Abstract
MLL is a common target for chromosomal translocations associated with acute leukemia resulting in its fusion with a large variety of nuclear or cytoplasmic proteins that may activate its oncogenic properties by distinct but poorly understood mechanisms. The MLL-AF6 fusion gene represents the most common leukemogenic fusion of mixed lineage leukemia (MLL) to a cytoplasmic partner protein. Here, we identified a highly conserved Ras association (RA1) domain at the amino-terminus of AF6 as the minimal region sufficient for MLL-AF6 mediated myeloid progenitor immortalization in vitro and short latency leukemogenesis in vivo. Moreover, the ability of RA1 to activate MLL oncogenesis is conserved with its Drosophila ortholog, Canoe. Although the AF6 RA1 domain has previously been defined as an interaction surface for guanosine triphosphate–bound Ras, single amino acid substitutions known to abolish the AF6-Ras interaction did not abrogate MLL-AF6–mediated oncogenesis. Furthermore, fusion of MLL to heterologous RA domains of c-Raf1 or RalGDS, or direct fusion of MLL to constitutively active K-RAS, H-RAS, or RAP1 was not sufficient for oncogenic activation of MLL. Rather, the AF6 RA1 domain efficiently mediated self-association, suggesting that constitutive MLL self-association is a more common pathogenic mechanism for MLL oncogenesis than indicated by previous studies of rare MLL fusion partners.
Introduction
The mixed lineage leukemia (MLL) gene encodes an epigenetic transcriptional regulator that is essential for definitive hematopoiesis.1,2 MLL is a frequent target of chromosomal translocations associated with particularly aggressive acute leukemias, including acute lymphoid (ALL), myeloid (AML), bi-phenotypic (ABL), and chemotherapy-related secondary leukemias that affect children and adults.3-5 These genetic events result in the consistent production of novel, dominant-acting oncogenic proteins in which MLL is fused with 1 of at least 50 distinct novel partner proteins dependent on the loci that participate in the translocation (reviewed in Ayton and Cleary6 ).
AF6 (ALL-1 fused on chromosome 6) was originally isolated as a novel MLL partner gene disrupted by the t(6;11)(q27;q23) chromosomal translocation associated with the M4/M5 subtype of adult AML.7 Additional studies have further associated the t(6:11)(q27;q23) with infant AML and pro-B ALL, and adult T-ALL.8-12 Thus, expression of an MLL-AF6 fusion protein is associated with transformation of progenitor populations from multiple hematopoietic lineages and developmentally distinct cellular targets. AF6 encodes a large, modular protein that localizes to regions of cell-cell contact such as tight and adherens junctions in epithelial cells.13,14 AF6 contains a single PDZ (Postsynaptic density-95/Discs large/Zonula occludens-1) domain, 2 motifs (RA1 and RA2) that interact with RAS family proteins and the ZO-1 tight junction protein, as well as regions related to myosin V-like and kinesin-like cargo binding domains, and an actin binding domain.7,15 AF6-deficient mouse embryos die at embryonic day (E) 10.5 with defective cell-cell junctions and reduced polarization within the embryonic ectoderm.16 It is currently unclear if AF6 function contributes to normal hematopoiesis.
MLL partner proteins have been broadly classified into 2 distinct groups, based on their structural features and normal cellular localization.6 These groups include nuclear partner proteins with features of putative transcriptional regulators or cytoplasmic partner proteins with various domains associated with intracellular signaling processes. Recent functional studies have proposed that different classes of partner proteins activate the oncogenic potential of MLL by distinct mechanisms. For the nuclear partners, minimal regions that contribute to transformation exhibit transcriptional activation functions in reporter assays, suggesting a common theme whereby components of the transcriptional machinery are inappropriately and constitutively recruited to MLL target genes.17-25 Several MLL-fusion partners, including AF4, AF9, AF10, and ENL, form a higher-order complex involved in transcriptional elongation and/or recruit the histone modifying enzyme hDOT1L (human DOT1-like), ultimately allowing aberrant expression of MLL target genes.26-28 In contrast, for cytoplasmic partners such as AF1p, GAS7, and GEPHYRIN, minimal regions contributing to MLL transformation do not possess intrinsic transcriptional activity but promote dimerization of the amino-terminal portion of MLL.29-31 Alternatively, aberrant recruitment of an epigenetic transcriptional regulator has been described for the rare cytoplasmic MLL fusion partner EEN.32
A limited number of nuclear proteins constitute the majority of fusion partners in MLL-associated acute leukemias, whereas cytoplasmic partner proteins are structurally more diverse but clinically rare. In contrast, AF6 is the only cytoplasmic MLL fusion partner to occur with significant clinical frequency.10 In this report, we investigated mechanisms of oncogenic transformation by MLL-AF6. Our study identifies the small, highly conserved RA1 domain of AF6 to be sufficient for myeloid transformation, but its oncogenic role is mediated by self-association, not RAS recruitment.
Methods
Expression constructs
Murine stem cell virus (MSCV) retroviral constructs encoding in-frame fusions between MLL and partner proteins (AF6, c-Raf, RalGDS, K-Ras10G11, H-RasV12, and Rap1E63) were created by cloning polymerase chain reaction–generated cDNA fragments (flanked by a 5′ NruI site and a 3′ stop codon followed by an XhoI restriction site) into the NruI and XhoI sites of the MSCV Neo 5′-MLL vector.18 For the MLL-CanoeRA1 fusion, a similar strategy was used to fuse a Canoe cDNA fragment (encoding amino acids 39-133) with MLL. Site-directed mutagenesis of the RA1 domain of AF6 was performed with an overlap extension polymerase chain reaction and Pfu Turbo (Stratagene), followed by cloning of NruI and XhoI flanked products into the MSCV 5′-MLL vector. Heterologous RA domains consisted of residues 51 to 131 of murine c-Raf1 and residues 730 to 823 of murine RalGDS. Portions of AF6 cDNAs were excised from MSCV MLL-AF6 constructs and transferred in-frame to a pCMV5 derivative containing a carboxy terminal FLAG or HA epitope tag, respectively. For MLL-AF6 constructs a similar strategy was used to add an amino terminal FLAG epitope tag. To generate cytomegalovirus (CMV) GAL4-AF6 N-terminal conserved region (NCR), the DNA binding domain of GAL4 (residues 1-147) was fused in frame to the NCR of AF6 (residues 35-347).
Myeloid progenitor immortalization and leukemogenicity assays
All experiments on mice in this study were performed with the approval of and in accordance with Stanford University's Administrative Panel on Laboratory Animal Care. Transduction of murine bone marrow (BM) cells, methylcellulose replating assays, and generation of immortalized cell lines were performed as previously described.33 For primary transplantations, 6-week-old syngeneic C57BL/6 mice were sublethally irradiated (4.5 Gy [450 rad]) and received a transplant with 106 immortalized cells via the retro-orbital plexus. Animals were routinely monitored for signs of disease and were killed when morbid. Secondary transplantations were performed under identical conditions except that 106 BM cells isolated from primary transplant recipients showing signs of morbidity were transplanted into sublethally irradiated syngeneic recipients.
Coimmunoprecipitation assays
Subconfluent 293 cells cultured in 6-cm dishes were transfected with 4 μg of each CMV-driven expression plasmid with the use of Lipofectamine Plus (Invitrogen). After 48 hours, cells were harvested and washed once in phosphate-buffered saline. Nuclei were prepared by suspension of washed cells in isotonic buffer (150mM NaCl, 1.5mM MgCl2, 10mM Tris-HCl [tris(hydroxymethyl)aminomethane] at pH 7.5, 0.5% NP-40, EDTA [ethylenediaminetetraacetic acid]–complete protease inhibitor; Roche) on ice for 5 minutes, followed by sedimentation at 300g for 5 minutes. The nuclei were suspended in 1 mL of ice-cold lysis buffer (250mM NaCl, 20mM sodium phosphate at pH 7.0, 30mM sodium pyrophosphate, 5mM EDTA, 10mM NaF, 0.1% NP-40, 10% glycerol, 1mM dithiothreitol, EDTA-complete protease inhibitor cocktail), and the resulting extract was clarified by ultracentrifugation at 30 000 rpm for 30 minutes at 4°C. For preparation of whole-cell lysate, washed cells were immediately suspended in 1 mL of ice-cold lysis buffer, followed by ultracentrifugation as described earlier. The cleared lysate was incubated with anti-FLAG M2 agarose beads (Sigma), and the sample was rotated for 2 hours at 4°C. The beads were pelleted and washed 5 times with 1 mL of ice-cold lysis buffer. Proteins were eluted by boiling in sodium dodecyl sulfate (SDS) sample buffer, separated by SDS–polyacrylamide gel electrophoresis, and electroblotted onto Hybond enhanced chemiluminescence (ECL) membranes (Amersham). Membranes were probed with anti-FLAG (Sigma-Aldrich), anti-HA 3F10 (Roche), or anti-MLL (N4.4) primary antibodies, followed by secondary anti–mouse or anti–rabbit immunoglobulin G–conjugated to horseradish peroxidase (Accurate Antibodies). Bands were visualized with ECL (Amersham).
Western blot detection of MLL fusion protein expression
MSCV retroviral constructs encoding MLL fusion proteins were transiently transfected into virus producing Phoenix E cells with the use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After harvesting the virus-containing supernatant, cells were lysed in SDS sample buffer, boiled, and electrophoresed through 4% SDS–polyacrylamide gel electrophoresis. Gels were electroblotted onto Hybond ECL membranes, which were subsequently probed with primary anti-MLL N4.4 monoclonal antibody, followed by secondary goat anti–mouse conjugated to horseradish peroxidase (Accurate Antibodies). Bands were visualized with ECL (Amersham).
Transcriptional transactivation assays
293 cells plated into 24-well plates were transfected with expression plasmids with the use of Lipofectamine Plus (Invitrogen). For all GAL4-based assays, transfections included 0.1 μg of Gal4 effector plasmid, 0.2 μg of CMV β-galactosidase, and a reporter construct containing 5 copies of the GAL4 consensus binding site and the luciferase gene driven by the thymidine kinase promoter. Twenty-four hours later, luciferase and β-galactosidase activities were determined with Luciferase Assay System (Promega) and Galacto-Light (Tropix), respectively. The expression of GAL4 fusion constructs was detected by immunoblot with the use of an anti-GAL4 RK5C1 monoclonal antibody (Santa Cruz Biotechnology Inc).
Microscopy
Cytospin images were obtained with an Eclipse microscope (Nikon) equipped with a 60×/0.95 DIC M numeric aperture objective, and a SPOT digital camera using acquisition software (Diagnostic Instruments). Images were processed using Adobe Photoshop CS2.
Sequence alignments
Alignments were performed with the use of Clustal W analysis with the MegAlign software (DNA Star Inc), followed by manual adjustment.
Results
The N-terminal conserved region of AF6 is sufficient for myeloid immortalization by MLL-AF6
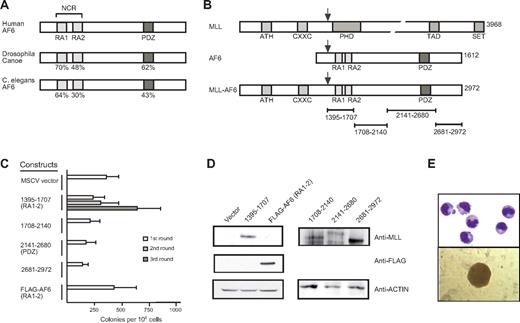
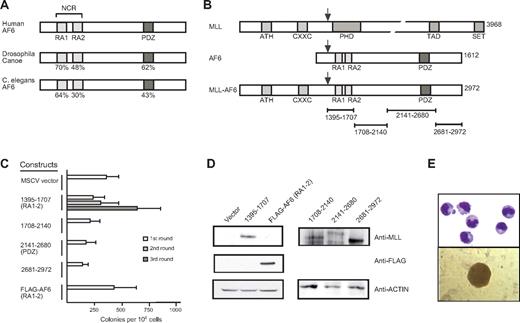
AF6 proteins exhibit a distinct domain structure conserved through evolution from Caenorhabditis elegans to mammals.7,34,35 Despite their large size, they possess only 2 regions with a high degree of sequence conservation. These include an NCR containing 2 conserved copies of the RAS association (RA) domain separated by a non–conserved intervening linker region and a carboxy-terminal PDZ domain (Figure 1A). All MLL-AF6 fusion proteins contain both the NCR and PDZ domain because MLL is consistently fused in-frame to residue 36 of the AF6 protein (Figure 1B).7,12
The N-terminal conserved region of AF6 is sufficient for myeloid immortalization by MLL-AF6. (A) Schematic diagram showing the positions of highly conserved domains within the AF6 protein family. The degree of identity between AF6 and orthologs from Drosophila and C. elegans is indicated. RA1 and RA2 indicates RAS association domains 1 and 2, respectively; NCR, N-terminal conserved region; PDZ, PSD-95/Dlg/ZO-1 domain. (B) Schematic diagram depicting MLL, AF6, and MLL-AF6 fusion protein. ATH indicates AT hook motifs; PHD, plant homeo-domain related; TAD, transcriptional activation domain; SET, Suvar3-9/enhancer-of-zeste/trithorax motif. Total number of amino acids comprising each protein is indicated on the right. Black arrows above each protein indicate the typical position of protein fusion after chromosomal translocations. Specific AF6 protein segments fused with MLL are indicated by brackets below the schematic. (C) Myeloid immortalization assay with various retroviral constructs encoding proteins indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells derived from at least 4 replicates. (D) Western blot analysis of MLL-AF6 and AF6 proteins expressed in transiently transfected virus-producing Phoenix cells. (E) Typical morphology of cells (top; May-Grünwald-Giemsa stain) and blast colony (bottom) in the third round of the myeloid progenitor immortalization assay after transduction with MLL-AF6NCR.
The N-terminal conserved region of AF6 is sufficient for myeloid immortalization by MLL-AF6. (A) Schematic diagram showing the positions of highly conserved domains within the AF6 protein family. The degree of identity between AF6 and orthologs from Drosophila and C. elegans is indicated. RA1 and RA2 indicates RAS association domains 1 and 2, respectively; NCR, N-terminal conserved region; PDZ, PSD-95/Dlg/ZO-1 domain. (B) Schematic diagram depicting MLL, AF6, and MLL-AF6 fusion protein. ATH indicates AT hook motifs; PHD, plant homeo-domain related; TAD, transcriptional activation domain; SET, Suvar3-9/enhancer-of-zeste/trithorax motif. Total number of amino acids comprising each protein is indicated on the right. Black arrows above each protein indicate the typical position of protein fusion after chromosomal translocations. Specific AF6 protein segments fused with MLL are indicated by brackets below the schematic. (C) Myeloid immortalization assay with various retroviral constructs encoding proteins indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells derived from at least 4 replicates. (D) Western blot analysis of MLL-AF6 and AF6 proteins expressed in transiently transfected virus-producing Phoenix cells. (E) Typical morphology of cells (top; May-Grünwald-Giemsa stain) and blast colony (bottom) in the third round of the myeloid progenitor immortalization assay after transduction with MLL-AF6NCR.
Because the resulting MLL-AF6 fusion gene is approximately 9000 nucleotides in length, which exceeds the upper size limit for efficient retroviral packaging capacity (Figure 1B), we tested smaller portions of AF6 that separately spanned the entire AF6 fusion partner protein for their oncogenic potential when fused with MLL. MSCV retroviral constructs encoding synthetic fusions of MLL with sequential parts of AF6 were used to transduce murine BM progenitor cells and perform serial replating assays. Among all constructs tested, only MLL-AF6NCR enhanced the self-renewal potential of primary myeloid progenitors (Figure 1C). Expression of the AF6 NCR region alone did not confer enhanced self-renewal. All fusion constructs efficiently expressed proteins of the predicted sizes (Figure 1D), indicating that the lack of myeloid immortalization by some constructs was not due to lack of expression. Myeloid progenitors transduced by MLL-AF6NCR readily adapted to cytokine-dependent growth in liquid culture media containing interleukin-1. MLL-AF6 immortalized cell lines exhibited an immunophenotype of myeloid progenitor populations coexpressing Mac-1 and Gr-1, with variable expression of c-kit and the B-cell marker B220 (data not shown). Morphologically, the cells resembled immature myeloid progenitors (Figure 1E). These results show that, despite the large (1577 residue) contribution of AF6 to MLL-AF6 fusion proteins, no more than 312 residues spanning the NCR are sufficient for myeloid progenitor immortalization by MLL-AF6.
The highly conserved RA1 domain within the NCR of AF6 mediates myeloid immortalization by MLL-AF6
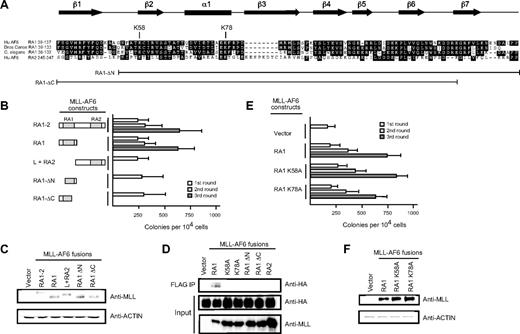
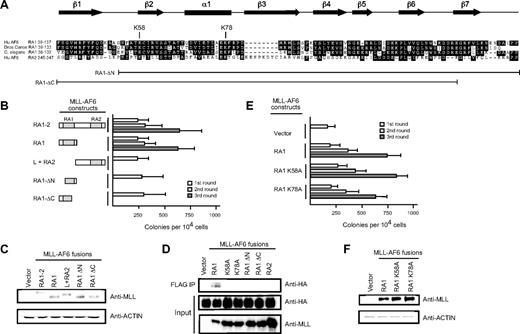
Deletion analysis was performed to further delineate the minimal domain within the AF6 NCR sufficient to confer myeloid immortalization activity to MLL-AF6. Retroviral constructs encoding fusions of MLL to the RA1 domain spanning residues 35 to 137 of AF6 (Figure 2A) behaved in an identical manner to MLL-AF6NCR in enhancing the self-renewal of myeloid progenitors in vitro and the generation of blastlike colonies giving rise to factor-dependent cell lines exhibiting a cell morphology and immunophenotype of a myeloid progenitor population (Figure 2B; data not shown). In contrast, deletions removing 15 conserved residues from either the amino- or carboxy-terminal ends of the RA1 domain abrogated enhancement of myeloid progenitor self-renewal. Similarly, retroviral constructs encoding fusions of MLL to the non–conserved intervening linker region followed by the weakly conserved RA2 domain of the AF6 NCR were unable to confer enhanced self-renewal to myeloid progenitors. All fusion constructs efficiently expressed proteins of the predicted sizes in immunoblot analysis (Figure 2C). These data define the RA1 domain spanning AF6 residues 35 to 137 to be the minimal region sufficient for myeloid transformation by MLL-AF6.
The RA1 domain within the AF6 NCR mediates myeloid immortalization by MLL-AF6. (A) Sequence alignment of the RA1 domains of AF6 family proteins. Dark shading defines residues of identity or similarity. Amino acid numbers are indicated on the left. The positions of conserved residues targeted for mutation are highlighted above the sequence. The conserved secondary structures are depicted above the sequence, where filled arrows represent β sheet and the filled box represents α helix. Brackets below indicate the RA1 amino acids present in deletion constructs identified on the left. (B) Myeloid immortalization assay resulting from transduction of various MLL-AF6 mutant proteins shown on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells (≥ 4 replicates). (C) Western blot analysis of MLL-AF6 fusion proteins expressed in transiently transfected virus-producing Phoenix cells. (D) Immunoprecipitation–-Western blot analysis shows that only the intact MLL-AF6RA1 is capable of coprecipitating HA-tagged RasV12 in transiently transfected 293 cells. (E) Myeloid immortalization assay with retroviral constructs encoding MLL-AF6RA1 point mutants indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells (≥ 4 replicates). (F) Western blot analysis of MLL-AF6 mutant proteins expressed in transiently transfected virus-producing Phoenix cells.
The RA1 domain within the AF6 NCR mediates myeloid immortalization by MLL-AF6. (A) Sequence alignment of the RA1 domains of AF6 family proteins. Dark shading defines residues of identity or similarity. Amino acid numbers are indicated on the left. The positions of conserved residues targeted for mutation are highlighted above the sequence. The conserved secondary structures are depicted above the sequence, where filled arrows represent β sheet and the filled box represents α helix. Brackets below indicate the RA1 amino acids present in deletion constructs identified on the left. (B) Myeloid immortalization assay resulting from transduction of various MLL-AF6 mutant proteins shown on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells (≥ 4 replicates). (C) Western blot analysis of MLL-AF6 fusion proteins expressed in transiently transfected virus-producing Phoenix cells. (D) Immunoprecipitation–-Western blot analysis shows that only the intact MLL-AF6RA1 is capable of coprecipitating HA-tagged RasV12 in transiently transfected 293 cells. (E) Myeloid immortalization assay with retroviral constructs encoding MLL-AF6RA1 point mutants indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells (≥ 4 replicates). (F) Western blot analysis of MLL-AF6 mutant proteins expressed in transiently transfected virus-producing Phoenix cells.
The RA1 domain of AF6 exhibits a similar potential secondary structure to the RA domains of Ras binding proteins RalGDS, c-Raf-1, and Byr2. The RA domains of the latter possess a common tertiary structure related to the ββαββα ubiquitin superfold, despite a lack of extensive primary sequence conservation.36-38 Complex formation takes place by the generation of an intermolecular antiparallel β-sheet between β1 and β2 of the RA domain and β2 and β3 of the switch 1 effector region of Ras. The AF6 RA1 domain is predicted to form 7 distinct β-sheets (β1-β7) and a single central α helix (α1) (Figure 2A).39 In particular, specific residues (including K58 and K78) within RA domains that perform critical contacts with the Ras effector region are highly conserved within the RA1 domains of AF6 family proteins. Indeed, biochemical studies have shown that single amino acid substitutions of alanine for lysine (K58A and K78A) result in a significant reduction of binding affinity between the RA1 domain of AF6 and Ras, confirming the functional importance of these residues.40
To further define the contribution of Ras interaction through K58 and K78 to transformation, these highly conserved residues were subjected to single amino acid substitutions to alanine, and their effects on Ras association (Figure 2D) and myeloid transformation (Figure 2E) by MLL-AF6 were then determined. Coimmunoprecipitation studies performed in 293 cells coexpressing FLAG-tagged MLL-AF6 proteins and HA-tagged H-RasV12 showed that MLL-AF6RA1 coprecipitated H-RasV12, whereas none of the mutant MLL-AF6 proteins containing mutated RA1 domains or RA2 coprecipitated H-RasV12 (Figure 2D). In contrast, residues K58 and K78 were dispensable for transformation despite their absolute conservation within the AF6 family and requirement for Ras binding. Both mutants, K58A and K78A, generated blast colonies indistinguishable from those derived from MLL-AF6RA1. Thus, single amino acid substitutions of 2 highly conserved residues, K58 and K78, known to significantly reduce binding affinity of the AF6RA1 domain to Ras, had no effect on myeloid transformation. These results strongly suggest that the transforming ability of the corresponding MLL fusion proteins is not mediated by Ras interaction.
Fusions of the amino terminus of MLL to heterologous RA domains or constitutively active mutants of Ras or Rap1 do not confer myeloid immortalization
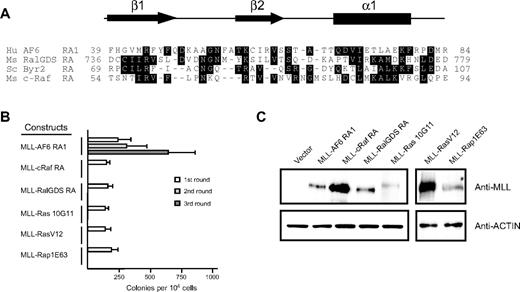
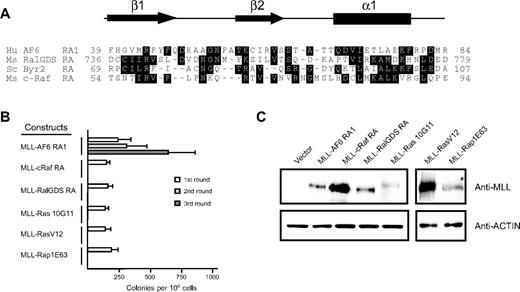
To determine whether recruitment of guanosine triphosphate–bound Ras to MLL target genes was sufficient to transform myeloid progenitors, we generated retroviral constructs encoding synthetic fusions between MLL and the minimal Ras binding domains of c-Raf and Ral-GDS.36,41 In addition, we generated constructs encoding synthetic fusions between MLL and the constitutively active mutants K-Ras10G11, H-RasV12, and Rap1E63.42,43 In contrast to MLL-AF6RA1, fusions of MLL to the heterologous Ras binding domain of c-Raf and Ral-GDS were unable to maintain myeloid clonogenic potential in vitro (Figure 3B). Furthermore, covalent fusions of constitutively active mutants of K-Ras, H-Ras, or Rap1 to MLL were also nontransforming. All fusion proteins were efficiently expressed at their predicted sizes in immunoblot analysis (Figure 3C), indicating that the lack of myeloid immortalization was not due to lack of expression of the constructs. These results indicate that oncogenic transformation by MLL-AF6 does not involve recruitment of Ras to the nucleus.
Fusion of N-terminal MLL to heterologous Ras association domains from c-Raf1, RalGDS, or constitutively active mutants of K-Ras, H-Ras, or Rap1 does not activate the oncogenic potential of MLL. (A) Sequence alignment of the N-terminal regions of various RA domains. Dark shading defines residues of identity or similarity. Amino acid numbers are indicated on the left. The conserved secondary structures are depicted above, where arrows represent β sheets and the filled box represents an α helix. Crystal structures for c-Raf1, RalGDS, and Byr2 have been previously described.36,38,41 Putative AF6 RA1 domain secondary structure was derived from the PHD prediction algorithm.39 (B) Myeloid immortalization assay with various retroviral constructs encoding MLL fusions to heterologous RA domains or proteins are as indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells (≥ 4 replicates). (C) Western blot analysis of MLL fusion proteins expressed in transiently transfected virus-producing Phoenix cells.
Fusion of N-terminal MLL to heterologous Ras association domains from c-Raf1, RalGDS, or constitutively active mutants of K-Ras, H-Ras, or Rap1 does not activate the oncogenic potential of MLL. (A) Sequence alignment of the N-terminal regions of various RA domains. Dark shading defines residues of identity or similarity. Amino acid numbers are indicated on the left. The conserved secondary structures are depicted above, where arrows represent β sheets and the filled box represents an α helix. Crystal structures for c-Raf1, RalGDS, and Byr2 have been previously described.36,38,41 Putative AF6 RA1 domain secondary structure was derived from the PHD prediction algorithm.39 (B) Myeloid immortalization assay with various retroviral constructs encoding MLL fusions to heterologous RA domains or proteins are as indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells (≥ 4 replicates). (C) Western blot analysis of MLL fusion proteins expressed in transiently transfected virus-producing Phoenix cells.
The oncogenic activity of the AF6 RA1 domain is highly conserved and retained by the Drosophila ortholog Canoe
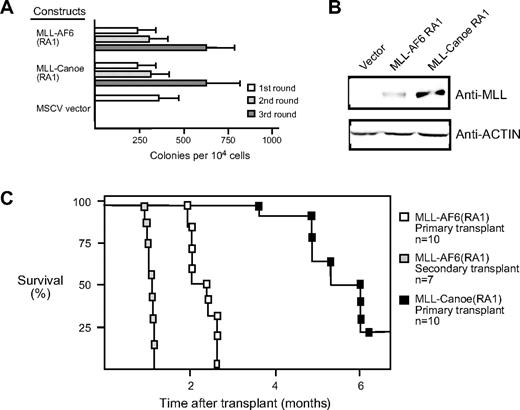
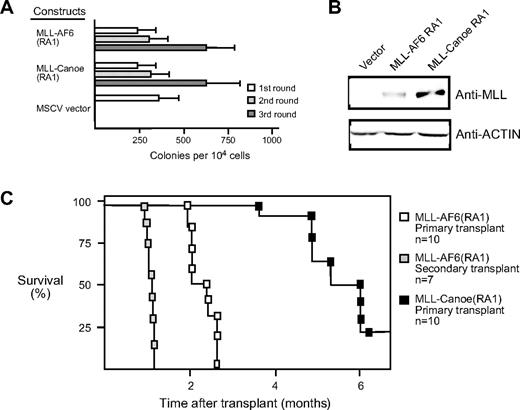
The RA1 domain of the Drosophila ortholog of AF6, termed Canoe, exhibits 72% sequence identity with that of the human AF6 RA1,34,35 suggesting that the function of this domain may have been evolutionarily conserved. To address whether the oncogenic contributions of RA1 were conserved, we generated a retroviral construct encoding a synthetic fusion between MLL and the corresponding RA1 domain of Drosophila Canoe. In serial replating assays, the MLL-CanoeRA1 fusion was able to maintain myeloid clonogenic potential in vitro and to generate factor-dependent cell lines in a manner identical to that of MLL AF6RA1 (Figure 4A; data not shown). Thus, the oncogenic potential of RA1 is highly conserved.
The RA1 domain of Canoe, the fly ortholog of AF6, activates the oncogenic potential of MLL. (A) Myeloid immortalization assay was performed with MLL fusion constructs indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells. Data are derived from at least 4 cultures from 2 or more independent experiments. (B) Western blot analysis of MLL fusion proteins expressed in transiently transfected virus-producing Phoenix cells. (C) Survival curves are shown for sublethally irradiated C57BL/6 mice that received a transplant with various cell lines immortalized by MLL fusion genes.
The RA1 domain of Canoe, the fly ortholog of AF6, activates the oncogenic potential of MLL. (A) Myeloid immortalization assay was performed with MLL fusion constructs indicated on the left. Each bar represents the mean ± SD of the total number of myeloid colonies per 104 plated cells. Data are derived from at least 4 cultures from 2 or more independent experiments. (B) Western blot analysis of MLL fusion proteins expressed in transiently transfected virus-producing Phoenix cells. (C) Survival curves are shown for sublethally irradiated C57BL/6 mice that received a transplant with various cell lines immortalized by MLL fusion genes.
MLL-AF6 induces short latency AML in vivo
To determine the leukemogenic potential of MLL-AF6, a cytokine-dependent cell line transduced by MLL-AF6RA1 was transplanted into sublethally irradiated syngeneic C57BL/6 mice (n = 10). Animals that received a transplant developed a fully penetrant AML within 3 months, with a mean latency period of 75.8 (± 8.2) days (Figure 4C). Animals typically presented with myeloid leukemias uniformly expressing Mac1 and variably B220 and with tumor infiltration of various tissues, including the liver, kidneys, and lungs. Tumor cells consistently infiltrated additional secondary lymphoid organs such as the thymus and multiple lymph nodes. When BM cells from 2 independent primary tumors were transplanted into secondary recipients (n = 7), all developed AML with a significantly accelerated mean latency of 32.5 (± 2.63) days. Thus, myeloid progenitors transduced by MLL-AF6RA1 are highly leukemogenic in vivo.
Transplantation of an MLL-CanoeRA1–immortalized cell line led to the induction of AML with identical clinical features to that seen with MLL-AF6RA1, although these tumors were observed with only 80% penetrance and a more protracted mean latency of 167.12 (± 28.81) days (Figure 4C). Thus, although the in vitro immortalizing activity of the AF6 RA1 domain seems to be fully conserved with Canoe, subtle functional differences exist between the orthologous domains in their ability to promote the self-renewal and/or expansion of myeloid progenitors in vivo.
The NCR of AF6 lacks intrinsic transactivation potential but mediates self-association
MLL partner proteins that normally localize to the nucleus contribute transcriptional effector domains to their respective oncogenic MLL fusion proteins that are essential for transformation. Despite the normal cytoplasmic localization of AF6, it remains plausible that its NCR may contain a cryptic transactivation domain that contributes to transformation by MLL-AF6. To investigate this possibility, the AF6 NCR was fused to the GAL4 DNA binding domain and tested for transcriptional activity in transient transfections. Under conditions in which the acidic transactivator Gal4-VP16 showed robust activation, Gal4-AF6 NCR did not exhibit transcriptional activation above background levels (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Gal4 fusion constructs were efficiently expressed at their predicted sizes in immunoblot analysis, indicating that the lack of transcriptional activation was not due to lack of construct expression. These results suggest that the AF6 NCR does not possess inherent transcriptional activation properties and most probably contributes to activation of MLL-transforming activity by a mechanism distinct from nuclear MLL partner proteins.
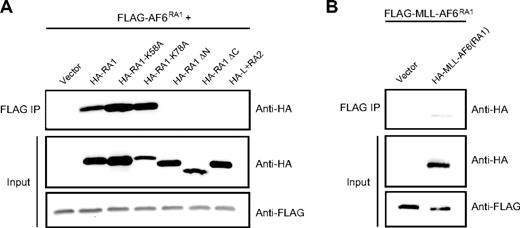
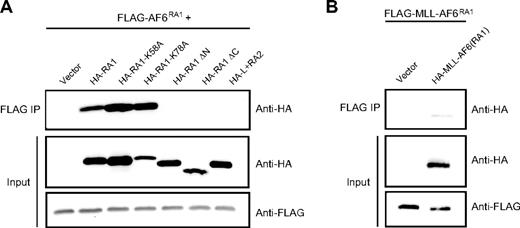
An alternative mechanism for MLL-mediated leukemogenesis involves constitutive self-association of amino terminal MLL mediated by cytoplasmic MLL partner proteins as shown for AF1p, GAS7, and GEPHYRIN.29-31 To determine whether AF6 acts to confer oncogenic properties to MLL through a similar mechanism, we evaluated the ability of AF6RA1 to mediate self-association. 293 cells were transiently cotransfected with FLAG-tagged AF6RA1 and various HA-tagged AF6 mutants. Proteins were immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-HA or anti-FLAG antibodies. These experiments showed that AF6RA1 is capable of mediating self-association in vitro (Figure 5A). Furthermore, the ability of AF6RA1 to induce self-association in the context of the MLL-AF6 fusion protein was verified (Figure 5B). To confirm the importance of self-association for transformation by MLL-AF6, we tested the correlation between transforming ability and self-association of different point or deletion mutants. Amino acid substitutions at residues K58 or K78 that do not affect the transforming potential of MLL-AF6RA1 also did not interfere with the ability of RA1 to self-associate. In contrast, deletion mutants lacking 15 conserved residues at the amino-terminal or carboxy-terminal ends of RA1 that result in loss of transforming ability by MLL-AF6RA1 were unable to self-associate. Similarly, the weakly conserved AF6RA2 region that does not confer MLL-transforming ability was unable to mediate self-association. Table 1 summarizes these results that show a complete correlation between transforming potential and ability to self-associate, suggesting that self-association is required for transformation by MLL-AF6.
The RA1 domain of AF6 mediates self-association in vitro. (A) Self-association was assessed by anti-HA Western blot analysis after anti-FLAG immunoprecipitation from lysates of 293 cells cotransfected with various HA-tagged AF6 constructs together with FLAG-tagged AF6RA1 (top). Expression levels of the relevant input proteins were determined by immunoblotting with either anti-HA (middle) or anti-FLAG (bottom) antibodies. (B) Self-association of MLL-AF6RA1 was shown by anti-HA Western blot analysis after anti-FLAG immunoprecipitation from lysates of cells cotransfected with HA-tagged MLL-AF6RA1 and FLAG-tagged MLL-AF6RA1 (top). Expression levels of the input proteins were determined with either anti-HA (middle) or anti-FLAG (bottom) antibodies.
The RA1 domain of AF6 mediates self-association in vitro. (A) Self-association was assessed by anti-HA Western blot analysis after anti-FLAG immunoprecipitation from lysates of 293 cells cotransfected with various HA-tagged AF6 constructs together with FLAG-tagged AF6RA1 (top). Expression levels of the relevant input proteins were determined by immunoblotting with either anti-HA (middle) or anti-FLAG (bottom) antibodies. (B) Self-association of MLL-AF6RA1 was shown by anti-HA Western blot analysis after anti-FLAG immunoprecipitation from lysates of cells cotransfected with HA-tagged MLL-AF6RA1 and FLAG-tagged MLL-AF6RA1 (top). Expression levels of the input proteins were determined with either anti-HA (middle) or anti-FLAG (bottom) antibodies.
Discussion
The t(6;11)(q27;q23) is the most common translocation in acute leukemias that fuses MLL with a cytoplasmic partner protein.10 In this study, we defined a critical region of AF6 that is sufficient for both in vitro immortalization of primary myeloid progenitors and the rapid induction of AML in vivo. Although AF6 routinely contributes 1577 amino acids to the MLL-AF6 fusion protein, our structure/function analysis identified the small N-terminal RA1 domain of AF6 encoding just 103 residues that, when fused to MLL, was sufficient for hematopoietic transformation and leukemogenesis. Consistent with our structure/function studies, all cases of the t(6;11)(q27;q23) in human leukemias thus far described display a fusion point between MLL and AF6 that localizes to the amino-terminal boundary of the RA1 domain, thereby consistently retaining the RA1 in MLL-AF6 fusion proteins.
In addition to the highly conserved RA domains 1 and 2, AF6 family proteins also contain a PDZ domain. The PDZ domain of AF6 mediates interactions with a variety of proteins, including negative regulators of Ras family proteins, such as Rap1 guanosine triphosphatase (GTPase)–activating protein, Spa-1, as well as Bcr, a Rac1 GTPase-activating protein.44,45 Thus, it has been proposed that AF6 may serve as a molecular scaffold for the recruitment of negative regulators of Ras family GTPases. Interestingly, Spa-1 deficiency promotes myeloproliferation in a mouse model by activation of Rap1.37 However, our structure/function analysis showed that the PDZ domain of AF6 is dispensable for myeloid transformation, suggesting that MLL-AF6 may promote leukemogenesis without disrupting Spa-1 or Bcr function. It remains to be determined whether the reduced availability of AF6 to serve as a scaffold in the cytoplasm of human t(6;11) leukemia cells may partially compromise Spa-1 function.
Our study is the first example whereby the oncogenic activity of the minimal transforming domain of an MLL partner protein has been shown to be conserved in an evolutionarily distant ortholog. When fused to MLL, the RA1 domain of the Drosophila ortholog of AF6, termed Canoe, also exhibited potent myeloid-immortalizing activity in vitro. Despite its potency, the ability of MLL-Canoe RA1 to promote leukemogenesis in vivo was modestly attenuated, suggesting that self-renewal/expansion of the immortalized myeloid progenitor population in vivo was somewhat compromised relative to AF6 RA1. It is currently unclear whether these subtle functional differences reflect quantitative and/or qualitative functional defects.
Having identified the RA1 domain as the minimal region of AF6 contributing to MLL-AF6 transformation, we investigated potential molecular mechanisms that could account for its critical oncogenic function. Independent biochemical and genetic approaches previously identified the RA1 domain of the AF6 family as a preferential binding motif for constitutively active oncogenic Ras family GTPases, including H-Ras, Rap1A, and M-Ras.35,43,46,47 Furthermore, in Drosophila, genetic interactions between Canoe and Ras1 in the developing compound eye, or alternatively between Canoe and Rap1 during dorsal closure, suggest bona fide functional interactions.48,49 We used 2 approaches to test the potential role of Ras family GTPases in mediating the oncogenic properties of MLL-AF6. First, individual highly conserved residues known to be critical for direct binding to the effector region of Ras family members were targeted for mutation. We found that both of these residues, K58 and K78, were dispensable for MLL-AF6–mediated transformation, suggesting that RA1 domain–mediated transformation may occur by a Ras family–independent mechanism. However, we cannot exclude the remote possibility that a certain threshold of Ras binding is critical and that K58 and K78 mutants retain sufficient affinity to mediate a transforming effect. Second, we generated synthetic fusions of MLL to 2 other Ras-binding domains from c-Raf and RalGDS, as well as a direct fusion of a constitutively active mutant of K-Ras, H-Ras, and Rap1. None of these were able to substitute for AF6 RA1 transforming function. These data strongly suggest that simple recruitment of Ras to MLL targets in the nucleus is unlikely to account for RA1 domain–dependent transformation.
Previous studies have proposed that fusion of MLL to rare cytoplasmic partner proteins leads to constitutive self-association and that this activity is sufficient for hematopoietic transformation.29-31 Our current study of AF6 further supports this model, because the RA1 domain was sufficient for both myeloid transformation and self-association. Furthermore, the high frequency of MLL-AF6 translocations suggests that self-association of the amino-terminus of MLL may represent a common pathogenic mechanism. We note that our study does not exclude the possibility that the AF6 RA1 domain recruits other yet unidentified essential cofactors by a dimerization-dependent mechanism to activate MLL oncogenesis. Although there seems to be consensus about the critical role of MLL self-association, it is currently unclear how such a mechanism promotes aberrant hematopoietic self-renewal. One possibility is that wild-type MLL normally self-associates to generate an active conformation, but that such a conformation is under tight regulation. Fusion of MLL to cytoplasmic partner proteins such as AF6 may result in constitutive self-association that is no longer subject to negative regulation, resulting in the production of a constitutively active gain-of-function mutant. Presumably dimeric forms of MLL recruit a coregulatory complex to MLL target genes to deregulate their expression. A possible candidate is the AEP complex, which constitutively co-occupies target gene chromatin due either to covalent fusion with MLL or, in the case of MLL-AF6, by a non–covalent mechanism that is currently undefined.50 Alternatively, the dimerization activity of MLL-AF6 may be independent of transformation activity but simply may reflect appropriate folding of the RA1 domain for recruiting a cofactor essential for transformation. Further understanding of the mechanistic basis of dimerization-dependent and -independent MLL transformation will require additional studies of the role of specific coregulatory complexes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Maria Ambrus and Cita Nicolas for excellent technical support, and Akihiko Yokoyama for scientific advice. We thank Eli Canaani for providing full-length AF6, Kevin Shannon for K-Ras10G11, and Linda van Elst for providing H-RasV12 and Rap1E63 constructs.
This work was supported by funds from the National Institutes of Health (M.L.C. and grant K08 CA120349; M.L.), the Children's Health Initiative of the Lucile Packard Foundation for Children's Health (M.L.C.), Hope Street Kids (M.L.), and the ASCO Foundation (M.L.).
National Institutes of Health
Authorship
Contribution: M.L., P.M.A., K.S.S., and T.C.P.S. designed and performed the experiments, analyzed the data, and wrote the manuscript; and M.L.C. designed experiments, analyzed the data, and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael L. Cleary, Stanford University School of Medicine, 300 Pasteur Dr, Stanford, CA 94305; e-mail: mcleary@stanford.edu.
References
Author notes
M.L. and P.M.A. contributed equally to this study.