Abstract
Diamond Blackfan anemia (DBA) is an inherited erythroblastopenia associated with mutations in at least 8 different ribosomal protein genes. Mutations in the gene encoding ribosomal protein S19 (RPS19) have been identified in approximately 25% of DBA families. Most of these mutations disrupt either the translation or stability of the RPS19 protein and are predicted to cause DBA by haploinsufficiency. However, approximately 30% of RPS19 mutations are missense mutations that do not alter the stability of the RPS19 protein and are hypothesized to act by a dominant negative mechanism. To formally test this hypothesis, we generated a transgenic mouse model expressing an RPS19 mutation in which an arginine residue is replaced with a tryptophan residue at codon 62 (RPS19R62W). Constitutive expression of RPS19R62W in developing mice was lethal. Conditional expression of RPS19R62W resulted in growth retardation, a mild anemia with reduced numbers of erythroid progenitors, and significant inhibition of terminal erythroid maturation, similar to DBA. RNA profiling demonstrated more than 700 dysregulated genes belonging to the same pathways that are disrupted in RNA profiles of DBA patient cells. We conclude that RPS19R62W is a dominant negative DBA mutation.
Introduction
Josephs1 and Diamond and Blackfan2 reported the first cases of Diamond Blackfan anemia (DBA; OMIM #205900), a rare (4-7 per 1 million live births)3-5 inherited bone marrow failure syndrome almost exclusively affecting the erythroid lineage.6 DBA is usually diagnosed in the first 3 months of life, and 99% of DBA patients are diagnosed before the age of 5 years.3,6 DBA is characterized by a moderate to severe anemia with normal neutrophil and platelet counts and a marked reduction in number of red cell precursors (0%-< 5%) in an otherwise normocellular bone marrow.6-9 DBA patients may exhibit elevated fetal hemoglobin,6 elevated erythrocyte deaminase adenosine levels,6,10 and a small increased risk of malignancy.3,11 Approximately 40% of patients have cranial/facial, limb, cardiac, or urogenital abnormalities.3,6,7,12 Although most DBA cases are sporadic, the familial cases show an inheritance pattern of autosomal dominant with incomplete penetrance.6,7
The first mutation associated with DBA was a deletion of Ribosomal Protein S19 (RPS19) gene,13 and subsequent studies have shown that approximately 25% of DBA patients carry mutations in the RPS19 gene.3,14,15 Recent studies have shown that 1% to 10% of DBA patients have mutations in other ribosomal protein genes, including: RPS24,16,17 RPS17,18,19 RPS10, RPS26,20 RPL5, RPL11,21-23 and RPL35a.24 Altogether, mutations in genes encoding ribosomal protein subunits account for approximately 50% of the genetic defects identified in DBA patients.25
The RPS19 gene encodes one of the proteins that make up the small 40S subunit of the ribosome. More than half of the RPS19 mutations are either deletions of one RPS19 allele or insertional, frame shift, splice site, or nonsense mutations that result in premature termination of RPS19 protein synthesis, resulting in a deficiency of RPS19 protein in patient cells, which is hypothesized to cause DBA.3,26-30 In support of this model, siRNA knockdown of RPS19 in erythroid TF-1 cells or in human CD34+ progenitor cells caused a decrease in proliferation.30 Growth retardation, defective erythroid differentiation, and hypoplastic anemia with increased apoptosis have been described in RPS19-deficient zebrafish.31,32 Ectopic expression of RPS19 in hematopoietic progenitor cells from RPS19-deficient DBA patients rescued erythroid colony-forming activity in vitro,33 leading to the conclusion that haploinsufficiency of RPS19 is responsible for DBA.27,28
The remaining DBA-associated mutations in the RPS19 gene are missense mutations that alter a single amino acid in the RPS19 protein. Similarly, the dark skin (Dsk3/+) mouse, which has a mild anemia, has a missense mutation in Rps19.34 Gregory et al introduced 16 missense mutations associated with DBA into the yeast homolog of RPS19 and analyzed the crystal structure of the mutant RPS19 proteins.35 More than half of the missense mutations (termed class I) caused the RPS19 protein to be folded improperly; and when expressed in human cells, these mutant RPS19 proteins were rapidly targeted for degradation, consistent with the haploinsufficiency model.35-37 The remaining mutations (termed class II mutations) did not affect protein folding and were stable when expressed in Cos-7, HEK293, and HeLa cells.35-38 These observations have led to the hypothesis that the class II mutations may act by a dominant negative mechanism.35,36
Dominant negative mutations were described by Herskovitz as “mutant polypeptides that when overproduced inhibit the function of the wild-type gene products.”39 To function as dominant negative proteins, the mutant polypeptides must interfere with the function of dimeric or higher-order complexes. Similar to the clinical spectrum of DBA patients with RPS19 mutations,3,14 dominant negative mutations are predicted to range from severe to mild.40
To test the hypothesis that class II RPS19 mutations cause DBA by a dominant negative mechanism, we developed mouse models that express either wild-type RPS19 or an RPS19 gene with a missense mutation that substitutes a tryptophan residue for an arginine residue at the highly conserved position 62 (RPS19R62W).14,35 In this report, we show that mice expressing wild-type RPS19 are normal, whereas mice expressing the RPS19R62W protein have a mild anemia that correlates with the level of RPS19R62W expression. Mice expressing RPS19R62W have decreased numbers of both burst-forming units-erythroid (BFU-E) and colony-forming units-erythroid (CFU-E) and a failure of the erythroblasts to condense their chromatin and enucleate. Microarray profiling of wild-type and mutant erythroblasts demonstrated that the most significantly affected pathways were posttranscriptional modification, protein synthesis, organismal injury (including p53), cell growth and proliferation, and iron metabolism. These data are consistent with a model in which the RPS19R62W mutation causes a DBA-like phenotype by a dominant negative mechanism.
Methods
RPS19 constructs
The plasmid Ins-CMV-C-B-A (developed by Jun Cheng of the National Human Genome Research Institute [NHGRI] Embryonic Stem Cell and Transgenic Mouse Core Facility) contains the CMV enhancer/chicken β-actin promoter, flanked by 1.2-kb chicken β-globin HS4 insulators (cHS4).41 The human wild-type RPS19 or RPS19R62W cDNAs (483 bp) were linked to a 1503-bp fragment containing the human γ-globin IVS2 and 3′-untranslated region. For constitutive expression, the RPS19/γ-globin transgene was inserted between the β-actin promoter and the 3′ insulator. To express the RPS19 genes conditionally, a 3.2-kb fragment containing the PGK-neomycin resistance gene (with stop codons in all 3 reading frames) flanked by Lox P sites was inserted between the β-actin promoter and the RPS19 cDNA.
The 6.4-kb constitutive or 9.6-kb conditional constructs were excised with Pac I and Pvu I and prepared for microinjection into fertilized FVB/N (Taconic Farms) eggs as described.42 Founder animals were identified by Southern blot analysis of DNA by probing with a chicken HS4 probe.42 Copy number was determined by comparing the cHS4 signals of F1 animals to known copy number controls using a Molecular Dynamics PhosphorImager. Heterozygous F1 females were crossed to Prion-Cre (Cre)43 males, and Cre-positive offspring were identified by polymerase chain reaction (PCR) using Cre-specific primers (Table 1). All mouse experiments were approved by the NHGRI.
RNA analyses
Total cellular RNA was extracted from adult bone marrow, spleen, thymus, brain, lung, heart, liver, muscle, kidney, and ovary/testis using TRIZOL reagent according to the manufacturer's specifications (Invitrogen). Quantitative reverse-transcriptase PCR was performed using oligonucleotide primers located in the β-actin promoter (forward) and the RPS19 cDNA (reverse) and the β2-microglobulin mRNA (Table 1). PCR reactions were performed in an iCycler (Bio-Rad) with SYBR Green, and the fluorescence intensities for each reaction were normalized to the intensity of β2-microglobulin.
A 1.3-kb fragment containing RPS19 and γ-globin sequences was cloned into pSP73. A linear DNA template was prepared and 32P-labeled RNA probes were transcribed using the MAXIscript in vitro transcription kit (Ambion). Hybridization of the probe and RNA (0.5 μg) and the RNase A/RNase T1 digestion were carried out according to standard procedures (RPA II, Ambion). The protected fragments were separated on an 8% polyacrylamide gel, and the relative amounts of human RPS19 (584 bp) and mouse Rps19 (93 bp) mRNA were determined on a Molecular Dynamics PhosphorImager by the following formula: (human RPS19 mRNA/mouse Rps19 RNA).
Colony-forming cell assays
Unfractionated cells from day 13.5 fetal livers were suspended at 1 × 104 cells/mL in Methocult M3334 (StemCell Technologies). CFU-E colonies were counted 2 to 3 days after plating. To analyze BFU-E and granulocyte-macrophage colony-forming units (CFU-GM), day 13.5 fetal liver cells were suspended at 2 × 104 or 1 × 105 cells/mL in Methocult GF M3434 (StemCell Technologies), and colonies were counted 10 to 12 days after plating. Cells were plated in triplicate, and each experiment was performed on a minimum of 3 RPS19R62W/Cre, 3+/Cre, 3 RPS19R62W/+, and 3 wild-type embryos. All experiments were analyzed using a Student t test.
Cells from adult (> 4 weeks) bone marrow and spleen were depleted of cells expressing nonerythroid lineage markers using a cocktail of antibodies (anti-CD4, anti-CD8, anti-B220, anti–Gr-1, and anti–Mac-1; BD Biosciences PharMingen) coupled with magnetic bead depletion as described.45 Lineage-negative (Lin−) cells were suspended at 2 × 102 cells/mLin Methocult M3334 (CFU-E) or 1 × 104 cells/mL in Methocult GF M3434 (BFU-E, CFU-GM), and colonies were counted as described. DNA was prepared from individual colonies and analyzed for Cre-mediated excision of the PGK Neo sequences by PCR amplification with primers specific for Slc4a1 (+ control),46 Neo (unexcised), and β-actin/RPS19 (excised; Table 1). The data described were similar in the 1- and 4-copy RPS19R62W transgenic lines and were combined for analysis.
FACS analyses
Fetal livers from day 13.5 embryos and Lin− bone marrow and spleen cells were stained with either anti-CD71 and anti-Ter119 antibodies,47 or anti-CD44 and anti-Ter119 antibodies48 (BD Biosciences PharMingen) and sorted using a FACSAria (BD Biosciences) using 488-nm argon and 633-nm helium-neon lasers. Cell-cycle analysis was performed using 7-amino-actinomycin D staining solution (BD Biosciences PharMingen) on a FACSCalibur flow cytometer (BD Biosciences) running FlowJo software Version MAC 9.0.2 (TreeStar). Each experiment was performed a minimum of 4 times, and the data were analyzed using a Student t test. The data described herein were similar in the 1- and 4-copy RPS19R62W transgenic lines and were combined for analysis.
Microarray analyses
RNA was prepared from Lin− CD71+ Ter119+ bone marrow cells from adult RPS19R62W/+ and RPS19R62W/Cre animals using TRIzol reagent. RNA quality and quantity were ensured using the Bioanalyzer (Agilent) and NanoDrop (Thermo Scientific), respectively. A total of 5 μg of total RNA was labeled with the Affymetrix One-Cycle Target Labeling Reagents (catalog no. 900493). The labeled cRNAs were hybridized to Affymetrix Mouse 430A GeneChips, washed, and stained using the standard format and protocols. An Affymetrix Gene Chip Scanner 3000 and Gene Chip Operating software, Version 1.2 were used to calculate the gene expression intensities. GeneSifter software Version 3.3 (VizX Labs) was used to calculate the significance of differences after application of the Benjamin and Hochberg correction. Differentially expressed genes were analyzed using the GeneSifter or Ingenuity Pathway Analysis (IPA) software Version 7.0 (Ingenuity Systems, Winter 2004 Release containing 20 000 genes). A IPA score of 3 indicates that there is a 1 in 1000 chance that the focus genes are placed into a network resulting from random chance. All microarray data are publicly available in the Gene Expression Omnibus (GEO; National Center for Biotechnology Information) database under accession number GSE22645.
Results
Generation of transgenic mice carrying a mutated RPS19 gene
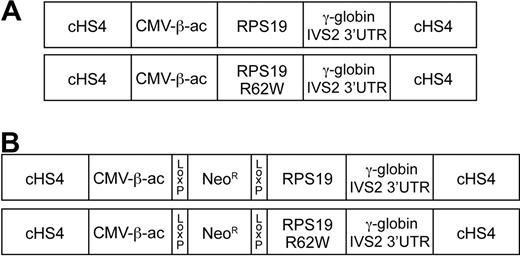
The RPS19R62W mutation has been identified in at least 4 DBA pedigrees.14 To test whether the RPS19R62W mutation could cause DBA by a dominant negative mechanism, transgenic mice were generated that constitutively express the human RPS19 and RPS19R62W genes. Wild-type and mutant RPS19 cDNAs were fused to the ubiquitously expressed CMV/chicken β-actin promoter. To provide introns for RNA processing and a stable 3′-untranslated region, human γ-globin sequences, including intron 2 and the 3′-UTR were placed 3′ to the RPS19 cDNAs. Finally, to prevent gene silencing, the constructs were flanked with the well-described chicken β-globin hypersensitive site 4 insulator41,42 (Figure 1A).
Diagrams of the constructs used to generate transgenic mice. All constructs are flanked by the chicken β-globin HS4 insulator (cHS4). The CMV enhancer/chicken β-actin promoter (CMV-β-ac) is used to express either the wild-type RPS19 cDNA (RPS19) or an RPS19 cDNA with the R62W point mutation (RPS19R62W). After the RPS19 stop codon, the 3′ end of the human γ-globin gene, including 52 bp of exon 2 (out of frame), IVS2, exon 3, and the 3′untranslated region (γ-globin IVS2 3′LTR) are attached to provide sequences for splicing, poly A addition, and mRNA stability. (A) Constructs that allow constitutive expression of the RPS19 transgenes. (B) Constructs that allow conditional expression of the RPS19 transgenes after Cre-mediated excision of the PGK-Neo sequences (NeoR) flanked by Lox P sites that were inserted between the CVM enhancer/chicken β-actin promoter (CMV-β-ac) and RPS19 cDNAs.
Diagrams of the constructs used to generate transgenic mice. All constructs are flanked by the chicken β-globin HS4 insulator (cHS4). The CMV enhancer/chicken β-actin promoter (CMV-β-ac) is used to express either the wild-type RPS19 cDNA (RPS19) or an RPS19 cDNA with the R62W point mutation (RPS19R62W). After the RPS19 stop codon, the 3′ end of the human γ-globin gene, including 52 bp of exon 2 (out of frame), IVS2, exon 3, and the 3′untranslated region (γ-globin IVS2 3′LTR) are attached to provide sequences for splicing, poly A addition, and mRNA stability. (A) Constructs that allow constitutive expression of the RPS19 transgenes. (B) Constructs that allow conditional expression of the RPS19 transgenes after Cre-mediated excision of the PGK-Neo sequences (NeoR) flanked by Lox P sites that were inserted between the CVM enhancer/chicken β-actin promoter (CMV-β-ac) and RPS19 cDNAs.
Nine independent lines of transgenic mice carrying 1 to approximately 25 copies of the constitutive wild-type RPS19 construct were identified, 8 of which transmitted the RPS19 gene to their F1 progeny (Table 2). Real-time reverse transcriptase PCR analysis demonstrated human RPS19 mRNA in every tissue tested, including bone marrow, spleen, thymus, brain, heart, lung, muscle, kidney, liver, and ovary/testis (data not shown). Regardless of the transgene copy number, complete blood counts of animals expressing wild-type RPS19 were similar to age- and sex-matched littermate controls (Table 3). We conclude that ectopic expression of wild-type RPS19 is not detrimental in transgenic mice.
Thirteen founder animals carrying the constitutive RPS19R62W construct were identified. Three of these animals were anemic and died between 2 and 4 months of age with hematocrits of less than 30%. In contrast to the wild-type animals, none of the RPS19R62W founder animals transmitted the transgene to their progeny (χ2 = 107.7; P < .001). To test whether animals were dying in utero, we examined a total of 54 E13.5 embryos from 4 founder lines. No RPS19R62W-positive embryos were detected (Table 2). Cotransfection of embryonic stem (ES) cells with a neomycin resistance gene and the RPS19R62W construct generated no colonies, whereas more than 50 ES cell clones carrying the wild-type RPS19 construct were isolated. These data suggest that ectopic expression of RPS19R62W is lethal during an early stage of embryogenesis.
To circumvent embryonic lethality, we inserted a PGK-neomycin resistance (PGK-Neo) gene with 3 stop codons flanked by Lox P sites between the β-actin promoter and the RPS19 sequences for conditional expression of RPS19 (Figure 1B). In these constructs, Cre recombinase/LoxP-mediated excision of the PGK-Neo gene allows RPS19 expression. Eleven conditional wild-type RPS19 and 11 conditional RPS19R62W founder animals were identified. Eight of 9 wild-type RPS19 lines transmitted the construct to F1 progeny, whereas germline transmission was observed in 4 of 11 RPS19R62W lines (Table 4). Two of the 3 RPS19R62W lines transmitted one and 4 unrearranged copies of the transgene, respectively, whereas the other 2 lines transmitted a rearranged RPS19 transgene. We bred female RPS19/+ and RPS19R62W/+ F1 mice to male Prion-Cre (Pn-Cre) transgenic mice that express Cre at high levels in early embryonic cells.44 Because the male genome is not expressed in the earliest embryonic stages,42 excision occurs later in development, which we hypothesized would bypass the early embryonic lethality.
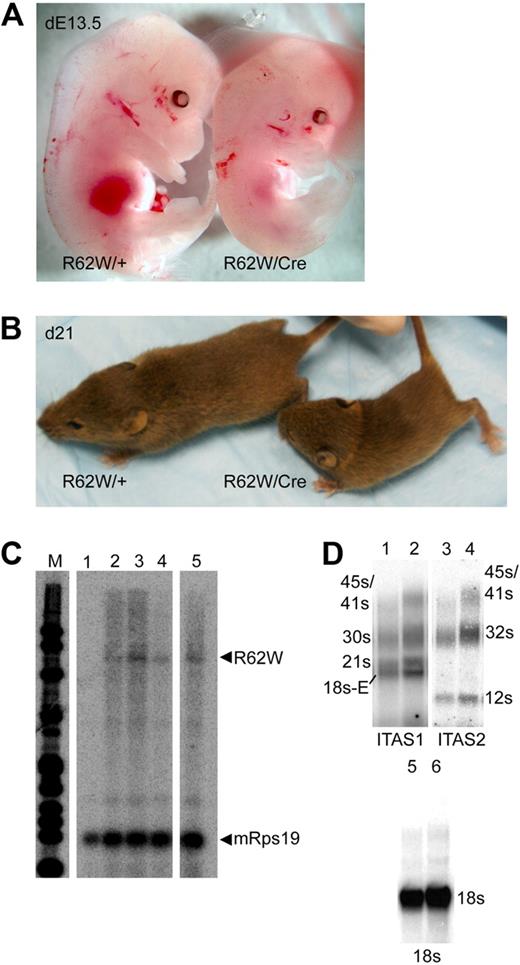
Mice carrying both the wild-type RPS19 (RPS19/+) and the Pn-Cre (+/Cre) transgenes were born at the expected frequency (25%) and were normal in all respects. In contrast, E13.5 embryos carrying both the RPS19R62W and the Pn-Cre transgenes (R62W/Cre) were found at lower than expected frequencies (< 15%), and were severely anemic (Figure 2A). R62W/Cre mice were born at a frequency of less than 10% (8 of 109; (χ2 = 27.68; P < .001), were smaller than their littermates (Figure 2B), and rarely lived beyond weaning (28 days). RNase protection analysis of RNA extracted from bone marrow cells of R62W/Cre animals demonstrated the expected 584-bp protected fragment, indicating normal splicing of the γ-globin IVS2 and 3′ processing of the RPS19R62W/γ-globin mRNA. The RPS19R62W mRNA was expressed at levels that were approximately 10% the level of the endogenous mouse Rps19 mRNA (Figure 2C). No skeletal, craniofacial, or pigmentation abnormalities were observed.
Analysis of transgenic mice carrying the conditional RPS19R62W construct with (R62W/Cre) and without (R62W/+) the Prion-Cre transgene. (A) Day 13.5 embryos. (B) Three-week-old pups. (C) RNase protection analysis of fetal liver RNA from day 13.5 embryos. Lane 1 indicates RNA from R62W/+ embryo; and lanes 2 to 5, RNA from R62W/Cre embryos. The transgenic RPS19 mRNA protects a 584-bp fragment of the probe. The mouse Rps19 mRNA protects 2 93-bp fragments of the probe, providing an internal control. (D) Northern blot analysis of rRNA processing. The individual rRNA products are indicated at the side of the figure. Lanes 1 and 2 indicate bone marrow RNA from R62W/+ (lane 1) R62W/Cre (lane 2) animals probed with an ITS1 probe; lanes 3 and 4, bone marrow RNA from R62W/+ (lane 3) R62W/Cre (lane 4) animals probed with an ITS2 probe; lanes 5 and 6, bone marrow RNA from R62W/+ (lane 5) R62W/Cre (lane 6) animals probed with an 18s rRNA probe. The apparent increases in the level of 21s (ITS1) and 12s (ITS2) are the result of the increased amount of R62W/Cre RNA in lanes 2, 4, and 6 as demonstrated by the level of 18s hybridization.
Analysis of transgenic mice carrying the conditional RPS19R62W construct with (R62W/Cre) and without (R62W/+) the Prion-Cre transgene. (A) Day 13.5 embryos. (B) Three-week-old pups. (C) RNase protection analysis of fetal liver RNA from day 13.5 embryos. Lane 1 indicates RNA from R62W/+ embryo; and lanes 2 to 5, RNA from R62W/Cre embryos. The transgenic RPS19 mRNA protects a 584-bp fragment of the probe. The mouse Rps19 mRNA protects 2 93-bp fragments of the probe, providing an internal control. (D) Northern blot analysis of rRNA processing. The individual rRNA products are indicated at the side of the figure. Lanes 1 and 2 indicate bone marrow RNA from R62W/+ (lane 1) R62W/Cre (lane 2) animals probed with an ITS1 probe; lanes 3 and 4, bone marrow RNA from R62W/+ (lane 3) R62W/Cre (lane 4) animals probed with an ITS2 probe; lanes 5 and 6, bone marrow RNA from R62W/+ (lane 5) R62W/Cre (lane 6) animals probed with an 18s rRNA probe. The apparent increases in the level of 21s (ITS1) and 12s (ITS2) are the result of the increased amount of R62W/Cre RNA in lanes 2, 4, and 6 as demonstrated by the level of 18s hybridization.
The number of circulating nonerythroid cells and the red cell indices were similar in age- and sex-matched wild-type, +/Cre, R62W/+, and R62W/Cre mice (Table 3; and data not shown). In contrast, compared with wild-type, R62W/+, or Cre/+ mice, R62W/Cre mice had a mild anemia with significantly decreased red blood cell numbers, hematocrit, and hemoglobin (Table 3; and data not shown). Partial compensation for the anemia was provided by an increase in Ter119+ erythroid cells in R62W/Cre bone marrow (increased 1.3-fold; 21.8% ± 7.7% to 29.1% ± 4.8%; P < .01) and spleen (3.7-fold; 4.9% ± 1.3% to 18.3% ± 5.7%; P < .001) compared with wild-type, R62W/+, or Cre/+ animals. R62W/Cre animals had 10-fold fewer circulating reticulocytes than wild-type, R62W/+, or Cre/+ animals, similar to DBA patients (Table 3). In the most severely affected animals, Northern blot analysis of ribosomal RNA (rRNA) processing with an ITS1 probe44 demonstrated increased levels of the 45/41s, 30s, and 18s-E rRNA intermediate products. Similarly, the ITS2 probes demonstrated increased levels of the 45/41s and 32s rRNA intermediate products (Figure 2D).
Effects of expression of RPS19R62W on hematopoietic progenitor cells
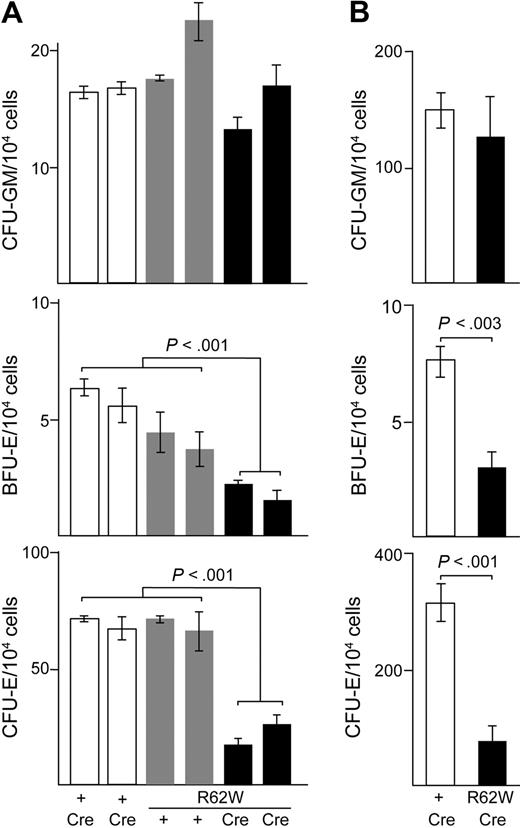
Hematopoietic colony-forming assays were performed to determine whether the anemia in RPS19R62W/Cre mice was related to a decrease in the number of erythroid precursors and progenitors. Nonerythroid CFU-GM were present in similar numbers in both fetal liver (Figure 3A) and bone marrow cells (Figure 3B) of wild-type, R62W/+, +/Cre, and R62W/Cre animals. In contrast, there were significant decreases in the number and frequency of BFU-E and CFU-E in R62W/Cre fetal liver and bone marrow cells compared with control animals (Figure 3). We conclude from these data that ectopic expression of RPS19R62W inhibits erythroid colony formation.
Analysis of hematopoietic colony formation in RPS19R62W transgenic mice. (A) Analysis of CFU-GM, BFU-E, and CFU-E from individual day 13.5 fetal livers. Cells were plated in triplicate. The genotypes of the embryos appear below the bars. The Prion-Cre transgene alone = + Cre (□); the R62W transgene alone = Tg + (□); the R62W transgene plus Prion-Cre = Tg Cre (■). (B) Analysis of CFU-GM, BFU-E, and CFU-E from 4- to 6-week-old mice. The data are compiled from 4 to 6 individual mice; cells from each mouse were plated in duplicate. The genotypes of the embryos appear below the bars. The Prion-Cre transgene alone = + Cre (□); the R62W transgene plus Prion-Cre = Tg Cre (■).
Analysis of hematopoietic colony formation in RPS19R62W transgenic mice. (A) Analysis of CFU-GM, BFU-E, and CFU-E from individual day 13.5 fetal livers. Cells were plated in triplicate. The genotypes of the embryos appear below the bars. The Prion-Cre transgene alone = + Cre (□); the R62W transgene alone = Tg + (□); the R62W transgene plus Prion-Cre = Tg Cre (■). (B) Analysis of CFU-GM, BFU-E, and CFU-E from 4- to 6-week-old mice. The data are compiled from 4 to 6 individual mice; cells from each mouse were plated in duplicate. The genotypes of the embryos appear below the bars. The Prion-Cre transgene alone = + Cre (□); the R62W transgene plus Prion-Cre = Tg Cre (■).
DNA was extracted from individual CFU-GM and BFU-E colonies and assayed for excision of the PGK-Neo stop sequence by PCR. Loss of the PGK-Neo sequence was analyzed using primers in the PGK-Neo sequence. Functional β-actin/RPS19 transcriptional units were identified using primers located in the β-actin promoter and the RPS19 gene. No R62W/+ colonies had any evidence of PGK-Neo excision. More than 90% (12 of 13) of the colonies from single copy R62W/Cre mice had no PGK-Neo sequence and a functional β-actin/RPS19 transcriptional unit. In the 4-copy line, Lox P recombination can delete one or more transgenes in addition to the PGK-Neo sequences. More than 90% (49 of 53) of the colonies from 4-copy R62W/Cre mice had at least one functional β-actin/RPS19 transcriptional unit, and 28 of 53 (52.8%) of colonies had no PGK-Neo sequences and at least one functional β-actin/RPS19 transcriptional unit. Semiquantitative real-time PCR demonstrated various numbers of functional β-actin/RPS19 transcriptional units among individual colonies from 4-copy R62W/Cre mice.
FACS analysis of erythropoiesis in transgenic mice expressing RPS19R62W
To compare the maturation of erythroid cells in R62W/Cre and control mice, we analyzed bone marrow and spleen cells depleted of nonerythroid cells (Lin−)45 by fluorescence-activated cell sorter (FACS) analysis using antibodies against Ter119 and CD71 (transferrin receptor)47 or Ter119 and CD44.48 In day 13.5 fetal liver, which is primarily developing erythroid cells, analysis was performed with Ter119 and CD71. These methods identify populations of proerythroblasts (R2; Ter119− CD71/CD44+), basophilic erythroblasts (R3; Ter119+ CD71/CD44+), and more mature orthochromic erythroblasts and reticulocytes (R4 and 5; Ter119+ CD71/CD44LO and Ter119+ CD71/CD44−; Figure 4). The most striking difference between R62W/Cre and control animals was the increase in the relative number of R4 and R5 cells in R62W/Cre animals (Figure 4). Consistent with the decrease in the number of BFU-E and CFU-E we observed in R62W/Cre animals, the relative number of cells in the Ter119− CD71/CD44+ (R2) gate was significantly decreased in R62W/Cre bone marrow (4-fold, P < .01) and spleen (40-fold, P < .001) cells compared with cells from control animals (Figure 4).
FACS analysis of erythropoiesis in R62W transgenic mice. (Left) Analysis of Lin− bone marrow cells (top) Lin− spleen cells (center) and fetal liver cells (bottom), stained with antibodies against CD71 and Ter119. (Right) Analysis Lin− bone marrow (top) and Lin− spleen cells (center) stained with antibiodies against CD44 and Ter119. (Bottom right) Gates used for sorting. Cells from mice carrying the RPS19R62W transgene alone = RPS19R62W; cells from mice carrying the RPS19R62W transgene and the Prion-Cre transgene = RPS19R62W; Cre.
FACS analysis of erythropoiesis in R62W transgenic mice. (Left) Analysis of Lin− bone marrow cells (top) Lin− spleen cells (center) and fetal liver cells (bottom), stained with antibodies against CD71 and Ter119. (Right) Analysis Lin− bone marrow (top) and Lin− spleen cells (center) stained with antibiodies against CD44 and Ter119. (Bottom right) Gates used for sorting. Cells from mice carrying the RPS19R62W transgene alone = RPS19R62W; cells from mice carrying the RPS19R62W transgene and the Prion-Cre transgene = RPS19R62W; Cre.
Transcriptional profiling of basophilic erythroblasts from control and R62W transgenic mice
To identify the effects of RPS19R62W expression during erythroid maturation, we compared the expression profiles of RNA extracted from basophilic erythroblasts (R3) sorted from Lin− control and R62W/Cre bone marrow on Affymetrix Mouse 430A GeneChips. The basophilic erythroblast population was chosen because the relative number of the wild-type and R62W/Cre R3 cells were homogeneous compared with the R2, R4, and R5 populations. At the P < .05 level of significance, we found that 716 genes were dysregulated: 394 genes that were down-regulated in R62W/Cre R3 cells and 322 genes that were up-regulated in R62W/Cre R3 cells (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
The mRNA levels of both the transferrin receptor (CD71) and CD44 genes were significantly down regulated by 2.14-fold (P < .01) and 1.8-fold (P < .01), respectively, which was confirmed by the lower mean fluorescence of CD71 and CD44 on R62W/Cre cells (Figure 4). Sieff et al observed a significant decrease in the level of c-kit mRNA levels in mouse cells rendered haploinsufficient by mRNA knockdown of Rps19 mRNA.28 We also observed a significant 1.7-fold (P < .02) reduction of c-kit mRNA, which was confirmed by FACS analysis (data not shown). Our data showed significant decreases in the levels of the ribosomal protein subunit genes Rpl13a (1.6-fold; P < .001) and Rpl14 (1.9-fold; P < .002) mRNA as well as significant increases in the level of Rps4Y2 (1.7-fold; P < .01), Rpl21 (1.5-fold; P < .02), and Rpl27a (1.6-fold; P < .001) mRNA. We conclude that ectopic expression of RPS19R62W causes changes in the mRNA profile of erythroid cells.
Bioinformatic analysis of the RPS19R62W transcriptional profile
For an unbiased analysis of the biologic significance of the differences in mRNA levels, the set of dysregulated genes was analyzed using IPA software Version 7.0, which maps genes to genetic networks. The networks identified involve 30 to 35 genes with significantly dysregulated expression, and a score of 3 indicates that there is a 1 in 1000 chance that the genes are assigned to a network by random chance. Consistent with the current understanding of DBA, the 5 highest scoring networks were RNA posttranscriptional and posttranslational modification (score = 50); hematologic disease, organismal injury, and abnormalities, including apoptosis (score = 47), cellular growth and proliferation (score = 43), protein synthesis (score = 40), and DNA replication and repair (score = 36; Table 5). An independent analysis of the microarray data using GeneSifter software Version 3.3 (VizX Labs) identified similar networks.
We hypothesized that the genes and gene products that were shared among the significantly affected pathways would represent the critical processes perturbed by the expression of RPS19R62W. Of the 987 genes in significantly affected networks (not every gene in a network needs to be dysregulated to receive a significant network score), 37 genes were present in more than one network (Table 5). Two genes, thioredoxin reductase and transferrin receptor (CD71), were parts of 3 highly scored networks. An analysis of these 37 genes indicated that they were associated with metabolism, oxidative metabolism, apoptosis, cell proliferation, transcription, RNA processing, and protein translation. We conclude that expression of RPS19R62W perturbs the critical gene products and networks associated with DBA.
Morphologic analysis of maturing erythroid cells from control and RPS19R62W transgenic mice
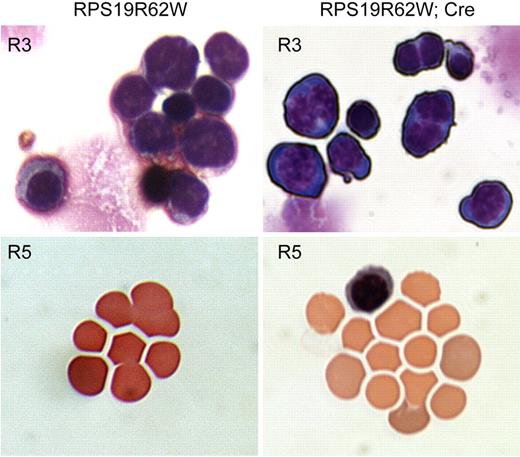
Because both CD71 and CD44 are down-regulated in R62W R3 cells, we compared the morphology of sorted R3, R4, and R5 cells from control and R62W/Cre bone marrow after staining with Wright-Giemsa to determine whether the accumulation of R4 and R5 cells represented delayed erythroid maturation or simply loss of CD71/CD44 expression on normally maturing cells. Control R3 cells had basophilic erythroblast morphology, with less than 2% of the cells containing 2 nuclei. Whereas the basophilic erythroblast morphology of the R62W/Cre R3 population was normal, more than 50% of the cells had 2 nuclei (P < .001; Figure 5). Mitotic figures were observed among the R62W/Cre R3 cells, suggesting that they were capable of undergoing mitosis. Control R4 and R5 cells had the typical morphology associated with orthochromic erythroblasts and reticulocytes with less than 0.2% nucleated cells. In contrast, more than 2% of R62W/Cre R4 and R5 cells were uncondensed, nucleated cells that resemble basophilic erythroblasts (P < .001; Figure 5). In addition, the reticulocytes in the R62W/Cre R5 population were poorly hemoglobinized (Figure 5). We conclude that R62W/Cre erythroid cells fail to undergo proper erythroid maturation.
Morphologic analysis of Wright-Giemsa-stained Lin− bone marrow cells from R62W transgenic mice in the basophilic erythroblast (R3) and reticulocyte (R5) gates. The analysis was performed on a Nikon Eclipse TE300 microscope with a 40×/0.75 NA oil objective under Cargill Immersion oil (type 5). Images were taken with a Nikon N70 camera. No subsequent manipulations were performed. (Left) Analysis of cells from mice carrying the RPS19R62W transgene alone (RPS19R62W). (Right) Analysis of cells from mice carrying the RPS19R62W transgene and the Prion-Cre transgene (RPS19R62W; Cre). Note the binucleate cells among the RPS19R62W, Cre basophilic erythroblasts (R3), and the poorly hemoglobinized and nucleated cells among the RPS19R62W, Cre reticulocytes (R5).
Morphologic analysis of Wright-Giemsa-stained Lin− bone marrow cells from R62W transgenic mice in the basophilic erythroblast (R3) and reticulocyte (R5) gates. The analysis was performed on a Nikon Eclipse TE300 microscope with a 40×/0.75 NA oil objective under Cargill Immersion oil (type 5). Images were taken with a Nikon N70 camera. No subsequent manipulations were performed. (Left) Analysis of cells from mice carrying the RPS19R62W transgene alone (RPS19R62W). (Right) Analysis of cells from mice carrying the RPS19R62W transgene and the Prion-Cre transgene (RPS19R62W; Cre). Note the binucleate cells among the RPS19R62W, Cre basophilic erythroblasts (R3), and the poorly hemoglobinized and nucleated cells among the RPS19R62W, Cre reticulocytes (R5).
Cell-cycle analysis of proerythroblasts and basophilic erythroblasts from control and RPS19R62W transgenic mice
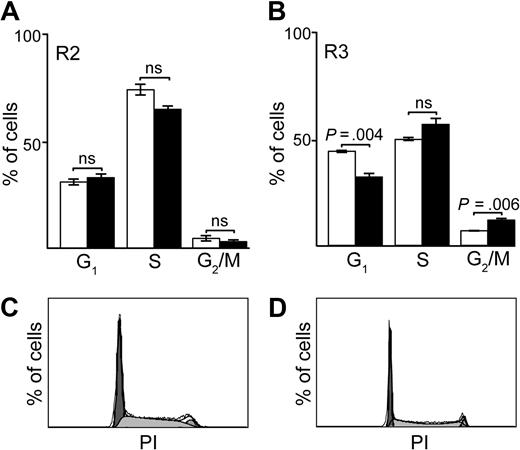
To confirm defects in the cellular growth and proliferation and the DNA replication and repair pathways, we performed cell-cycle analysis of sorted Lin− bone marrow proerythroblasts (Ter119− CD71+; R2) and basophilic erythroblasts (Ter119+ CD71+ R3) from control and R62W/Cre mice. In the R2 population, the cell-cycle profiles of control and R62W/Cre cells were similar (Figure 6A), despite the decreased number of R2 cells (Figure 4) and CFU-E (Figure 3) in R62W/Cre mice. In the R3 population, the cell-cycle profiles of R62W/Cre cells showed a significant decrease in the percentage of cells in the G1 (2n DNA content; P = .004) phase of the cell cycle, and a corresponding significant increase in the percentage of cells in the G2/M (4n; P = .006) phase of the cell cycle (Figure 6B-D). These findings are consistent with a defect in mitosis or cytokinesis and confirm the high percentage of binucleate cells we observed in the basophilic erythroblast population (Figure 5).
Cell-cycle analysis of erythroid progenitors (R2) and basophilic erythroblasts (R3) in day 13.5 fetal liver cells from R62W transgenic mice. (A) Cell-cycle analysis of fetal liver R2 cells from embryos carrying the RPS19R62W transgene alone (□), and from embryos carrying the RPS19R62W transgene and the Prion-Cre transgene (■). (B) Cell-cycle analysis of fetal liver R3 cells from embryos carrying the RPS19R62W transgene alone (□), and from embryos carrying the RPS19R62W transgene and the Prion-Cre transgene (■). The percentage of cells in each phase of the cell cycle is indicated on the y-axis, and the cell-cycle phases (G1, S, G2/M) are on the x-axis. Bars represent data from 4 to 6 individual embryos. (C) Cell-cycle profile of fetal liver R3 cells from embryos carrying the RPS19R62W transgene alone. (D) Cell-cycle profile of fetal liver R3 cells from embryos carrying the RPS19R62W transgene and the Prion-Cre transgene.
Cell-cycle analysis of erythroid progenitors (R2) and basophilic erythroblasts (R3) in day 13.5 fetal liver cells from R62W transgenic mice. (A) Cell-cycle analysis of fetal liver R2 cells from embryos carrying the RPS19R62W transgene alone (□), and from embryos carrying the RPS19R62W transgene and the Prion-Cre transgene (■). (B) Cell-cycle analysis of fetal liver R3 cells from embryos carrying the RPS19R62W transgene alone (□), and from embryos carrying the RPS19R62W transgene and the Prion-Cre transgene (■). The percentage of cells in each phase of the cell cycle is indicated on the y-axis, and the cell-cycle phases (G1, S, G2/M) are on the x-axis. Bars represent data from 4 to 6 individual embryos. (C) Cell-cycle profile of fetal liver R3 cells from embryos carrying the RPS19R62W transgene alone. (D) Cell-cycle profile of fetal liver R3 cells from embryos carrying the RPS19R62W transgene and the Prion-Cre transgene.
Discussion
Haploinsufficiency of RPS19 leading to defective rRNA processing and abnormal ribosome assembly and function is a well-accepted mechanism for how RPS19 deletions and nonsense mutations cause DBA.27,28 However, the mechanism by which RPS19 missense mutations that do not affect protein folding (eg, class II mutations) cause DBA is not as clear, and several groups have proposed that these mutations cause DBA by a dominant negative mechanism.35,36 The transgenic mouse model we have developed specifically tests whether the RPS19R62W point mutation associated with DBA can poison the normal functions of mouse RPS19. We chose the RPS19R62W mutation because it is seen in multiple pedigrees14 and when expressed in monkey or human cells the RPS19R62W protein is stable.36-38 Our data show that ectopic expression of low levels of RPS19R62W are sufficient to cause a mild anemia in mice that is associated with defects in rRNA processing, erythroid proliferation, and erythroid maturation. These effects were specific to the expression of RPS19R62W as mice expressing wild-type RPS19, Cre, or the conditional construct with intact stop sequences were normal in all respects.
Gazda et al have reported dysregulation of multiple genes in human progenitor cells from DBA patients with RPS19 mutations in clinical remission.29 The overlap of the dysregulated genes in our study and the study by Gazda et al29 was less than 10%. We think that the differences in the specific dysregulated genes are the result of differences in the stage of the cells analyzed, CD34+ progenitor cells in the Gazda et al study,29 and basophilic erythroblasts in our analysis. Additional differences could be related to our analysis of mice expressing RPS19R62W, whereas the Gazda et al data29 were from patients with 3 different RPS19 mutations: an insertion that leads to premature termination, a point mutation at position 131, which is proposed to be a class I mutation, and a substitution of a glutamine for arginine at position 62 (RPS19R62Q).29 Finally, the presence of binucleate cells in our RPS19R62W R3 cells may have confounded the comparison with the RNA from the control, mononucleate R3 cells. Despite these differences, at the level of pathways, our IPA analysis identified the same significantly affected pathways as the Gazda et al study.29 RPS19 has been shown to interact with nonribosomal proteins.49 Of the 37 dysregulated genes we identified that were common to 2 or more pathways, 2 gene products (splicing factor arginine/serine rich10 [SFRS10] and β4-integrin [ITGB4]) have been shown by Orrù et al to interact with wild-type RPS19.49 We propose that, in addition to ribosome assembly and function, the interactions of RPS19R62W with SFRS10 and ITGB4 may also contribute to the defects we have observed.
Although sharing many characteristics of DBA, the phenotype we observed in our conditional RPS19R62W/Cre mice differs from DBA in several important respects. First, the anemia in our animals is considerably milder than patients with severe DBA caused by either RPS19R62W or mutations in other ribosomal protein subunits.3,6,14,15 Second, whereas erythropoiesis in DBA patients does not progress beyond the proerythroblast stage,28,50 we observed a decreased number of proerythroblasts and a population of basophilic erythroblasts. Many of these erythroblasts were binucleate, similar to what is seen in congenital dyserythropoietic anemia type II (HEMPAS), a related congenital bone marrow failure syndrome with defects in erythropoiesis.6 Finally, whereas DBA erythroid cells arrest at the G1/S phase of the cell cycle, RPS19R62W erythroblasts were arrested in the G2/M phase of the cell cycle. Our results are consistent with either a mitotic block or a failure of cytokinesis, neither of which has been reported in DBA.
We think that many of the differences between the phenotype we have observed in our mice and DBA patients are the result of the relatively low ratio of RPS19R62W mRNA to endogenous mouse Rps19 mRNA expressed in our animals. In our conditional RPS19R62W/Cre mice, the relative level of mutant RPS19R62W mRNA is approximately 10% of the level of endogenous mouse Rps19 mRNA, whereas in the cells of patients carrying one RPS19R62W allele, the level of RPS19R62W mRNA and protein is predicted to be closer to 50%.36-38 We propose that the low level of RPS19R62W permits erythropoiesis to proceed past a block at the progenitor stage, allowing extramedullary erythropoiesis to compensate for the anemia, but that high levels of RPS19R62W are lethal to differentiating erythroid progenitor cells.
Our results support the hypothesis that the RPS19R62W mutation acts by a dominant negative mechanism. Dominant negative mutations are often more severe than loss of function mutations because the abnormal protein can “poison” otherwise normal protein complexes.39,40 In support of this prediction, we have shown that relatively low levels of RPS19R62W can have a profound effect on rRNA processing. At this point, it is not clear whether RPS19R62W interferes with ribosome assembly or whether incorporation of RPS19R62W into ribosomes renders them dysfunctional. Because expression of RPS19R62W in both ES cells and early embryos results in significant fetal loss, it is possible that fetal loss is also an unreported feature of DBA, particularly in pedigrees with dominant negative mutations.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Jun Cheng, who developed the InsCMV C-B-A vector.
This work was supported by the NHGRI intramural funds (D.M.B.), National Institutes of Health (grant HL79565; N.M.), Fondation Médicale pour la Recherche, Agence Nationale de la Recherche (“maladies rares” RIBODBA; L.D.C.), the Daniella Arturi Foundation (L.D.C.), and the Diamond Blackfan Anemia Foundation (E.E.D.).
E.E.D. is the recipient of 2008 American Society of Hematology Merit Award.
National Institutes of Health
Authorship
Contribution: E.E.D., G.E., and D.M.B. performed experiments; E.E.D. and D.M.B. wrote the paper; and L.D.C., N.M., and D.M.B. designed the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David M. Bodine, Genetics and Molecular Biology Branch, Hematopoiesis Section, NHGRI, 49 Convent Dr, Rm 4A04, MSC 4442, Bethesda MD 20892-4442; e-mail: tedyaz@mail.nih.gov.