mL for 5 μg/mL). For all CD37-SMIP experiments, a group of samples with the same concentration of trastuzumab treatment was applied as isotype control. In addition, a group of samples with no treatment was collected as media control.
Cell viability
Cell viability was determined with the use of a fluorometric resazurin reduction method (CellTiter-Blue; Promega) following the manufacturer's instructions. The number of viable cells in each treated well was calculated 48 or 72 hours after treatment. Cells (100 μL; 105 cells) were plated in 96-well plates (8 replicates per condition), and 20 μL of CellTiter-Blue Reagent (Promega) was added to each well. After 1 hour of incubation with the dye, fluorescence (560Ex/590Em) was measured with the Polarstar Optima microplate reader (BMG Labtechnologies). The number of viable cells in each treated well was calculated, based on the linear least-squares regression of the standard curve. Cell viability was normalized in drug-treated cells to their respective untreated controls. Cells were counted manually and confirmed on the Countess automated cell counter (Invitrogen) according to the manufacturer's specifications.
Immunohistochemistry and flow cytometry for CD37
A previously assembled tissue microarray (TMA) that contained 14 cases of MCL confirmed by cyclin D1 protein expression through immunohistochemistry with duplicate cores was used. Four-micron thick sections were cut with the use of a traditional water bath technique and dried at room temperature overnight. TMA slides were heated at 60°C for 30 minutes to melt paraffin, followed by deparaffinization and rehydration through xylene, graded ethanol, and dH20 washes. Endogenous peroxidases were blocked with Peroxidazed (Biocare Medical), and nonspecific binding was minimized with Background Terminator (Biocare Medical). Endogenous biotin was blocked with the use of the Avidin and Biotin kit (Biocare Medical). Staining was performed by applying monoclonal mouse anti-CD37 (clone IPO-24; Santa Cruz Biotechnology) diluted 1:200 followed by biotinylated secondary antibody, streptavidin–horseradish peroxidase conjugate, the chromogen diaminobenzidine, and hematoxylin counterstain. For flow cytometry, cells were washed once with RPMI and 10% FBS and incubated with mouse anti–human fluorescein isothiocyanate–conjugated anti-CD37 (clone M-B371; Becton Dickinson) at 4°C for 30 minutes. Then cells were spun down at 4°C at 300g for 5 minutes and resuspended in 300 μL of PBS with 2% FBS and were analyzed by flow cytometry.
Quantitative immunofluorescence by automated quantitative analysis
Quantitation of CD37 expression on TMA cores from MCL and benign lymphoid tissue (lymph node and tonsil) were assessed by automated quantitative analysis (AQUA) system (HistoRx). Briefly, a B-cell mask and CD37 expression within that mask were defined by labeling with a primary rabbit anti-CD20 antibody (polyclonal; NeoMarkers) and mouse anti-CD37 (clone IPO-24) respectively, followed by the addition of biotinylated goat antirabbit and Alexa 555 goat antimouse (Invitrogen), ending with incubation with streptavidin–horseradish peroxidase and Alexa Fluor 647–Tyramide. Automated image acquisition and analysis with the use of AQUA has been previously described,23 but, briefly, a B-cell binary mask was created through a pixel-based locale assignment for compartmentalization of expression algorithm, and CD37 expression was expressed as the average signal intensity per unit of B-cell mask area as the AQUA score. CD37 expression between groups was evaluated by nonpaired t test of AQUA scores of 28 tissue cores of MCL and 14 cores of benign lymphoid tissue.
Apoptosis assay, quantitative real-time PCR, and Western blot analysis are provided in supplemental Document 1.
Results
MCL primary samples have distinct genomic methylation profiles compared with naive B cells
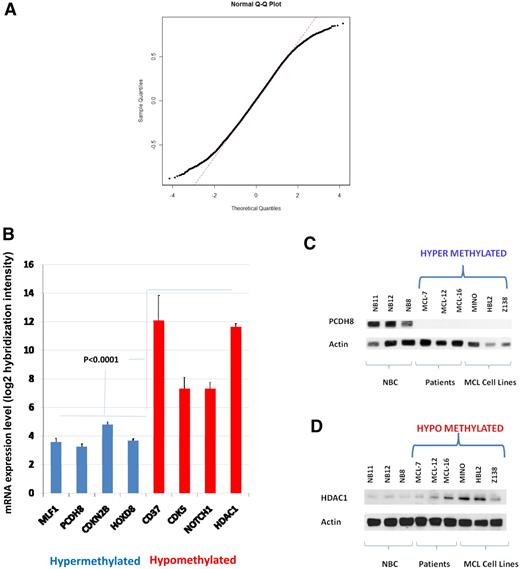
DNA methylation profiling was performed in 22 primary MCL patient samples and 10 control samples of purified NBCs from healthy donors was performed. DNA methylation was quantified for more than 50 000 CpGs over 14 000 gene promoter regions with the use of the HELP assay and a custom, high-density oligonucleotide microarray.14 All the methylation arrays passed a rigorous quality control and quantile normalization procedure.16 Principal component analysis was used to assess the underlying methylation differences between these groups of samples in an unsupervised manner. As seen in Figure 1A, DNA methylation profiles readily discriminate MCL patient samples and NBCs into discrete groups, indicating that the distribution of promoter DNA methylation is quite different between these genomes. The basis for the differences was predominantly hypomethylation in patient samples, as shown in a plot of fold-change versus P value for differentially methylated genes (Figure 1B). A heat map representation of DNA methylation ratios for the loci identified by the above cutoffs allows the predominant hypomethylation of gene promoters in patients with MCL versus NBCs to be readily visualized (Figure 1C; supplemental Tables 2-3).
The MCL genome is distinctly methylated compared with normal naive B cells. (A) Three-dimensional principal component analysis of DNA methylation data comparing MCL patient samples (red) and naive B cells (blue). (B) Plot of methylation difference (x-axis) vs significance (on y-axis) shows marked asymmetry with an overall tendency for hypomethylation in patients with MCL compared with NBCs. Blue dots mark probe sets that reached both criteria for differential methylation on our analysis (methylation difference of 1.5 and P < .001). (C) Methylation (HELP data) at the differentially methylated loci identified above from patients with MCL and NBCs represented in a heat map. Scale shows relative methylation (red indicates hypomethylation; blue indicates hypermethylation).
The MCL genome is distinctly methylated compared with normal naive B cells. (A) Three-dimensional principal component analysis of DNA methylation data comparing MCL patient samples (red) and naive B cells (blue). (B) Plot of methylation difference (x-axis) vs significance (on y-axis) shows marked asymmetry with an overall tendency for hypomethylation in patients with MCL compared with NBCs. Blue dots mark probe sets that reached both criteria for differential methylation on our analysis (methylation difference of 1.5 and P < .001). (C) Methylation (HELP data) at the differentially methylated loci identified above from patients with MCL and NBCs represented in a heat map. Scale shows relative methylation (red indicates hypomethylation; blue indicates hypermethylation).
Aberrant DNA methylation in MCL involves cancer-associated gene networks and is associated with aberrant gene expression
A supervised analysis was performed to identify aberrantly methylated genes in MCL versus NBCs. With the use of a moderated t test with P values less than .01 and a Benjamini-Hochberg correction for multiple comparisons (Figure 2A), 1168 differentially hypermethylated and 2598 hypomethylated probe sets were identified that discriminate between MCL cell lines and patients compared with NBCs (see supplemental Tables 4-5). This corresponded to 4110 differentially methylated genes, because some probe sets represented bidirectional promoters.
Differentially methylated genes chosen for validation. (A) Plot of methylation difference (x-axis) vs significance (on y-axis) showing selected differentially methylated loci between patients with MCL and MCL cell lines compared with NBCs. Probe sets that were differentially methylated are marked in blue (P < .01). (B) Ingenuity pathway core analysis of the top differentially methylated loci in patients with MCL compared with NBCs identifies HDAC1 and NFKB1 as central nodes in the most significantly enriched pathway. Differentially hypomethylated loci in patients with MCL are indicated in red, and differentially hypermethylated loci are indicated in green. (C) Methylation [HELP log(HpaII/MspI) ratios] at the 8 differentially methylated loci chosen for validation represented in a heat map. Scale shows relative methylation with blue indicating hypermethylation and red indicating hypomethylation.
Differentially methylated genes chosen for validation. (A) Plot of methylation difference (x-axis) vs significance (on y-axis) showing selected differentially methylated loci between patients with MCL and MCL cell lines compared with NBCs. Probe sets that were differentially methylated are marked in blue (P < .01). (B) Ingenuity pathway core analysis of the top differentially methylated loci in patients with MCL compared with NBCs identifies HDAC1 and NFKB1 as central nodes in the most significantly enriched pathway. Differentially hypomethylated loci in patients with MCL are indicated in red, and differentially hypermethylated loci are indicated in green. (C) Methylation [HELP log(HpaII/MspI) ratios] at the 8 differentially methylated loci chosen for validation represented in a heat map. Scale shows relative methylation with blue indicating hypermethylation and red indicating hypomethylation.
To identify pathways and functional groups enriched by differentially methylated loci, the 4110 genes were entered into the Ingenuity Pathway Analysis database and the Database for Annotation, Visualization and Integrated Discovery (see “Methods”). The top pathways identified by the differentially methylated loci revolved around key cellular proteins such as histone deacetylase 1 (HDAC1) and nuclear factor κB 1 (NFKB1) transcription factor (Figure 2B) and, hence, were involved in critical cellular regulatory processes of transcription, regulation of transcription, and gene expression (Table 1).
A step-wise approach was used to integrate data from DNA methylation and gene expression arrays to select genes of potential biologic importance for validation. Affymetrix U133 arrays were used to interrogate 14 500 genes from the same pretreatment biopsy samples that were used for methylation profiling by HELP. We found 1413 loci to have inverse changes in mRNA levels above and below the expression array mean, ie, hypermethylated genes having expression less than array mean and vice versa (explained in more detail below and listed in supplemental Table 6). From this set, 8 genes (Figure 2C) having the following characteristics were selected as candidates for further studies. (1) Hypomethylated genes involved in pathways controlling biologic processes with known oncogenic activity in cancer, ie, cell cycle control, apoptosis: CD37,24,25 HDAC1,26 NOTCH1,27 and CDK5.28 (2) Genes that function as tumor suppressors in the context of MCL or other tumors and that were hypermethylated in MCLs compared with NBCs: CDKN2B, HOXD8, MLF-1, and PCDH8.
Six of the 8 genes (PCDH8, HOXD8, CDKN2B, CDK5, HDAC1, NOTCH1) met the threshold for differential methylation (P < .01) in both patients and MCL cell lines (supplemental Figure 4A) with CD37 and MLF1 as exceptions. However, single-locus DNA methylation assays in our preliminary studies29 also showed concordance in promoter methylation status between patients and cell lines for CD37 and MLF1. CD37 is of particular interest because it is strongly expressed on the surface of B cells, making it an attractive target in B-cell NHL.24,25 Myeloid leukemia factor 1 (MLF1) is differentially hypermethylated in gastric cancer patient samples,30 suggesting a possible role beyond aberrant transcriptional control in leukemia. Cyclin dependent kinase inhibitor 2b (CDKN2B) inhibits cyclin-dependent kinases 4 and 6 and has been reported to be epigenetically silenced in MCL.6 Protocadherin 8 (PCDH8) and homeobox D8 (HOXD8) have been found to be inactivated by methylation in breast cancer.31,32 Cyclin-dependent kinase 5 (CDK5) plays a role in the DNA damage response and cell cycle checkpoint activation in breast cancer.28 HDAC inhibitor HDAC126 and the transcription factor NOTCH127 are known therapeutic targets for the treatment of B- and T-cell malignancies.
Quantitative promoter methylation sequencing confirms aberrant regulation of candidate genes
To confirm that DNA methylation of the 8 candidate genes was indeed consistent with the predictions of the HELP analysis, we performed EpiTyper mass spectrometric quantitative DNA methylation sequencing of the promoter regions (supplemental Table 7). Twenty-two patient samples were from the same set used for HELP analysis, and 14 additional MCL lymph node samples were used as an independent confirmatory cohort. In Figure 3, the percentage methylation is plotted in the form of a heat map with each column representing one CpG and each row representing one sample. The heat maps show the distinct patterns of methylation between MCL cell lines, MCL patient samples, and NBCs. PCDH8 (Figure 3A) and MLF1 (Figure 3B) are examples of hypermethylated loci, and CD37 (Figure 3C) and HDAC1 (Figure 3D) are examples of hypomethylated loci (additional heat maps are in supplemental Figure 1). We found statistically significant (P < .001) differences in methylation between MCL patient samples compared with NBCs in promoter regions of all 8 genes, confirming our HELP results (supplemental Table 7). The additional 14 primary samples confirmed the methylation changes seen in the initial 22 patient samples in each of the 8 candidate gene promoters sequenced.
MassArray confirms differential methylation in patients with MCL and MCL cell lines compared with NBC controls. (A-D) Heat maps of MassArray data comparing methylation in patients with MCL, MCL cell lines, and NBC controls at individual CpGs for hypermethylated genes PCDH8 and MLF1 and for hypomethylated genes HDAC1 and CD37. Rows correspond to individual samples, and columns correspond to individual CpGs. The area covered by the primer, HpaII sites, CpG islands, direction of transcription, and gene start are all indicated as tracks in the University of California Santa Cruz Browser. Scale shows relative methylation (green indicates hypomethylation; purple indicates hypermethylation).
MassArray confirms differential methylation in patients with MCL and MCL cell lines compared with NBC controls. (A-D) Heat maps of MassArray data comparing methylation in patients with MCL, MCL cell lines, and NBC controls at individual CpGs for hypermethylated genes PCDH8 and MLF1 and for hypomethylated genes HDAC1 and CD37. Rows correspond to individual samples, and columns correspond to individual CpGs. The area covered by the primer, HpaII sites, CpG islands, direction of transcription, and gene start are all indicated as tracks in the University of California Santa Cruz Browser. Scale shows relative methylation (green indicates hypomethylation; purple indicates hypermethylation).
Inverse correlation of DNA methylation and gene expression in MCL
We examined the genomewide correlation of methylation to gene expression values from 10 patients with MCL and 10 MCL cell lines by comparing HELP ratios with mRNA expression in 12 731 genes represented in common between the 2 array platforms. The distribution of correlation coefficients shows nonrandom skewing of tails by genes having a high correlation coefficient (R > 0.5) compared with a standard normal distribution (Figure 4A). There were 3528 genes with a correlation coefficient greater than 0.5, corresponding to 11.3% of the loci compared. This indicates a weak but nonrandom correlation genomewide between methylation and gene expression. We next compared the mRNA expression levels with HELP methylation status in patients with MCL for the 8 genes in our panel (Figure 4B). The expression of all 4 hypermethylated loci was significantly lower (P < .001) than the expression of hypomethylated loci. With the use of Western blotting, the protein expression of candidate hypermethylated (PCDH8) and hypomethylated (HDAC1) genes were shown to be reciprocal to methylation status (Figure 4C-D) in 3 MCL patient samples and 3 MCL cell lines (MINO, HBL-2, and Z138) compared with 3 NBC samples. Similar validation was performed for NOTCH1 and CD37 by Western blot and AQUA, respectively, and by quantitative real-time PCR for HOXD8 and MLF1 (Figure 5A-C; supplemental Figure 5A-C).
Gene and protein expressions are reciprocal to methylation. (A) Correlation of HELP ratios and mRNA levels in 12 731 genes represented as a quantile-normal plot. Distribution of correlation coefficients shows nonrandom skewing of tails by genes having a high correlation coefficient (R > 0.5) compared with a standard normal distribution. (B) mRNA expression levels from 10 patients with MCL and 10 cell lines plotted as a function of signal intensity. Log 2 hybridization intensity for each probe set represents the relative level of mRNA expression of a gene transcript (y-axis). Hypermethylated loci are color coded in blue, and hypomethylated loci are color coded in red. Error bars indicate SD. (C-D) Western blot for protein expression of HDAC1 and PCDH8 in 3 NBCs, 3 patients, and 3 cell lines (HBL2, MINO, Z138) along with actin as a loading control.
Gene and protein expressions are reciprocal to methylation. (A) Correlation of HELP ratios and mRNA levels in 12 731 genes represented as a quantile-normal plot. Distribution of correlation coefficients shows nonrandom skewing of tails by genes having a high correlation coefficient (R > 0.5) compared with a standard normal distribution. (B) mRNA expression levels from 10 patients with MCL and 10 cell lines plotted as a function of signal intensity. Log 2 hybridization intensity for each probe set represents the relative level of mRNA expression of a gene transcript (y-axis). Hypermethylated loci are color coded in blue, and hypomethylated loci are color coded in red. Error bars indicate SD. (C-D) Western blot for protein expression of HDAC1 and PCDH8 in 3 NBCs, 3 patients, and 3 cell lines (HBL2, MINO, Z138) along with actin as a loading control.
CD37 is expressed in patients with MCL and MCL cell lines, and CD37-SMIP treatment is cytotoxic in MINO. (A) Representative medium-power (20× original magnification) view of normal tonsil control tissue showing CD37 expression by the B cells of a B-cell follicle and lack of expression in the interfollicular lymphocytes. The intensity of membranous and cytoplasmic staining is greater in the B cells found within the mantle zone compared with those in the germinal center. (B) Representative high-power (40×) and low-power (10×, inset) view of a 2-mm MCL core from a tissue microarray. Images were captured with an Olympus DP25 digital camera using DP2-BSW Version 1.3 software mounted on an Olympus BX41 microscope with 10×/0.30, 20×/0.40, and 40×/0.65 objectives. Images were resized without color or contrast manipulation in Adobe Photoshop CS2 Version 9.0.2. (C) CD37 expression assessed by AQUA of immunofluorescence in CD20+ B cells from cases of MCL and benign lymphoid tissue represented on a tissue microarray. Column indicates average from 28 tissue cores representing 14 cases of MCL and 14 tissue cores from 8 cases of benign lymphoid tissue (6 lymph node and 2 tonsil); bars, 1 SD 2-tailed, unpaired t test, P < .001. (D) Flow cytometric assessment of CD37 surface expression in 6 MCL cell lines with fluorescein isothiocyanate (FITC)–conjugated anti-CD37 antibody and IgG1 isotype control antibody. (E) Viability of MINO cells 72 hours after treatment with CD37-SMIP, rituximab, and trastuzumab in the presence of a cross-linking, Fc-specific, goat anti–human IgG. Viability was measured by Annexin V/propidium iodide staining and normalized to control. Data are representative of 3 separate experiments.
CD37 is expressed in patients with MCL and MCL cell lines, and CD37-SMIP treatment is cytotoxic in MINO. (A) Representative medium-power (20× original magnification) view of normal tonsil control tissue showing CD37 expression by the B cells of a B-cell follicle and lack of expression in the interfollicular lymphocytes. The intensity of membranous and cytoplasmic staining is greater in the B cells found within the mantle zone compared with those in the germinal center. (B) Representative high-power (40×) and low-power (10×, inset) view of a 2-mm MCL core from a tissue microarray. Images were captured with an Olympus DP25 digital camera using DP2-BSW Version 1.3 software mounted on an Olympus BX41 microscope with 10×/0.30, 20×/0.40, and 40×/0.65 objectives. Images were resized without color or contrast manipulation in Adobe Photoshop CS2 Version 9.0.2. (C) CD37 expression assessed by AQUA of immunofluorescence in CD20+ B cells from cases of MCL and benign lymphoid tissue represented on a tissue microarray. Column indicates average from 28 tissue cores representing 14 cases of MCL and 14 tissue cores from 8 cases of benign lymphoid tissue (6 lymph node and 2 tonsil); bars, 1 SD 2-tailed, unpaired t test, P < .001. (D) Flow cytometric assessment of CD37 surface expression in 6 MCL cell lines with fluorescein isothiocyanate (FITC)–conjugated anti-CD37 antibody and IgG1 isotype control antibody. (E) Viability of MINO cells 72 hours after treatment with CD37-SMIP, rituximab, and trastuzumab in the presence of a cross-linking, Fc-specific, goat anti–human IgG. Viability was measured by Annexin V/propidium iodide staining and normalized to control. Data are representative of 3 separate experiments.
CD37 is expressed in MCL patient samples and cell lines and CD37-SMIP treatment is cytotoxic in MINO cells
Methylation and gene expression array analyses identified the B-cell surface antigen CD37 as hypomethylated and overexpressed in patients with MCL and MCL cell lines. The expression of CD37 in primary MCLs was further evaluated by immunohistochemistry in an independent cohort of 14 patients with MCL. The anti-CD37 (clone IPO-24) antibody was optimized on formalin-fixed paraffin-embedded benign tonsil control tissue to show staining on mature B cells, without staining on T cells and macrophages. B-cell follicles showed a dual pattern of CD37 expression with mantle zone B cells having a greater intensity of membranous and cytoplasmic CD37 expression than germinal center B cells (Figure 5A). The number of cells (frequency) and intensity of CD37 staining was assessed by a trained hematopathologist (D.T.Y.). Staining was considered positive if greater than 80% of tumor cells showed a cytoplasmic/membrane staining pattern of 3+ or greater on a 0 to 4+ subjective intensity scale when averaged between 2 cores. Thirteen of 14 MCL specimens (93%), represented in duplicate on a tissue microarray, expressed CD37. A combined cytoplasmic and membrane expression pattern was seen in all positive cases (Figure 5B). Expression appears to be an all-or-none phenomenon with the single negative case showing no detectable staining and with all positive cases having strong expression in the tumor cells. With the use of quantitative assessment of protein expression in CD20+ B cells in formalin-fixed paraffin-embedded tissue cores on a TMA by AQUA, CD37 expression in MCL samples was significantly greater than that found in benign lymphoid tissue biopsies (Figure 5C). CD37 expression was positive in 6 MCL cell lines (Figure 5D) assayed by flow cytometry. Seventy-two hours after treatment of MINO cells with 5 μg/mL CD37-SMIP, in the presence of a cross-linking, Fc-specific, goat anti–human IgG, there was 75% reduction in viability assessed by Annexin V/propidium iodide staining (Figure 5E). Treatment with control antibodies rituximab and trastuzumab resulted in cell death in greater than 70% and 0% cells, respectively. These results suggest that CD37 is strongly expressed in most patients with MCL at levels higher than in normal NBCs and support a potential therapeutic role for CD37-SMIP in MCL.
DAC synergizes with SAHA in the induction of methylated tumor suppressor genes and cytotoxicity in MCL cell lines
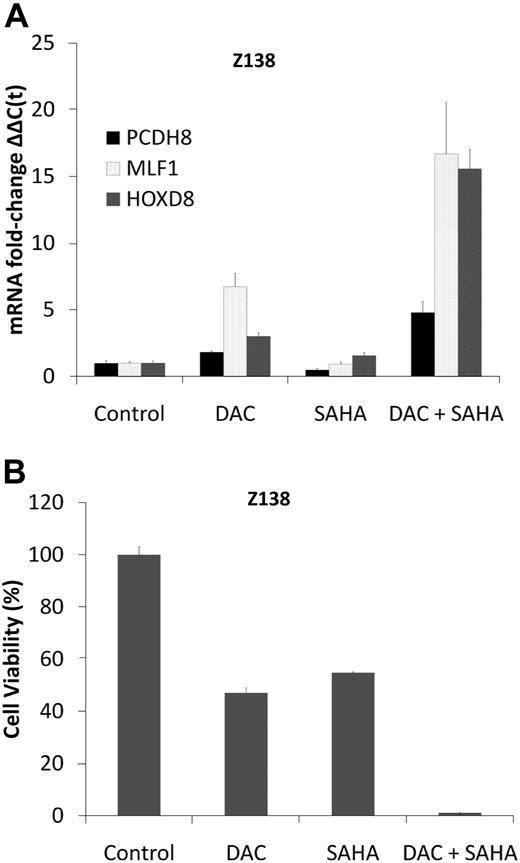
DNA methyltransferase (DNMT) inhibitors such as DAC and HDAC inhibitors such as SAHA cooperate and even synergize in reactivating epigenetically silenced tumor suppressor genes.33 The effect of these drugs on the re-expression of aberrantly methylated and silenced tumor suppressor genes was assessed in 2 well-characterized MCL cell lines, MINO and Z138, in which MLF1, PCDH8, HOXD8, and CDKN2B genes are hypermethylated (Figure 3A-B; supplemental Figure 1A-B) and repressed (Figure 4B), using a low concentration (0.5μM) of DAC for 3 days with and without 1μM SAHA. This concentration of DAC causes extensive DNA hypomethylation and induces expression of hypermethylated genes (supplemental Figure 3A-D) without causing DNA damage.9,34 The combination of DAC plus SAHA increased MLF1, PCDH8, and HOXD8 mRNA levels 5- to 15-fold in Z138 cells and PCDH8 and CDKN2B to a similar extent in MINO (Figure 6A; supplemental Figures 3B and 5D-F). Treatment of the MCL cells with DAC led to significant demethylation of CCND1, p27, and p21 in both MINO and Z138 cell lines and BIM alone in Z138 cells (supplemental Table 8). In Z138 cells, 0.5μM DAC for 3 days alone decreased cell viability by 40%, and 1μM SAHA reduced viability by 60%, whereas the combination decreased cell viability by greater than 90% (Figure 6B). Synergy in cytotoxicity and gene expression was quantified by the Chou-Talalay35 equation with the use of the Calcusyn software, with combination indices less than 1 for SAHA and DAC in both cell lines. Although not as pronounced in MINO cells, treatment with DAC and SAHA also resulted in greater cell kill compared with either drug alone (supplemental Figures 3E and 6A-C). Similar synergy in cytotoxicity was found for one more MCL cell line HBL2 (supplemental Figure 6A-C).
DAC synergizes with SAHA in gene induction and cytotoxicity in Z138 cells. (A) Real-time PCR for mRNA expression of PCDH8, HOXD8, and MLF1 in Z138 cells before and after treatment with DAC 0.5μM daily for 3 days, SAHA 1μM (single dose on day 0), and their combination. Data are representative of 3 separate experiments. Error bars indicate SD. (B) Z138 cells were treated with DAC 0.5μM daily for 3 days and SAHA 1μM (single dose on day 0) alone, and in combination. Cell viability (y-axis) as measured by reduction of resazurin 48 hours after drug treatment (performed in 8 replicates).
DAC synergizes with SAHA in gene induction and cytotoxicity in Z138 cells. (A) Real-time PCR for mRNA expression of PCDH8, HOXD8, and MLF1 in Z138 cells before and after treatment with DAC 0.5μM daily for 3 days, SAHA 1μM (single dose on day 0), and their combination. Data are representative of 3 separate experiments. Error bars indicate SD. (B) Z138 cells were treated with DAC 0.5μM daily for 3 days and SAHA 1μM (single dose on day 0) alone, and in combination. Cell viability (y-axis) as measured by reduction of resazurin 48 hours after drug treatment (performed in 8 replicates).
Discussion
Here, we show that the distribution of cytosine methylation is profoundly perturbed in MCL cells. The HELP platform used in this study allowed query of more than 50 000 CpGs located in more than 14 000 promoters, providing a more in-depth view of DNA methylation than single-locus assays or more limited array platforms. DNA methylation was inversely correlated with gene expression, and candidate genes of interests were validated with the use of MassArray quantitative DNA methylation sequencing. Precise identification of what constitutes aberrant DNA methylation was possible by comparison of MCL methylation profiles with those obtained from purified populations of tonsillar NBCs, the normal counterpart of MCLs.10
The data indicate a surprising predominance of gene promoter hypomethylation in MCL compared with NBCs. Aberrant hypermethylation of promoters and subsequent silencing of tumor suppressor genes such as p14ARF and CDKN2A has been described in MCL and other tumors.5,6 In contrast, aberrant hypomethylation of gene promoters can lead to increased transcription of genes with oncogenic potential. Liu et al36 have shown that the CCND1 promoter is hypomethylated, and this is associated with RNA polymerase II recruitment and increased expression of CCND1 mRNA in MCL. DNA hypomethylation can also occur in intergenic and noncoding repetitive DNA segments where it may contribute to cancer pathogenesis by causing genomic instability or altering euchromatin-heterochromatin interactions.37 For example, DNA hypomethylation in DNMT1 hypomorphic mice has been associated with genomic instability and development of T-cell lymphomas.38,39 Our findings suggest that promoter hypomethylation and hypermethylation occurs concurrently and that each may contribute independently to MCL pathogenesis. Acute myeloid leukemia40 and ovarian and gastric tumors have been shown to have similar patterns of aberrant methylation.37,41
DNMTs are enzymes that add methyl groups to DNA or RNA. DNMT1 is predominately responsible for hemimethylated CpG island methylation, DNMT2 transfers methyl groups to RNA not DNA and hence has been renamed to tRNA aspartic acid methyltransferase 1 (TRDMT1), and DNMT3 is responsible for unmethylated CpG island methylation.42 In our analysis only DNMT3A was significantly hypomethylated and overexpressed in patients with MCL and MCL cell lines (supplemental Table 6). An increased expression of DNMT3A has been associated with aberrant gene silencing in myelodysplastic syndrome43 and acute myeloid leukemia44 and can be targeted with DNMT inhibitors such as DAC. DNMT3a may similarly mediate aberrant gene silencing in MCL and is amenable to inhibition by DAC in MCL cell lines (supplemental Figure 6B).
Several hypomethylated and up-regulated genes, including HDAC1, CD37,25 NOTCH1,27 and CDK5,28 were validated in single-locus assays. CD37 is a member of the tetraspanin transmembrane family of molecules with expression on B cells but not resting T cells, natural killer cells, or macrophages.45,46 During B-cell development, CD37 is expressed in cells progressing from pre-B to mature B-cell stages and is absent on terminal differentiation to plasma cells.47 The lower expression of CD37 in NBCs, in the presence of greater promoter methylation, is consistent with a model in which NBCs appear poised to enter the germinal center reaction but have not yet completely switched off CD37. The CD37-SMIP was developed with the use of variable regions from the G28-1 hybridoma and engineered constant regions encoding human IgG1 domains.25 CD37-SMIP was engineered to have a molecular size above that filtered by the glomerulus, extending its half-life and conferring a potential advantage over monoclonal antibodies.25 CD37-SMIP has shown significant activity in vitro and in vivo in a murine severe combined immunodeficient xenograft model of chronic lymphocytic leukemia.25 Mice lacking CD37 do not exhibit any defects in development or constitution of lymphoid organs but do show a reduced IgG1 level in the serum and an alteration in T-cell antigen–dependent B-cell response.48 Treatment of patients with unselected B-cell NHL with CD37 conjugated with radioactive iodine-131 caused only a transient lymphopenia, without infectious complications, and did not result in changes in immunoglobulin levels.49,50 We observed hypomethylation of CpGs in the CD37 promoter in patients with MCL and MCL cell lines compared with NBCs, suggesting that loss of promoter hypermethylation contributes to high CD37 expression in MCL. Our results may be used to stratify patients in clinical trials of MCL on the basis of their CD37 promoter methylation and gene expression to identify a group at higher likelihood to respond to CD37-SMIP therapy. Further investigation into this and other hypomethylated/overexpressed targets in MCL may expand the opportunities to therapeutically target this disease.
Numerous aberrantly hypermethylated genes were identified in MCL. We focused on the CDKN2B,6 MLF-1,30 PCDH8,31 and HOXD832 genes that have tumor suppressor functions and were found to be hypermethylated with lower expression in both patients with MCL and MCL cell lines compared with NBCs. There have been large differences in the reported proportions of patients with MCL having hypermethylated CDKN2B, from 9%7 to 62%6 despite the use of identical PCR primers. In the current study, CDKN2B was clearly hypermethylated compared with NBCs as assessed by both HELP analysis (P < .001) and mass spectrometry (P < .001), providing independent confirmation of CDKN2B promoter hypermethylation in most of the patients with MCL studied, consistent with the report by Hutter et al.6 Treatment of Z138 MCL cell line with DAC, SAHA, or both led to induction of HOXD8, PCDH8, and MLF1 expression and a reduction in cell viability. The combination of DAC and SAHA was synergistic, which could reflect multiple anti-MCL mechanisms: (1) reversing hypermethylation of tumor suppressor genes (supplemental Figure 3), (2) inhibition of HDAC1 (which was one of the hypomethylated and overexpressed genes identified), and (3) inhibition of CCND151 translation. Even low doses of these drugs had potent cytotoxic synergy in MCL cells, suggesting the clinical utility of this combination in the treatment of MCL.
In summary, this study shows prominent and aberrant methylation of the MCL genome and indicates novel gene targets in samples from patients with MCL and MCL cell lines. The identification of aberrant methylation may have implications for investigation and treatment of malignancies beyond MCL. Hypomethylated and overexpressed genes such as CD37 may be directly targeted for therapy. We also demonstrate anti-MCL potential for the combination of 2 epigenetic drugs, DAC and SAHA. Given the urgent need for expanding therapeutic options in MCL and the established safety profile of DAC and SAHA in clinical trials, epigenetic therapy with the use of this combination may be a useful addition in the armamentarium against MCL. More generally, our data support a rationale for an integrative approach studying genomewide promoter methylation and gene expression to yield novel insights into the biology and treatment of human disease.
The publisher or recipient acknowledges right of the US government to retain a nonexclusive, royalty-free license in and to any copyright covering the article.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Einstein Epigenomics Core, especially Suzuki Masako, PhD, John Greally, MD, PhD, Paraic Kenny, PhD, and Kenny Ye, PhD, for help with array analysis. We also thank Robert Gallagher, MD, Barbara Birshtein, PhD, and David Goldman, MD, for reading and commenting on the manuscript.
This work was supported by a grant from the Chemotherapy Foundation and by a Paul Calabresi Career Development Award (K12 CA132783-01; S.P.); by the National Cancer Institute (R01 CA104348 to A.M.; R01 HL082946 to A.V.; P01 CA95426 and P01 CA813534 to J.C.B. and N.M.); by the Chemotherapy Foundation and the Samuel Waxman Research Foundation (A.M.); by the Gabrielle Angel Foundation, the Leukemia & Lymphoma Society, and the Hershaft Family Foundation (A.V.); and by the D. Warren Brown Foundation and the Leukemia & Lymphoma Society (J.C.B. and N.M.). A.M. is a Leukemia & Lymphoma Scholar and Burroughs-Wellcome Clinical Translational Scientist.
National Institutes of Health
Authorship
Contribution: V.V.L., P.-Y.K., D.T.Y., and S.P. designed and performed the research, analyzed and interpreted data, and wrote the paper; R.S. performed the experiments and contributed vital reagents; T.G. and A.P. contributed to data analysis; Y.Y. provided support for statistical analysis; W.W., I.B., N.M., B.S.K., and J.C.B. designed and performed research and analyzed data; Y.R., M.A.W., and S.R. designed and performed the experiments; K.S.S., A.G., and A.W. contributed vital reagents and designed the research; and A.V., and A.M. contributed in the design of the research and performed the critical review of the manuscript. All the authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Samir Parekh, Albert Einstein College of Medicine of Yeshiva University, 1300 Morris Park Ave, Chanin 302-D2, Bronx, NY 10461; e-mail: samir.parekh@einstein.yu.edu.
References
Author notes
V.V.L. and P.-Y.K. contributed equally to this study.


![Figure 2. Differentially methylated genes chosen for validation. (A) Plot of methylation difference (x-axis) vs significance (on y-axis) showing selected differentially methylated loci between patients with MCL and MCL cell lines compared with NBCs. Probe sets that were differentially methylated are marked in blue (P < .01). (B) Ingenuity pathway core analysis of the top differentially methylated loci in patients with MCL compared with NBCs identifies HDAC1 and NFKB1 as central nodes in the most significantly enriched pathway. Differentially hypomethylated loci in patients with MCL are indicated in red, and differentially hypermethylated loci are indicated in green. (C) Methylation [HELP log(HpaII/MspI) ratios] at the 8 differentially methylated loci chosen for validation represented in a heat map. Scale shows relative methylation with blue indicating hypermethylation and red indicating hypomethylation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/7/10.1182_blood-2009-12-257485/4/m_zh89991055060002.jpeg?Expires=1762130397&Signature=jFv--yr0c2BMfNBswg2KdpoNA6MGxD7UeWsMFjv-RLUAuoJmpwOt5eKyiWppaQ4JFqyerSEEnRUGVarHGHQrmDBiWWu95vJqyC0IMyARk15D5-IBammcJL8x6R4HCKfZJqqWkcp7SXpr1LxFJ1SbThjpxaB0lWDobL4jSC8cIIhBcZy~juYxX8ursqpPB-TTiTKrxzvyQ65FuTFRDKvi2yuhlGJOgaRM5zGlbrFVo~RFEPvEAxQ-67zxeQAh1eSz8SwHQ02BxMbeDRMwIUbq6nsZpt9FkAjwFnoyDmSip3eS6c1qcNbYrqIrCFOw40XzEh--isxMaQwjcu2WgUAJbQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)






![Figure 2. Differentially methylated genes chosen for validation. (A) Plot of methylation difference (x-axis) vs significance (on y-axis) showing selected differentially methylated loci between patients with MCL and MCL cell lines compared with NBCs. Probe sets that were differentially methylated are marked in blue (P < .01). (B) Ingenuity pathway core analysis of the top differentially methylated loci in patients with MCL compared with NBCs identifies HDAC1 and NFKB1 as central nodes in the most significantly enriched pathway. Differentially hypomethylated loci in patients with MCL are indicated in red, and differentially hypermethylated loci are indicated in green. (C) Methylation [HELP log(HpaII/MspI) ratios] at the 8 differentially methylated loci chosen for validation represented in a heat map. Scale shows relative methylation with blue indicating hypermethylation and red indicating hypomethylation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/7/10.1182_blood-2009-12-257485/4/m_zh89991055060002.jpeg?Expires=1763234624&Signature=QR34-0KOJyi8s0k8BO-JqEMUq0LS2ZPJu3f~giocgQ5BNJ~t2qCOJ7mN16uI-3MTQikFjwKfocJiACQWa2h0ylldfSl-Hndvcybl01peMWpkENkmhk2NwncnoU1VbWO4s6r6p15MMVM1XXC9mLafrTH3h26GQeGNxe4g29bUvqSjg~qXN5YX8M30vJHD4CSaVjLq4Uhl~yEGRrJVCBMozmL1FSXQmbKiDvYz6zsB-0~LAuFYjmL6kFxZUmSV9G5uV4ITsPzdgFnIbWF~QLNP0Zs67DqH1DvIjKDXAdUThpxG80HghqtvpXqNRML2R-MhFrk9rJ2Ebma5cyiFeHyWIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)