Abstract
Previously, we have shown that overexpression of an activated mutant of signal transducer and activator of transcription-5 (STAT5) induces erythropoiesis, impaired myelopoiesis, and an increase in long-term proliferation of human hematopoietic stem/progenitor cells. Because GATA1 is a key transcription factor involved in erythropoiesis, the involvement of GATA1 in STAT5-induced phenotypes was studied by shRNA-mediated knockdown of GATA1. CD34+ cord blood cells were double transduced with a conditionally active STAT5 mutant and a lentiviral vector expressing a short hairpin against GATA1. Erythropoiesis was completely abolished in the absence of GATA1, indicating that STAT5-induced erythropoiesis is GATA1-dependent. Furthermore, the impaired myelopoiesis in STAT5-transduced cells was restored by GATA1 knockdown. Interestingly, early cobblestone formation was only modestly affected, and long-term growth of STAT5-positive cells was increased in the absence of GATA1, whereby high progenitor numbers were maintained. Thus, GATA1 down-regulation allowed the dissection of STAT5-induced differentiation phenotypes from the effects on long-term expansion of stem/progenitor cells. Gene expression profiling allowed the identification of GATA1-dependent and GATA1-independent STAT5 target genes, and these studies revealed that several proliferation-related genes were up-regulated by STAT5 independent of GATA1, whereas several erythroid differentiation-related genes were found to be GATA1 as well as STAT5 dependent.
Introduction
Signal transducer and activator of transcription-5 (STAT5) is a member of a family of transcription factors that is composed of 7 genes (STAT1 to STAT6, with STAT5 being encoded by 2 genes, STAT5A and STATB). The pathways mediated by these transcription factors are involved in many processes in hematopoietic cells, including regulation of proliferation, antiapoptosis, differentiation, and self-renewal.1,2 STAT transcription factors contain a conserved tyrosine residue in their SH2 domain, which on phosphorylation, gives rise to heterodimerization or homodimerization (or tetramerization3 ) with another STAT molecule, leading to nuclear translocation. Constitutively activated STAT5 has been found in blast cells of up to 70% of acute myeloid leukemia cases.4 Besides its function as a transcription factor, tyrosine phosphorylated STAT5 has also been described to interact with other partners to induce phosphoinositide-3 kinase activity, thereby contributing to leukemic transformation.5 STAT transcription factors can be activated by growth factors via JAK kinases, present at the cytoplasmic domains of growth factor receptors, as well as by intrinsic kinase activity of certain membrane receptors or cytoplasmic tyrosine kinases.6
STAT5 is expressed in a large variety of hematopoietic cells and involved in the signal transduction of many different growth factors and cytokines. Early acting cytokines, such as Fms-like tyrosine kinase 3 ligand (FLT-3L), thrombopoietin, and stem cell factor, all mediate at least part of their signals through STAT5.7-9 During erythropoiesis, STAT5 is critically involved in the signal transduction mediated by erythropoietin (EPO).10 Furthermore, signals induced by myeloid growth factors, such as interleukin-3 (IL-3), IL-5, granulocyte colony-stimulating factor, macrophage colony-stimulating factor, and granulocyte-macrophage colony-stimulating factor are all, at least in part, mediated by STAT5.1,11,12 Loss-of-function studies in mice have revealed a critical role for STAT5 in the maintenance of hematopoiesis as shown by a reduced repopulation ability in STAT5A−/−, STAT5B−/− double-knockout mice.13-16 We have shown that knockdown of STAT5 in CD34+ cells from cord blood (CB) resulted in reduced stem cell and progenitor frequencies,17 whereas overexpression of active STAT5A promoted stem cell self-renewal.18,19 STAT5 plays an important role in differentiation toward the erythropoietic lineage by up-regulating antiapoptotic pathways via Bcl-XL,10,20 but a more direct role in inducing erythroid differentiation probably exists as well.18,19,21-23
GATA1 is a transcription factor, originally identified by the ability to bind a distinct motif present in promoter and enhancer sequences of a variety of erythroid genes. This motif, WGATAR, was shown to be critical in the regulation of these genes.24 Apart from GATA1, 5 other members of the GATA family have been identified (named GATA2 to GATA6). Of the 6 GATA proteins, GATA1 to GATA3 are expressed in hematopoietic cells, whereas GATA4 to GATA6 are expressed in nonhematopoietic tissues, such as the heart, gut, lung, and liver.25 GATA1 and GATA2 are critically involved in erythroid and megakaryocytic cell development.26 It has been shown in transgenic mice carrying the GATA2 regulatory domain fused to green fluorescent protein (and therefore recapitulating the endogenous GATA2 expression) that GATA2 is detected very early in hematopoietic development.27 Analogous experiments with a lacZ reporter under the control of the GATA1 regulatory elements showed expression at somewhat later stages of erythroid development.28 Gene knockout studies have indicated that homozygous deletion of both GATA1 and GATA2 is embryonically lethal at days 10.5 to 12.5.29,30 Studies performed in GATA1 knockout embryonic stem cells have revealed that GATA1-null cells did not contribute to red blood cell formation, whereas white blood cell formation in chimeric animals was normal.31 Furthermore, GATA1-null cells were arrested at the proerythroblast stage in in vitro colony formation assays, underscoring the essential role of GATA1 in erythroid development.31 GATA1(low) mice, lacking a DNA hypersensitive site in the GATA1 promoter and therefore expressing only 10% to 25% of normal GATA1 levels, are thrombocytopenic but display initially normal hematocrit levels.32 After12 months, these mice, however, develop anemia accompanied by myelofibrosis.32
We have previously shown that overexpression of active STAT5A results in a profound burst of erythropoiesis, accompanied by impaired myelopoiesis and enhanced self-renewal and long-term expansion.18,19 The underlying mechanisms have not been elucidated yet. Because GATA1 has been shown to be essential for erythropoiesis, we decided to investigate the possible dependence of STAT5-induced phenotypes on GATA1 by a lentiviral GATA1 knockdown approach. We observed that depletion of GATA1 allowed the dissection of STAT5-induced differentiation phenotypes from the effects of STAT5 on long-term expansion of human hematopoietic stem/progenitor cells. This provided us with a tool to further study the molecular mechanisms and genes involved in these processes.
Methods
Cell culture and viral transductions
Neonatal CB was obtained from healthy full-term pregnancies from the Obstetrics departments of the Martini Hospital and University Medical Center in Groningen, The Netherlands, after informed consent. The protocol was approved by the Medical Ethical Committee of the University Medical Center Groningen. CD34+ cells were isolated with the use of a hematopoietic progenitor isolation kit from Miltenyi Biotec according to the manufacturer's instructions. The pMSCV-STAT5AwtER-IRES-tNGFR retroviral vector was described previously.19 An RNAi-resistant GATA1-expressing vector was made by introduction of silent mutations at the RNAi target site by polymerase chain reaction (PCR)–mediated site directed mutagenesis in the cDNA of human GATA1. The resulting RNAi-resistant GATA1 cDNA was fused to a myc tag and cloned into the pMSCV-IRES-EGFP vector in which the EGFP was replaced with cDNA encoding for dTomato, kindly provided by B. Giepmans (Department of Cell Biology, University of Groningen, Groningen, The Netherlands), resulting in pMSCV-mutGATA1myc-IRES-dTomato. Stable PG13 high titer retroviral producer cell lines were generated as described previously.19 GATA1 was knocked down by lentiviral transduction of CD34+ CB cells using a modified version of the pLVUT-tTRKRAB-shGATA1 vector, which was a kind gift of D. Trono (School of Life Sciences, Ecole Polytechnique Federale de Lausanne, Lausanne, Switzerland), in which the IRES-KRAB and TetO sequences were deleted, resulting in constitutive expression of the short hairpin against GATA1. A control vector was made by cloning a hairpin against firefly luciferase into the pLVUT vector. Lentiviral particles were made by transient transfection of 293T cells with the lentiviral expression vectors as described previously.33 Isolated CD34+ cells were cultured for 48 hours in serum-free medium (Lonza Netherlands) containing 100 ng/mL each of stem cell factor, thrombopoietin, and FLT-3L and subsequently transduced with the retroviral and lentiviral particles in 3 consecutive rounds as described previously.33
Stromal coculture assays, CFC assays
MS5 cocultures were performed as described previously.19 Half of the cultures were removed weekly and analyzed by fluorescence-activated cell sorter (FACS) or sorted to perform colony-forming cell (CFC) assays or cytospins. CFC assays were performed in MethoCult H4230 (StemCell Technologies) supplemented with 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL granulocyte colony-stimulating factor, 20 ng/mL c-Kit ligand, and 1 U/mL EPO. A total of 103 to 104 cells were plated in methylcellulose per plate in duplicate. Cultures were scored using a Leica DM-IL inverted microscope (Leica Microsystems; total original magnification ×40).
mRNA analysis
Total RNA was isolated using the RNeasy kit from QIAGEN according to the manufacturer's recommendations. For real-time reverse-transcribed (RT)–PCR, cDNA was prepared and amplified using iQ SYBR Green supermix (Bio-Rad) in a MyIQ thermocycler (Bio-Rad), and quantified using MyIQ software (Bio-Rad). Hypoxanthine phosphoribosyl transferase, ribosomal protein like 27 (RPL27), and RPL30 expression levels were used to normalize between samples and to calculate relative expression levels.34 Primers and conditions are available on request. Genome-wide expression analysis was performed on Illumina BeadChip Arrays Sentrix Human-6 (46k probe sets). Typically, 0.5 to 1 μg of RNA combined from 3 independent transduction experiments was used in labeling reactions and hybridization with the arrays according to the manufacturer's instructions. Data were analyzed using the BeadStudio, Version 3 Gene Expression Module (Illumina) and Genespring (Agilent Technologies).
Flow cytometric analysis
Antibodies against CD11b, CD14, CD15, CD34, CD38, CD45RA, CD71, and CD123 were obtained from BD Biosciences. Anti–NGFR-APC was obtained from Miltenyi Biotec. GlycophorinA-PE was obtained from Dako. Cells were incubated with antibodies at 4°C for 30 minutes. All FACS analyses were performed on an LSRII flow cytometer (BD Biosciences), and data were analyzed using WinList 3D (Topsham) or FlowJo (TreeStar) software.
Immunoblotting and cytospins
Sorted cells were boiled in Laemmli sample buffer for 5 minutes before separation on 12% sodium dodecyl sulfate–polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membrane (Millipore) by semidry electroblotting. Membranes were blocked in Odyssey blocking buffer (Westburg) before incubation with antibodies. Binding of antibodies was detected by incubating with Alexa680 or IRDye800-labeled secondary antibodies (Invitrogen) and scanning of the membrane on an Odyssey infrared scanner (Li-Cor Biosciences). Antibodies against STAT5 (C17), GATA1 (N6), and actin (C4) were obtained from Santa Cruz Biotechnology and were used in dilutions of 1:1000. May-Grünwald-Giemsa (MGG) staining was used to stain cytospins. Cytospin preparations were evaluated and photographed using a Leica DM3000 microscope (Leica Microsystems) equipped with a Leica DFC420C digital camera (total original magnification ×400). Images were cropped and reverted to grayscale using Corel Photopaint 12 (Corel Corp) software.
Results
STAT5-induced erythroid differentiation is GATA1-dependent
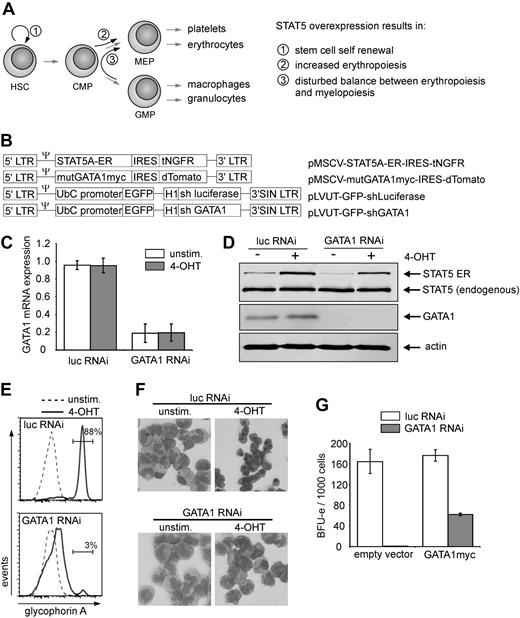
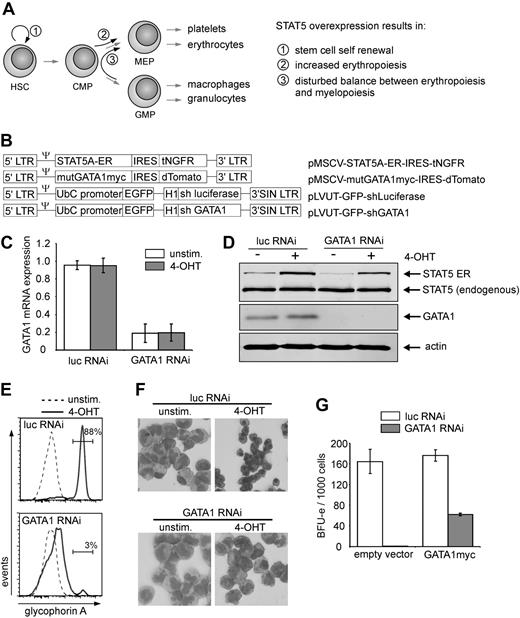
Previously, we observed that persistent activation of STAT5 in human CB CD34+ cells resulted in long-term self-renewal and expansion, while at the same time erythropoiesis was induced (Figure 1A).18,19 To assess whether GATA1 would be dispensable for the increased erythropoiesis induced by STAT5, GATA1 was knocked down in CB CD34+ cells expressing a 4-hydroxytamoxifen (4-OHT)–activatable form of STAT5 (Figure 1B).19 Both GATA1 mRNA levels (Figure 1C) as well as GATA1 protein levels (Figure 1D) were efficiently down-regulated in GATA1 shRNA-transduced cells. STAT5A activation did not change GATA1 mRNA or protein expression levels (Figure 1C-D).
STAT5-induced erythroid differentiation is GATA1 dependent. (A) Schematic overview of previously obtained results caused by active STAT5 overexpression. (B) Retroviral and lentiviral constructs used in this study. (C) Quantitative RT-PCR results of CB CD34+ cells that were double transduced with the STAT5A-ER vector in combination with the indicated lentiviral RNAi constructs. Cells were left untreated or stimulated for 24 hours with 4-OHT, sorted, and RNA was extracted. GATA1 mRNA expression was calculated after normalization against expression of hypoxanthine phosphoribosyl transferase, RPL27, and RPL30 mRNA. The graph represents the mean of 3 independent experiments. (D) Western blot of sorted cells, transduced and treated as in panel C. Extracts were blotted against STAT5, GATA1, and actin to show equal loading. (E) Representative FACS histograms of GPA expression on cells transduced with STAT5A-ER and the indicated lentiviral vectors expressing short hairpins against luciferase or GATA1. Cells were cultured on MS5 stromal cells, left untreated or stimulated with 4-OHT, and analyzed for GPA expression after 10 days. (F) MGG-stained cytospin preparations of suspension cells of cultures described in panel E and harvested after 2 weeks. (G) BFU-E numbers of methylcellulose culture of CB cells, transduced with lentiviral vectors for expression of short hairpins against luciferase and GATA1, together with an empty expression vector or a vector expressing RNAi resistant, myc-tagged GATA1. A total of 1000 double-transduced cells were sorted 1 day after transduction directly into CFC medium. BFU-E colonies were counted after 2 weeks.
STAT5-induced erythroid differentiation is GATA1 dependent. (A) Schematic overview of previously obtained results caused by active STAT5 overexpression. (B) Retroviral and lentiviral constructs used in this study. (C) Quantitative RT-PCR results of CB CD34+ cells that were double transduced with the STAT5A-ER vector in combination with the indicated lentiviral RNAi constructs. Cells were left untreated or stimulated for 24 hours with 4-OHT, sorted, and RNA was extracted. GATA1 mRNA expression was calculated after normalization against expression of hypoxanthine phosphoribosyl transferase, RPL27, and RPL30 mRNA. The graph represents the mean of 3 independent experiments. (D) Western blot of sorted cells, transduced and treated as in panel C. Extracts were blotted against STAT5, GATA1, and actin to show equal loading. (E) Representative FACS histograms of GPA expression on cells transduced with STAT5A-ER and the indicated lentiviral vectors expressing short hairpins against luciferase or GATA1. Cells were cultured on MS5 stromal cells, left untreated or stimulated with 4-OHT, and analyzed for GPA expression after 10 days. (F) MGG-stained cytospin preparations of suspension cells of cultures described in panel E and harvested after 2 weeks. (G) BFU-E numbers of methylcellulose culture of CB cells, transduced with lentiviral vectors for expression of short hairpins against luciferase and GATA1, together with an empty expression vector or a vector expressing RNAi resistant, myc-tagged GATA1. A total of 1000 double-transduced cells were sorted 1 day after transduction directly into CFC medium. BFU-E colonies were counted after 2 weeks.
Next, the short hairpin against GATA1 was expressed in CD34+ CB cells together with STAT5-ER, and the cells were cultured on MS5 bone marrow stromal cells. After 10 days of culture, part of the suspension cells were harvested and stained for glycophorin A (GPA) expression as a measure for erythroid differentiation. As indicated in Figure 1E, activation of STAT5 induced a strong erythroid differentiation within 10 days of MS5 coculture (Figure 1E top panel, 88% GPA+), which was strongly reduced by expression of the GATA1-specific hairpin (Figure 1E bottom panel, 3% GPA+). Erythroid differentiation was also monitored morphologically. As depicted in Figures 1F and 2A, STAT5A activation resulted in the appearance of erythroid cells at various stages of commitment, including erythroblasts and orthochromatic normoblasts with distinct pyknotic nuclei to the enucleated erythrocyte stage. These erythroid cells were virtually absent after knockdown of GATA1 (Figure 1F bottom panels). Furthermore, the efficacy as well as the specificity of the GATA1 knockdown were investigated by expressing the short hairpin in the absence and presence of an expression vector for mutant GATA1. This mutant GATA1 was genetically modified to be resistant to the used shRNA by introduction of silent mutations at the recognition site of the hairpin. The formation of burst-forming colony-erythroid (BFU-E) colonies was assayed after transduction of CB CD34+ cells with the shRNA vectors as well as an empty vector control or mutant GATA1-expressing vector and subsequent sorting of the double-transduced cells. It is shown in Figure 1G that expression of the short hairpin against GATA1 resulted in a robust reduction of the number of colonies, which could be partially rescued by expression of shRNA resistant GATA1.
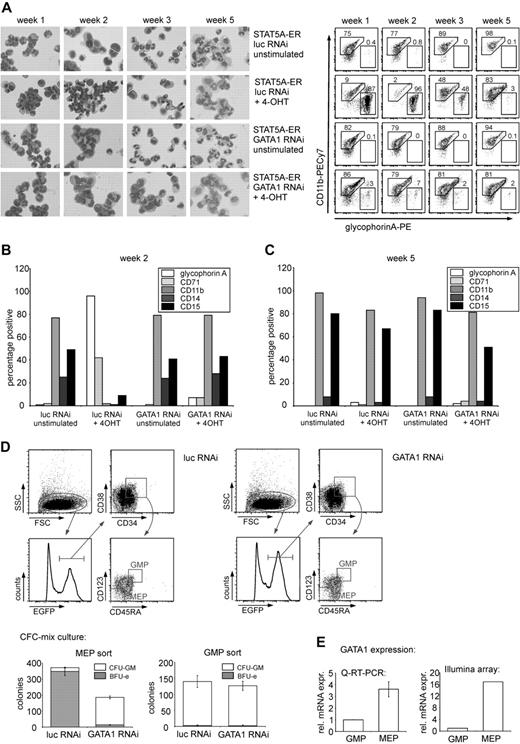
STAT5-induced block in myeloid differentiation is reversed after GATA1 depletion. (A) CD34+ CB cells were transduced with STAT5A-ER and the indicated lentiviral RNAi constructs. After transduction, cells were cultured on MS5 stromal cells and either left untreated or stimulated with 4-OHT. Each week, half of the medium was removed and double-transduced cells were sorted and cytospin preparations were made (left panel). The same cells were analyzed by flow cytometry for the expression of CD11b and GPA at weeks 1, 2, 3, and 5. Representative FACS plots are shown in the right panel with percentage positive cells indicated in the plots. (B) FACS analysis of the expression of GPA, CD11b, CD14 (monocyte/macrophage), CD15 (granulocyte), and CD71 (transferrin receptor) of cells cultured and stimulated as in panel A at week 2. (C) FACS data as in panel B from week 5. (B-C) One representative experiment of 3 experiments performed is shown. (D) CD34+ CB cells were transduced with the indicated lentiviral shRNA constructs. One day after transduction, the cells were stained for CD34, CD38, CD45RA, and CD123 and transduced (EGFP-positive) cells were sorted according to the shown gates to obtain MEP and GMP progenitor populations. The sorted cells were cultured in methylcellulose medium to enumerate the number of BFU-Es and CFU-GMs. After 2 weeks, progenitors were scored. (E) GATA1 expression in GMPs and MEPs as determined by quantitative RT-PCR and Illumina arrays.
STAT5-induced block in myeloid differentiation is reversed after GATA1 depletion. (A) CD34+ CB cells were transduced with STAT5A-ER and the indicated lentiviral RNAi constructs. After transduction, cells were cultured on MS5 stromal cells and either left untreated or stimulated with 4-OHT. Each week, half of the medium was removed and double-transduced cells were sorted and cytospin preparations were made (left panel). The same cells were analyzed by flow cytometry for the expression of CD11b and GPA at weeks 1, 2, 3, and 5. Representative FACS plots are shown in the right panel with percentage positive cells indicated in the plots. (B) FACS analysis of the expression of GPA, CD11b, CD14 (monocyte/macrophage), CD15 (granulocyte), and CD71 (transferrin receptor) of cells cultured and stimulated as in panel A at week 2. (C) FACS data as in panel B from week 5. (B-C) One representative experiment of 3 experiments performed is shown. (D) CD34+ CB cells were transduced with the indicated lentiviral shRNA constructs. One day after transduction, the cells were stained for CD34, CD38, CD45RA, and CD123 and transduced (EGFP-positive) cells were sorted according to the shown gates to obtain MEP and GMP progenitor populations. The sorted cells were cultured in methylcellulose medium to enumerate the number of BFU-Es and CFU-GMs. After 2 weeks, progenitors were scored. (E) GATA1 expression in GMPs and MEPs as determined by quantitative RT-PCR and Illumina arrays.
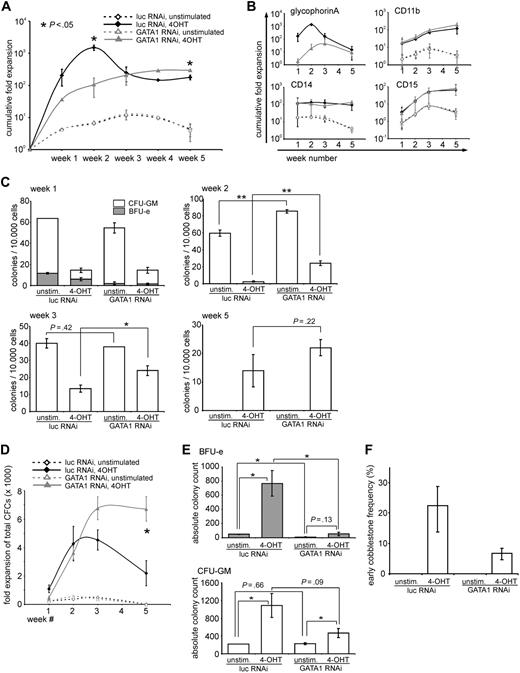
Restored balance between erythroid versus myeloid differentiation upon GATA1 down-modulation in cells expressing activated STAT5
Because the balance between erythroid and myeloid differentiation was disturbed in CB CD34+ cells expressing activated STAT5A (Figure 1A),18,19 we questioned whether down-modulation of GATA1 would modulate myelopoiesis. CB CD34+ cells were transduced with STAT5-ER together with vectors containing the short hairpin against GATA1 or a control shRNA vector. The cells were cultured on MS5 stromal cells, and the phenotype of the suspension cells resulting from these cultures was determined for a period of 5 weeks. Each week the double-transduced cells from the suspension fraction were sorted, analyzed morphologically by performing MGG staining on cytospin preparations, and analyzed phenotypically by flow cytometry for the presence of erythroid and myeloid markers. Figure 2A shows cytospin preparations of a representative experiment (left panel) as well as the expression of the myeloid marker CD11b and the erythroid marker GPA at the indicated time points (right panel). Activation of STAT5 by 4-OHT clearly resulted in a relative increase in erythroid cells in the first 3 weeks, as judged by enumeration of erythroid cells in the MGG-stained cytospins. The increase in erythroid differentiation relative to myeloid differentiation was also observed by FACS, whereby the expression of CD11b was dramatically decreased in the 4-OHT–stimulated cultures (from 75%, 77%, and 89% in the unstimulated cultures to 9%, 2%, and 48% in the stimulated cultures in the first 3 weeks). Down-modulation of GATA1 in STAT5-activated cells restored the balance between erythroid and myeloid differentiation as determined by MGG staining, as well as by FACS for CD11b, which was retained in the cultures in which GATA1 was down-regulated (from 82%, 79%, and 88% in the unstimulated cultures to 86%, 79%, and 81% in the 4-OHT–stimulated cultures; Figure 2A) The expression of other myeloid and erythroid markers at week 2 is shown in Figure 2B, and these data indicate that also the expression of CD14 and CD15 was restored to levels comparable with control groups upon down-regulation of GATA1.
The STAT5-induced erythropoiesis was most pronounced within the first 3 weeks of MS5 coculture. Thereafter, the erythroid cells had exhausted and the STAT5A cultures displayed a more myeloid phenotype, comparable with control cultures (Figure 2A,C). At this point, the suspension cells of the unstimulated cultures were almost exclusively granulocytic, with up to 90% of the cells expressing CD15 (Figure 2A,C). Regardless of GATA1 down-modulation, the activated STAT5-expressing cells displayed a clear CD11b-positive myeloid phenotype at week 5, although granulocytic maturation was less pronounced because less segmented nuclei were observed in the MGG-stained cytospins (Figure 2A). CD15 expression was slightly lower compared with control cultures (Figure 2C).
The reversal of the STAT5-induced misbalance between erythroid and myeloid differentiation by the depletion of GATA1 was further investigated by assessing the myeloid, erythroid, and megakaryocytic colony formation potential of CB progenitors in the presence or absence of GATA1. To obtain pure populations of transduced megakaryocytic/erythroid versus myeloid progenitors, the CB cells were sorted after transduction on the basis of EGFP+, CD34+, CD38+, CD123+, and CD45RA+ for the granulocyte-monocyte progenitor (GMP) progenitors and EGFP+, CD34+, CD38+ CD123,− and CD45RA− for the megakaryocyte-erythroid progenitor (MEP) fraction. As shown in the upper 2 graphs in Figure 2D, both the MEP- and the GMP-sorted cell populations were highly pure, resulting in 94% BFU-E and 98% colony-forming unit-granulocyte macrophage (CFU-GM) in the respective populations. After knockdown of GATA1, however, the proportion of BFU-Es was strongly reduced from 350 to 14 per 103 plated cells, whereas the CFU-GM in the MEP fraction was increased from 23 to 173 per 103 plated cells, corresponding to an increase from 6% to 93% in CFU-GM colony frequency. These data suggest that the lineage fate of MEPs can be redirected into GMPs on down-modulation of GATA1 expression. The proportion of CFU-GM colonies in the GMP fraction was not affected by GATA1 down-modulation, probably as a result of low expression levels of GATA1 in this population (Figure 2E). Intriguingly, the percentages of cells within the MEP and GMP gates had not yet changed within 24 hours after transduction with GATA1 shRNA vectors.
STAT5-induced long-term growth is enhanced by GATA1 depletion
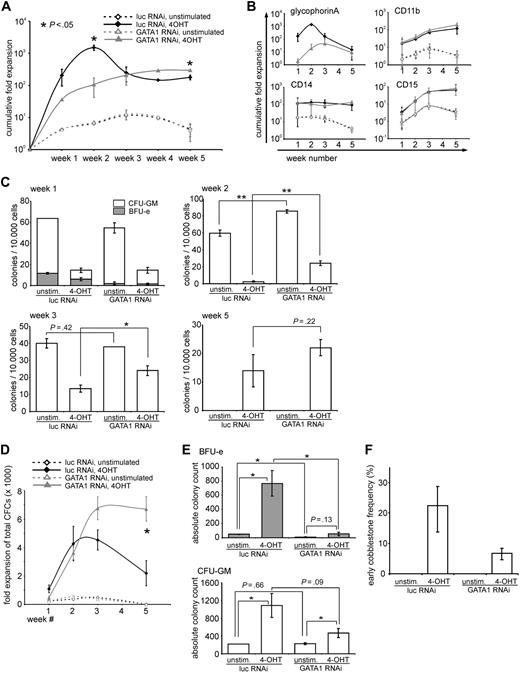
Besides differentiation, long-term expansion of hematopoietic stem/progenitor cells is markedly enhanced by expression of active STAT5 (schematically illustrated in Figure 1A).18 Because depletion of GATA1 had profound effects on the STAT5-induced differentiation of CB CD34+ cells, we next investigated the contribution of GATA1 on STAT5-induced long-term expansion. CB CD34+ cells were transduced with vectors expressing STAT5-ER in the presence of a control vector expressing luciferase shRNA or GATA1 shRNA. Cells were plated on MS5 stromal cells, and proliferation in the cultures was analyzed weekly. Progenitor frequencies in the double-transduced cells were analyzed by weekly CFC assays. The cumulative cell growth expressed as fold expansion and normalized with respect to the initial seeded cell numbers is shown in Figure 3A. Initially, in the first 2 weeks, the cultures with the active STAT5 and the control shRNA proliferated more strongly compared with the GATA1-depleted cultures. After 2 weeks, however, the cellular output of the control cultures dropped, whereas the output of the cultures expressing the short hairpin against GATA1 continued to increase, leading to a reproducibly higher cellular output at weeks 4 and 5. The specific expansion of erythroid (GPA) and myeloid (CD11b, CD14, and CD15) cells is shown in 4 separate panels in Figure 3B. Within the first few weeks of coculture on MS5, it is clear that particularly the STAT5-induced erythroid differentiation was blocked by down-modulation of GATA1, whereas the absolute numbers of myeloid cells were not affected at these time points. At week 5, no erythroid cells were observed anymore, and cocultures produced predominantly myeloid cells. In activated STAT5 cocultures, significantly more (myeloid) progeny was produced at weeks 4 and 5.
STAT5-induced long-term growth is enhanced in the absence of GATA1. (A) CD34+ CB cells were transduced with STAT5A-ER and the indicated lentiviral shRNA constructs. After transduction, cells were cultured on MS5 stromal cells and either left untreated or stimulated with 4-OHT. Each week, half of the medium was removed and the cells were counted and analyzed by flow cytometry to calculate the number of double-transduced cells. The cumulative expansion of double-transduced cells was calculated relative to the starting amount of double-transduced cells, which was set at 1. Data are mean ± SD of 2 experiments, which is representative of 4 experiments performed. (B) The same cells as in panel A were stained for GPA, CD11b, CD14, and CD15. The cumulative fold expansion of the cells, separated by the respective phenotype, is shown. (C) Double-transduced CD34+ CB cells as in panel A were sorted after 1, 2, 3, and 5 weeks, and progenitor frequencies per 10 000 plated cells were determined by culture in CFC-mix methylcellulose medium. (D) Cumulative expansion of progenitors was calculated based on the progenitor frequencies depicted in panel C and the cumulative expansion as shown in panel A. (E) The absolute amount of colonies at week 1, divided by BFU-E, and CFU-GM was calculated according to the expansion shown in panel A. (F) Early (day 10) cobblestone frequency of CB CD34+ cells, transduced as in panel A and cultured in limiting dilution on MS5 stromal cells. Significant differences (2-tailed t test): *P < .05, **P < .01.
STAT5-induced long-term growth is enhanced in the absence of GATA1. (A) CD34+ CB cells were transduced with STAT5A-ER and the indicated lentiviral shRNA constructs. After transduction, cells were cultured on MS5 stromal cells and either left untreated or stimulated with 4-OHT. Each week, half of the medium was removed and the cells were counted and analyzed by flow cytometry to calculate the number of double-transduced cells. The cumulative expansion of double-transduced cells was calculated relative to the starting amount of double-transduced cells, which was set at 1. Data are mean ± SD of 2 experiments, which is representative of 4 experiments performed. (B) The same cells as in panel A were stained for GPA, CD11b, CD14, and CD15. The cumulative fold expansion of the cells, separated by the respective phenotype, is shown. (C) Double-transduced CD34+ CB cells as in panel A were sorted after 1, 2, 3, and 5 weeks, and progenitor frequencies per 10 000 plated cells were determined by culture in CFC-mix methylcellulose medium. (D) Cumulative expansion of progenitors was calculated based on the progenitor frequencies depicted in panel C and the cumulative expansion as shown in panel A. (E) The absolute amount of colonies at week 1, divided by BFU-E, and CFU-GM was calculated according to the expansion shown in panel A. (F) Early (day 10) cobblestone frequency of CB CD34+ cells, transduced as in panel A and cultured in limiting dilution on MS5 stromal cells. Significant differences (2-tailed t test): *P < .05, **P < .01.
In Figure 3C and D, the progenitor frequency and fold expansion of progenitors in the harvested suspension fractions of the cultures at the different time points are shown. In the first 3 weeks, the progenitor frequency of the unstimulated cultures (with no active STAT5) was higher than in the cultures with active STAT5 as determined by the number of CFCs per 104 plated cells (Figure 3C). Because the total number of cells was strongly increased on STAT5 activation, the total expansion of CFCs in STAT5 cultures was strongly increased compared with control cultures (Figure 3D). At week 1, down-modulation of GATA1 significantly impaired the total amounts of BFU-Es that were produced, both in control as well as in STAT5-transduced cultures, whereas GATA1 down-modulation did not significantly change total CFU-GM numbers (Figure 3E). At week 5, the unstimulated cultures were depleted of any progenitor activity, whereas the cultures with active STAT5 were still producing progenitors (in line with previously obtained results18 ).
A striking feature of active STAT5-expressing cells is the potential to form cobblestone areas underneath MS5 bone marrow stromal cells. To enumerate the proportion of early cobblestone-forming cells, CB CD34+ cells were similarly transduced as in Figure 3A and were seeded in limiting dilution on MS5 stromal cells in 96-well plates. After 10 days, the wells positive for cobblestone formation were scored and the frequency was calculated. Figure 3F shows that approximately 1 in 5 plated cells (22%) formed early cobblestones in the presence of active STAT5, which was, although reduced, not abrogated in the absence of GATA1. Furthermore, after 5 weeks of culture, cobblestones were still present in both cultures with active STAT5, regardless of the presence of GATA1 (data not shown).
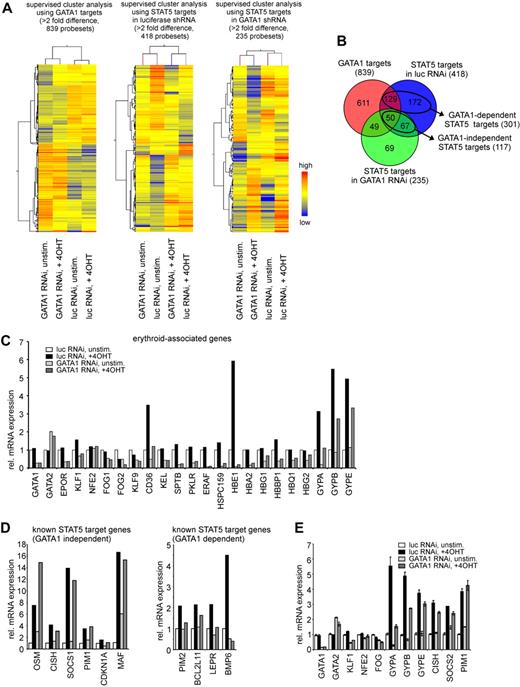
Identification of GATA1-dependent and GATA1-independent STAT5 target genes
Because depletion of GATA1 from CD34+ CB cells could dissect the phenotypes caused by STAT5 activation (differentiation vs long-term proliferation), the underlying molecular mechanisms were investigated at the level of gene expression. CD34+ CB cells were transduced with conditionally active STAT5 in the presence or absence of short hairpins against GATA1. After transduction, the transduced cells were cultured for 3 days in serum-free medium and either left untreated or stimulated with 4-OHT to induce STAT5 activity for 24 hours, after which the double-transduced cells were sorted and total RNA was extracted. Equal amounts of RNA of 3 independent experiments were pooled, reverse transcribed, and hybridized to human Illumina BeadChip Arrays that contained 46k probe sets. A total of 839 probe sets were differentially expressed between the luciferase and GATA1 short hairpin-transduced groups (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), and a supervised cluster analysis of these GATA1 targets is shown in Figure 4A. A total of 434 genes were down-regulated on GATA1 depletion, and gene ontology (GO) annotation35,36 revealed that this list was highly enriched for erythroid-associated genes (supplemental Table 1). Apparently, many of these erythroid genes are under direct control of GATA1, including EPOR, FOG1, CD36, ERAF, various hemoglobins, and GYPA. A total of 397 genes were up-regulated on GATA1 depletion, and GO annotation revealed that this list was enriched for, among others, genes associated with the plasma membrane, leukocyte activation, and apoptosis (supplemental Table 1). Although CD33 was also up-regulated on GATA1 depletion, no immediate up-regulation of a general myeloid gene expression program was observed within 24 hours after GATA1 down-modulation. A potentially important gene, GATA2, was up-regulated approximately 2-fold by GATA1 down-modulation, confirming the known reciprocal regulation of these genes.37 The key regulators of myeloid commitment, CCAAT/enhancer-binding protein-α (C/EBP-α) and PU.1, were up-regulated on down-modulation of GATA1, but the fold change did not yet reach the 2-fold significance level within the time frame of our analyses (1.5- and 1.8-fold up-regulated, respectively). Furthermore, when analyzing STAT5 target genes, it was observed that, in the presence of GATA1, 418 different probe sets were induced by STAT5 activation, whereas 235 probe sets were induced in the absence of GATA1 (supervised cluster analyses are shown in Figure 4A; data are available in supplemental Tables 2-3). These results led to the identification of 301 GATA1-dependent STAT5 target genes (defined as genes up-regulated more than 2-fold in the presence and less than 2-fold in the absence of GATA1) and 117 GATA1-independent STAT5 target genes (defined as genes up-regulated more than 2-fold irrespective of the presence of GATA1), as shown in the Venn diagram in Figure 4B and supplemental Tables 4 and 5. GO annotation revealed that the GATA1-independent up-regulated STAT5 targets were enriched for genes associated with growth regulation and the JAK-STAT pathway, and included several well-described STAT5 target genes, including OSM, PIM1, and CISH. Interestingly, several erythroid-associated genes were also up-regulated by STAT5 in the absence of GATA1, including CD36, HBE1, GYPA, GYPB, and GYPE. The GATA1-dependent STAT5 targets were enriched for genes associated with signal transduction and cell proliferation, but surprisingly no erythroid-associated genes were identified that were up-regulated by STAT5 in a GATA1-dependent manner. The Illumina expression data of several known erythroid-associated and known STAT5 target genes, divided in GATA1-dependent and GATA1-independent, are shown in Figure 4C and D, and the expression of several of these target genes was verified by quantitative PCR analysis (Figure 4E).
STAT5-induced genes can be divided as GATA1-dependent and GATA1-independent. (A) CB CD34+ cells, transduced with 4-OHT–inducible STAT5 and lentiviral vectors expressing short hairpins against luciferase and GATA1 were cultured for 3 days, STAT5 was induced by 4-OHT stimulation for 24 hours, and RNA was extracted. Equal amounts of RNA from 3 independent performed experiments were pooled and hybridized to Illumina arrays. Supervised cluster analysis: left panel, GATA1 targets with more then 2-fold difference; middle panel, STAT5 target genes in luciferase shRNA-transduced samples; and right panel, STAT5 target genes in GATA1 shRNA-transduced samples. (B) Venn diagram showing differences and overlap between the 3 groups analyzed in panel A. (C) Array-derived gene expression data of known erythroid-associated genes. (D) Array-derived gene expression data of several previously described STAT5 target genes, divided as GATA1-dependent and GATA1-independent STAT5 target genes. (E) Quantitative RT-PCR verification of several genes shown in panels C and D.
STAT5-induced genes can be divided as GATA1-dependent and GATA1-independent. (A) CB CD34+ cells, transduced with 4-OHT–inducible STAT5 and lentiviral vectors expressing short hairpins against luciferase and GATA1 were cultured for 3 days, STAT5 was induced by 4-OHT stimulation for 24 hours, and RNA was extracted. Equal amounts of RNA from 3 independent performed experiments were pooled and hybridized to Illumina arrays. Supervised cluster analysis: left panel, GATA1 targets with more then 2-fold difference; middle panel, STAT5 target genes in luciferase shRNA-transduced samples; and right panel, STAT5 target genes in GATA1 shRNA-transduced samples. (B) Venn diagram showing differences and overlap between the 3 groups analyzed in panel A. (C) Array-derived gene expression data of known erythroid-associated genes. (D) Array-derived gene expression data of several previously described STAT5 target genes, divided as GATA1-dependent and GATA1-independent STAT5 target genes. (E) Quantitative RT-PCR verification of several genes shown in panels C and D.
Discussion
We have shown previously that overexpression of (conditionally) active STAT5A in human CB-derived CD34+ cells results in at least 2 clearly distinguishable phenotypes in MS5 bone marrow stromal cocultures. First, a pronounced erythroid differentiation is induced within the first 2 weeks on activation of STAT5A. Second, long-term proliferation is induced, which is associated with the formation of cobblestone areas underneath the stroma capable of self-renewal as determined by their long-term serial replating capacity.18,19,21 In the present study, we aimed to dissect these different phenotypes and identified that the differentiation phenotypes imposed on CB CD34+ cells by active STAT5A could be uncoupled from the long-term growth phenotype by down-regulation of GATA1.
It remains unknown how activation of STAT5A can contribute to the induction of erythropoiesis. Mouse embryos that lack STAT5 expression are severely anemic because of enhanced apoptosis as a consequence of reduced expression levels of the antiapoptotic proteins Mcl1 and Bcl-XL.10,22 However, within the conditions of our assays, we found no evidence that these genes were strongly up-regulated by activation of STAT5. Furthermore, STAT5-deficient cells display reduced levels of IRP-2/IREB2, which regulates the translation of TfR122 ; but within the time frame of our gene expression studies, we also did not find evidence that IREB2 expression levels are enhanced by activated STAT5. Because GATA1 is well known for its role in erythropoiesis,26,31 the role of this protein in the STAT5-induced phenotypes was investigated in the present study. GATA1 expression was down-regulated using a lentiviral RNAi approach, and our data clearly indicate that GATA1 is required for STAT5-induced erythropoiesis. This result is in line with the present understanding that GATA1 is essential for erythropoiesis but does highlight that GATA1 is not dispensable for STAT5-induced erythroid commitment. We tried to address whether the STAT5-induced signals involved in erythroid commitment were all mediated through GATA1 or whether STAT5 and GATA1 could also act independently. We found no evidence that STAT5 could up-regulate the expression of GATA1 in CB CD34+ cells and thereby drive erythropoiesis (Figures 1, 4), and no experimental evidence was obtained that STAT5 overexpression enhances GATA1 transcriptional activity as determined in α-globin promoter-driven luciferase reporter assays (H. de Jong and J.J.S., unpublished observations, February 2004). Previously, we did observe that overexpression of another activating mutant of STAT5, STAT5A(1*6), did slightly enhance GATA1 expression levels in mobilized peripheral blood CD34+ cells.38 Whether differences in cell type or in levels of STAT5 activity underlie these observations is currently unclear, but also in a previous study in which we studied STAT5 target gene expression in relation to the dosage of STAT5 activity using the STAT5-ER vectors we did not observe any effects on GATA1 expression.19 Whether posttranslational modifications, such as acetylation of GATA1,39 could play a role in the STAT5-induced erythroid differentiation is at present unknown. To get more insight into the interplay between STAT5 and GATA1 in the process of erythroid commitment, we performed expression array analysis on CB CD34+ cells transduced with a conditional expression vector for STAT5 in the absence and presence of GATA1. Several conclusions can be drawn from these experiments. First, as expected, various erythroid-associated genes were down-regulated on depletion of GATA1, including EPOR, KLF1, FOG1, FOG2, CD36, ERAF, and several hemoglobin genes. These data are in line with previously published data.40 Interestingly, several of these erythroid-associated genes can also be activated by STAT5, even in the absence of GATA1, including CD36, HBE1, and GYPA. Apparently, STAT5 and GATA1 can act on these promoters independently, and we indeed indentified STAT5 response elements in their promoter sequences (data not shown). But because no erythroid commitment was observed in the absence of GATA1, it can be concluded that the STAT5-induced expression of these genes by itself is not sufficient for erythroid development. We also identified genes that were induced by STAT5 in a GATA1-dependent manner (supplemental Table 5), suggesting that STAT5 and GATA1 can cooperatively act on several genes as well. However, no genes were identified in this list that have directly been linked to erythroid development. GO analysis revealed that this group is enriched for genes associated with signal transduction, cell communication, and proliferation, and future studies will clarify the role of these genes in STAT5-induced erythroid differentiation.
It has been well documented that key regulators of myeloid/erythroid cell fate decisions, such as PU.1, C/EBP-α, and GATA1, are capable of directly affecting each other's activities.41-43 For instance, once erythroid commitment is initiated, an increase in GATA1 expression or activation levels will immediately impose negative feedback control on the myeloid transcription factors by negatively affecting their transcriptional activities and ultimately also their expression levels. We observed that down-regulation of GATA1 in the MEP compartment not only impaired BFU-E formation but also resulted in increased frequencies of CFU-GM progenitors (Figure 2D). Although our cloning efficiencies do not exceed 35% in these assays and therefore single-cell assays are required to study erythroid-to-myeloid lineage switching, our data do suggest that down-modulation of GATA1 is sufficient to convert BFU-E into CFU-GM progenitors. In line with these data, we find that down-modulation of GATA1 expression is sufficient to enhance expression of C/EBP-α and PU.1, although the fold change did not yet reach the 2-fold significance level within the time frame of our analyses (1.5- and 1.8-fold up-regulated, respectively).
Another marked feature of persistent STAT5 activation is the enhanced proliferation potential of stem and progenitor cells.18,19 The effective knockdown of GATA1 expression, thereby preventing erythropoiesis, provided us with the opportunity to study the effects of STAT5 overexpression in the absence of erythroid differentiation. Within the first 2 weeks of MS5 coculture, the proliferative advantage of STAT5 cells over controls was less dramatic in the absence of GATA1 compared with when GATA1 was present. After 3 weeks of culturing, the number of suspension cells as well as the number of progenitors in STAT5 cultures that were generated in the absence of GATA1 exceeded those that were generated in the presence of GATA1. Apparently, STAT5-induced erythroid differentiation resulted in exhaustion of the stem cell/progenitor pool, and inhibition of erythroid commitment by GATA1 down-modulation allowed a better long-term expansion and stem cell/progenitor maintenance. These data are in line with our previously published transcription factor dosage studies whereby we observed that intermediate STAT5 activity levels resulted in maximal long-term expansion, whereas high STAT5 activity levels resulted predominantly in erythroid commitment and exhaustion of the stem cell/progenitor compartment.19 As observed previously, early cobblestone formation was clearly enhanced in the presence of active STAT5, but the frequency was approximately 3-fold down-regulated in the absence of GATA1, despite the capacity to generate long-term expanding cultures under those conditions. Apparently, the 10-day cobblestones represent a mixture of erythroid-predisposed progenitor cells that lack long-term self-renewal capacity as well as cobblestone areas that represent more primitive cells that retain the capacity for long-term self-renewal and can sustain STAT5-induced long-term expansion for more than 5 weeks. Another gene that was up-regulated after GATA1 depletion was GATA2, confirming the known reciprocal regulation of these genes.37 GATA2 is known to be involved in regulation of stem cell quiescence, with high expression levels in the most quiescent stem cells.27 GATA2−/+ mice are described to have impaired competitive repopulation potential, accompanied by reduced CFC and long-term culture-initiating cell frequencies.44 Possibly, increased GATA2, in conjunction with the elevated expression of C/EBP-α and PU.1 in the cells transduced with GATA1 shRNA in the presence of active STAT5, facilitates further expansion of these cells. However, GATA2 up-regulation in the control cultures did not result in increased long-term proliferation. In addition, overexpression of GATA2 in human stem and progenitor cells has been described to result in quiescence and impaired function of these cells,45 so whether the degree of up-regulation of GATA2 is the cause of the increased long-term output in our setting is not clear.
The uncoupling of the STAT5-induced phenotypes with the erythroid lineage provided several interesting GATA1-independent STAT5 target genes. GO annotation revealed that the GATA1-independent up-regulated STAT5 targets were enriched for genes associated with growth regulation and the JAK-STAT pathway. Indeed, growth-related genes, such as PIM1 and MAF, were represented in this group. PIM1 is well known for its role in the response to hematopoietic growth factors.46,47 PIM1 might also play a role in the enhanced interaction between STAT5-expressing stem/progenitor cells and the formation of cobblestone areas because it was recently shown that FLT3-ITD–induced leukemogenesis depends on PIM1 expression to regulate CXCL12-CXCR4–mediated homing and migration.48 MAF is an important transcription factor involved in many different cell types, including myeloid hematopoietic cells.49 In multiple myeloma, the oncogene MAF has been shown to stimulate cell-cycle progression as well as to improve the interaction between tumor and stromal cells.50 The dissection of the precise roles that these genes fulfill in the process of STAT5-induced long-term stem cell/progenitor expansion is under current investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek-VENI, Nederlandse Organisatie voor Wetenschappelijk Onderzoek-VIDI (2008), and Koningin Wilhelmina Fonds (2009-4275).
Authorship
Contribution: A.T.J.W. designed and performed research, analyzed data, and wrote the paper; E.V. designed research and analyzed data; and J.J.S. designed research, analyzed data, and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Jacob Schuringa, Department of Hematology, University Medical Center Groningen, Hanzeplein 1, 9713 GZ Groningen, The Netherlands; e-mail: j.schuringa@int.umcg.nl.