Abstract
X-linked severe-combined immunodeficiency (SCID-X1) has been treated by therapeutic gene transfer using gammaretroviral vectors, but insertional activation of proto-oncogenes contributed to leukemia in some patients. Here we report a longitudinal study of gene-corrected progenitor cell populations from 8 patients using 454 pyrosequencing to map vector integration sites, and extensive resampling to allow quantification of clonal abundance. The number of transduced cells infused into patients initially predicted the subsequent diversity of circulating cells. A capture-recapture analysis was used to estimate the size of the gene-corrected cell pool, revealing that less than 1/100th of the infused cells had long-term repopulating activity. Integration sites were clustered even at early time points, often near genes involved in growth control, and several patients harbored expanded cell clones with vectors integrated near the cancer-implicated genes CCND2 and HMGA2, but remain healthy. Integration site tracking also documented that chemotherapy for adverse events resulted in successful control. The longitudinal analysis emphasizes that key features of transduced cell populations—including diversity, integration site clustering, and expansion of some clones—were established early after transplantation. The approaches to sequencing and bioinformatics analysis reported here should be widely useful in assessing the outcome of gene therapy trials.
Introduction
Therapeutic gene transfer has been used successfully to treat a variety of human genetic diseases, including X-linked severe-combined immunodeficiency (SCID-X1), adenosine deaminase deficiency, chronic granulomatous disease, adrenoleukodystrophy, and β-thalassemia (M.C.-C., E. Payen, O. Negre, G.P.W., K. Hehir, F. Fusil, J. Down, M. Denaro, T. Brady, R. Pawliuk, K. Westerman, R. Cavallesco, B. Gillet-Legrand, L. Caccavelli, F. Bernaudin, R. Girot, R. Dorazio, G.-J. Mulder, A. Polack, A. Bank, J. Soulier, J. Larghero, N. Kabbara, B. Dalle, B. Gourmel, G. Socié, S. Chrétien, N. Cartier, P. Aubourg, A.F., K. Cornetta, F. Galacteros, Y. Beuzard, E. Gluckman, F.D.B., S.H.-B.-A., P.L., manuscript submitted, January 27, 2010).1-6 Although these protocols showed positive clinical outcome in patients with few other therapeutic options, successes of the SCID-X1 trials were tempered by adverse events, in which integration of the therapeutic vectors increased transcription from cancer-related genes and thereby contributed to development of leukemia.3,6-8 Thus, there is intense interest in improving methods for following the fate of gene-corrected cells in patients to monitor for adverse events and aid in the development of safer vectors. Such studies also provide a unique window on human hematopoiesis because all progenitor cell clones contributing T cells and natural killer cells to the periphery are marked by unique integration sites.6,8-11 However, complicating such analysis is the finding that commonly used methods for integration site recovery are severely biased.12 Here we report the use of 454/Roche pyrosequencing12-14 and mathematical reconstruction to quantify evolution of gene-corrected cell populations based on extensive resampling of patient genomic DNA specimens.
Our results also help to assess mechanisms giving rise to preferential growth of cell clones. For the adverse events in trials using gammaretroviral vectors characterized to date, all appear to involve integration of enhancers in the therapeutic vectors near cancer-related genes, resulting in an increase in the rate of transcription initiation.3,6-8,11,15,16 Findings on clonal expansion after stem cell transduction with a lentiviral vector to treat β-thalassemia (M.C.-C. et al, manuscript submitted), and data for SCID-X1 patients reported here provide data relevant to assessing whether mechanisms in addition to enhancer/promoter insertion may also contribute to gene activation.
Here we report analysis of 203 805 integration site sequence reads (9767 unique sites) from 8 of the SCID-X1 patients and computational correction of the recovery bias to allow detailed modeling of the dynamics of cell populations. Diversity was positively correlated with the numbers of cells transduced initially. In some but not all patients, diversity slowly declined over time. We documented new examples of clonal expansion in healthy patients, including 2 examples associated with integration sites in the cancer-related genes CCND2 and another 2 in HMGA2. Because T cells are marked by unique vector insertion sites, the effectiveness of chemotherapy for adverse events could be documented by tracking the vector-marked blast cells. A central point emerging from the longitudinal study is that many of the key features of integration site populations–including diversity, correlations with genomic features, clustering, and expansion of some cell clones–were largely established by the earliest time points measured. These data provide a detailed longitudinal picture dynamic of cells marked by vector integration, and disclose that persistent patterns in populations of transduced cell clones were established early after infusion. A detailed summary of the clinical and immunologic status of these patients after 10 years of treatment has been submitted elsewhere (S.H.-B.-A., J. Hauer, A. Lim, C. Picard, G.P.W., C.C.B., C. Martinache, F. Rieux-Laucat, S. Latour, B. H. Belohradsky, L. Leiva, R. Sorensen, M. Debré, J. L. Casanova, S. Blanche, A. Durandy, F.D.B., A.F., M.C.-C., manuscript submitted, January 6, 2010).
Methods
Isolation of peripheral blood circulating cells and cell subsets
Peripheral blood samples were obtained at various time points from patients enrolled in the SCID-X1 gene therapy trial with approval from Hôpital Necker-Enfants Malades. Peripheral blood mononuclear cells (PBMC) were separated by density centrifugation on Ficoll-Hypaque (Nycomed; Pharmacia) and then the T lymphocyte population was selected by immunomagnetic columns using a monoclonal antibody against CD3 (Miltenyi Biotec). Unfractionated PBMC samples contained roughly 70% to 80% CD3+ cells. Genomic DNA was extracted from cells using proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. The DNA pellets were resuspended in ethylenediaminetetraacetic acid 10:1 buffer and stored at −20°C until use.
Recovering integration site sequences
Detailed methods for integration site analysis were as reported3,12,17 and can be found in the supplemental Reports (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Briefly, aliquots of genomic DNA extracted from patient samples were digested using up to 6 different cocktails of restriction enzymes (AvrII/NheI/SpeI, MseI, ApoI, BstYI, Tsp509I, or NlaIII). The digested DNA samples were ligated to linkers, then digested using SpeI or MscI to cleave the vector internal fragments, and amplified by nested polymerase chain reaction (PCR). Each second-round long-terminal repeat (LTR) specific primer contained a unique 8-nucleotide bar code that indexed the amplification products. The PCR products were gel purified, pooled, and sequenced using the 454/Roche GS FLX platform. All integration site sequences are available in GenBank under accession nos. GS901925 through GS923002.
Data processing and analysis
Pyrosequencing reads were decoded, trimmed to remove LTR and linker sequences, then mapped to the human genome (hg18) to yield integration sites using quality control criteria as previously described.3,12
Statistical methods for generating heat maps, corrected abundance, and integration site clustering are described18 and detailed in supplemental Reports. Random control integration sites matched to control for restriction bias were generated in silico for comparison to integration site data (details in supplemental Report 1). Statistical methods used in Figure 2 are described in the supplemental data of the integration target study.19 Gene ontology studies were carried out using DAVID (Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov/). Cluster analysis was carried out using Ingenuity (http://www.ingenuity.com/). Tables describing genomic landmarks were downloaded from the University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/). Comparisons among the different types of cell samples analyzed (specified in supplemental Table 1) did not yield any notable trends associated with sample type when integration frequencies were compared over sets of genomic annotation or in overlap studies among sample types (data not shown). Because of sample availability in some cases, we were not able to use 6 restriction enzymes for analysis, so in some of the statistical tests only the deeper data were used.
Cancer-related genes used in our experiment were defined in the allOnco database (http://microb230.med.upenn.edu/protocols/cancergenes.html), which is a collection of 1650 genes proposed to be involved in cancer from the Retroviral Tagged Cancer Gene Database, Sanger Cancer Gene list, from Retroviruses,20 and from other datasets summarized on the site. For genes called cancer-related in model organisms, the human homologs were identified and added to the list. In some of the analysis in the supplemental Reports, a more restrictive list (French Lymphoma) was also compared, which is a list of 38 genes commonly involved in lymphoid cancers. French Lymphoma is also available at the website listed above for cancer-related genes.
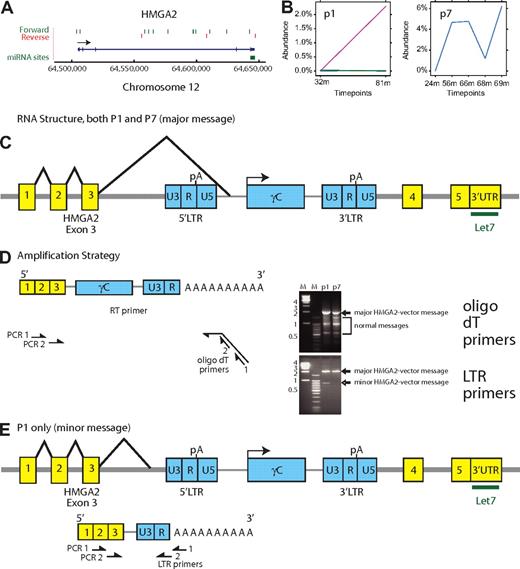
Analyzing the structure of the HMGA2-vector chimeric messages
To determine the structure of the HMGA2-vector fusion message, total cellular RNA was extracted, and the first-strand cDNA was synthesized using the OmniScript Reverse Transcriptase Kit (QIAGEN) and a polyT primer containing a 5′ extension. The cDNA was then amplified by 2 methods of nested PCR as illustrated in Figure 6D. In the first method, a primer bound to the 5′ extension was used with a second primer bound to the HMGA2 third exon. In the second round, PCR was carried out with nested third exon and 5′ extension primers. In a second method, HMGA2 primers were used with nested vector LTR primers. The PCR products were separated by agarose gel electrophoresis. Distinct bands were excised, purified, cloned, and sequenced, and the fusion message reconstructed.
Browsing integration sites on the human genome
Unique integration sites from this study can be viewed together with user-configurable annotation on the UCSC browser (http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&hgt.customText = http://microb230.med.upenn.edu/ucsc/allSCIDsites.bed&position=chr12:64447590-64652492).
To view integration sites near particular genes, follow the link at the UCSC integration website mentioned above, type the gene name in the “position/search” field, hit return, then click on the gene name. “SCIDintSites” indicates the positions of integration site, and “+” and “−” indicate the proviral orientation relative to the chromosomal numbering.
Results
Isolation of vector integration sites using 454/Roche pyrosequencing
Cells from blood or bone marrow were harvested from 8 gene-corrected SCID-X1 patients to yield the samples listed in supplemental Table 1. Genomic DNA was purified, cleaved with restriction enzymes, ligated to DNA linkers, amplified, and sequenced as described12,13 using DNA bar coding21,22 and the 454/Roche GS FLX system.14
The integration site recovery method used is known to be highly biased because of cleavage of genomic DNA using restriction enzymes.12 Quantification of the recovery bias and the procedure devised to correct it are presented in supplemental Reports 1 and 2. Integration sites are most commonly recovered when they are approximately 80 bases from a restriction enzyme cleavage site, resulting in sharp recovery biases when single restriction enzymes were used to cleave DNA. Cleavage of genomic DNA with 6 different restriction enzymes, however, resulted in much more even coverage, as illustrated by the comparison of integration site recovery probabilities between 1 versus 6 enzymes (supplemental Report 2, page 8). Using 6 enzymes per sample and the reported correction procedure, the abundance of cell clones could be estimated from integration site sequence data. A total of 120 patients/time point/enzyme combinations was studied (supplemental Table 1).
Analysis of the number and diversity of unique integration sites
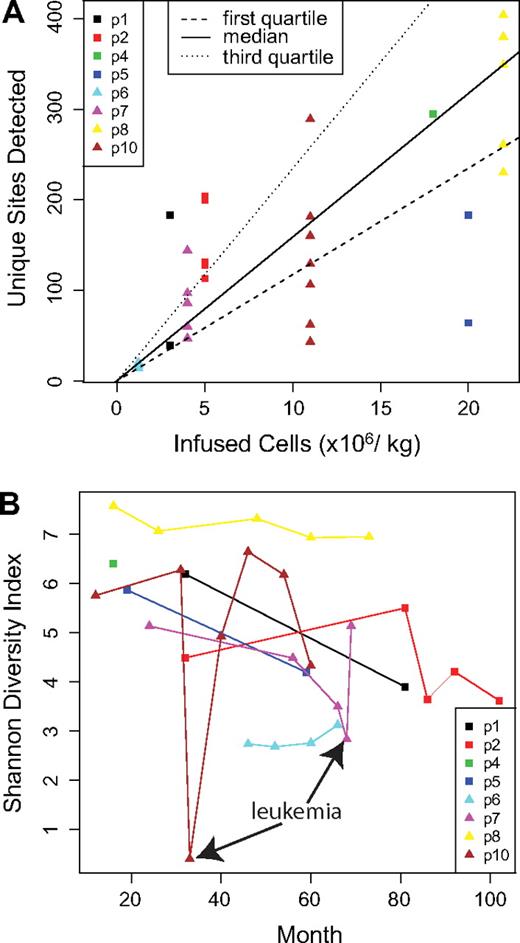
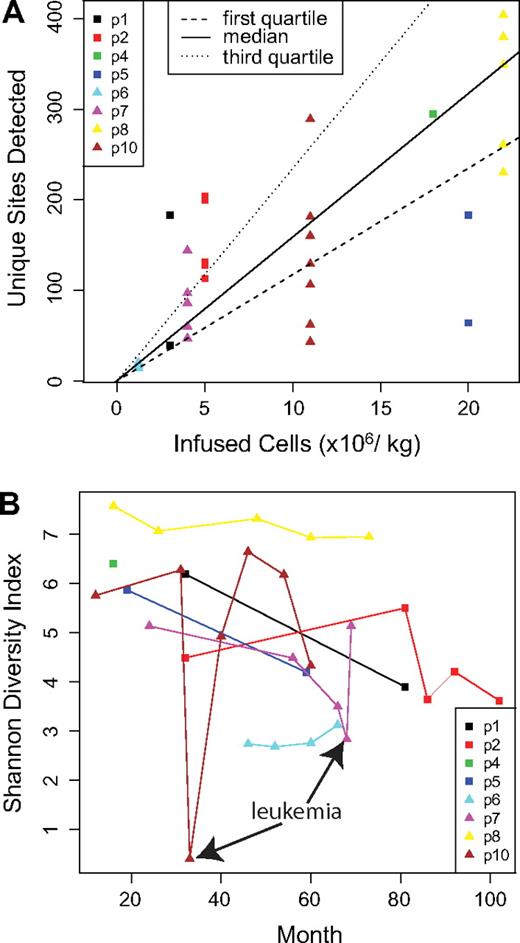
We first asked whether the number of gene-corrected cells initially infused into the patients correlated with the number of unique integration sites detected (Figure 1A and supplemental Report 3). The number of transduced cells ranged from 1 to 22 million cells/kg. A significant positive correlation was seen (P = .023, Pearson correlation; detailed analysis is presented in supplemental Report 3). A similar significant correlation was seen when the diversity of integration sites was compared with the number of infused cells using the Shannon Diversity Index (P = .013, Pearson correlation), which quantifies the number of different sites together with the evenness of distribution (data not shown). However, these trends were both driven by the 2 patients at the lowest and highest extremes. Thus, more infused cells did result in a greater number and diversity of marked cells after infusion, though it does not appear that precise predictions of these effects can be made.
Population structure of gene-corrected cells. (A) The number of infused cells per kilogram in each patient is shown on the x-axis. The number of unique integration sites detected at each time point is shown on the y-axis. For the comparison, only data obtained by cleaving the genome with MseI were used because this allowed a fair comparison among samples (complete data are summarized in supplemental Table 1). (B) The longitudinal trends in diversity are shown. The x-axis shows time after infusion of gene-corrected cells, and the y-axis shows diversity as quantified using the Shannon Diversity Index. Time points corresponding to the adverse events in patients no. 7 and no. 10 are marked.
Population structure of gene-corrected cells. (A) The number of infused cells per kilogram in each patient is shown on the x-axis. The number of unique integration sites detected at each time point is shown on the y-axis. For the comparison, only data obtained by cleaving the genome with MseI were used because this allowed a fair comparison among samples (complete data are summarized in supplemental Table 1). (B) The longitudinal trends in diversity are shown. The x-axis shows time after infusion of gene-corrected cells, and the y-axis shows diversity as quantified using the Shannon Diversity Index. Time points corresponding to the adverse events in patients no. 7 and no. 10 are marked.
How many unique cell clones, as reported by integration sites, are active in these patients? In any single sample, only a subset of all sites is recovered, but methods are available for estimating population size based on repeated resampling. We used a “capture-recapture” analytical approach, treating independent time points as separate samples from the pool of gene-corrected cells (supplemental Report 4). The estimated number of corrected cells ranged from 1784 (patient no. 2) to 9659 (patient no. 8). Comparison of this value to the number of cells initially infused in each patient (vector copy number per cell estimated to be 0.5-1.5) indicates that fewer than 1 in 100 of the infused cells gave rise to the circulating gene-corrected cells, consistent with long-term repopulating cells comprising a minority of the infused cell population.
Longitudinal trends in diversity were quantified by plotting Shannon Index values as a function of time after cell infusion (Figure 1B). The absolute values varied widely among patients. For patients no. 7 and no. 10, sharp reductions in diversity were seen at the times of adverse events, in which the leukemic cells expanded and so diminished the representation of other cell clones. However, for both patient no. 7 and patient no. 10, chemotherapy restored the population diversity, and the patients continue to benefit from the gene therapy treatment. For patient no. 5, the time point corresponding to the adverse event has been previously studied and shown to have low diversity.8 The postchemotherapy time point analyzed here showed that chemotherapy restored the population diversity for patient no. 5 as well. For several of the patients, a slow decline in diversity was observed over time (analysis not shown).
Integration near sites of epigenetic marks and genomic features
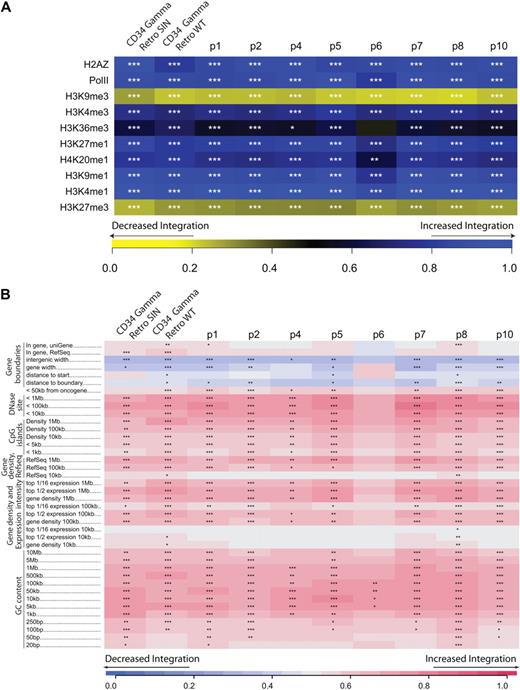
We next catalogued the distribution of the 9767 unique integration sites from the SCID-X1 patients relative to features mapped on the human genome. The color codes in Figure 2 compare the distribution of integration sites from each patient to random distributions (corrected for the recovery bias).
Integration site abundance near epigenetic marks and genomic features. (A) Integration frequency near sites of histone posttranslational modification or bound chromatin proteins. Integration frequency is quantified relative to genome-wide mapping data in CD34+ hematopoietic stem cells studied.23 The integration frequency scale is shown along the bottom of the panel. Increasingly intense shades of yellow indicate negative correlation of the experimental dataset with the matched random control, and increasing shades of blue indicate positive correlation. The scale is generated using the ROC (receiver operator characteristic) area method.18,19 CTCF is a DNA-binding protein proposed to be associated with chromatin boundaries. H2AZ is a histone variant associated preferentially with promoters. For both panels, the asterisks in each tile indicate the significance of any departures from random integration; *P < .05, **P < .01, ***P < .001). The datasets marked “Retro SIN” and “Retro WT” are for gammaretroviral integration in CD34+ cells reported.25 (B) Integration frequency near annotated sequence features is quantified using the ROC area method.18 Increased integration near the indicated feature compared with random distribution is shown in red, decreased integration in blue. For many of the features, the strength of the trend was examined over several genomic length intervals. The interval lengths are shown to the right of the feature name (eg, for GC content, 1 kb indicates intervals of 1 kb around each integration site were used for analysis). Intervals marked “<” indicate measures of integration within the indicated distance of that feature. Intergenic width indicates the length of intervals between transcription units for those sites outside transcription units. The short intergenic regions (gene dense regions) indicated in blue were favored for integration. Effects of gene activity are captured in the expression intensity measure. Affymetrix expression data for lymphoid cells were used to annotate genes, then density of genes with different expression levels used to annotate integration sites as in the gene density analysis. For example, for the top 1/2 expression, the density of genes was analyzed at each integration site or random control, but only the most active 50% of genes was scored. For the top 1/16 expression, the most active 1/16th of genes was used. Because the datasets are large, in a few cases statistically significant differences were achieved for tiles where little color is evident. One anomalous dataset was excluded from the analysis as an extreme outlier (BstYI for patient no. 6).
Integration site abundance near epigenetic marks and genomic features. (A) Integration frequency near sites of histone posttranslational modification or bound chromatin proteins. Integration frequency is quantified relative to genome-wide mapping data in CD34+ hematopoietic stem cells studied.23 The integration frequency scale is shown along the bottom of the panel. Increasingly intense shades of yellow indicate negative correlation of the experimental dataset with the matched random control, and increasing shades of blue indicate positive correlation. The scale is generated using the ROC (receiver operator characteristic) area method.18,19 CTCF is a DNA-binding protein proposed to be associated with chromatin boundaries. H2AZ is a histone variant associated preferentially with promoters. For both panels, the asterisks in each tile indicate the significance of any departures from random integration; *P < .05, **P < .01, ***P < .001). The datasets marked “Retro SIN” and “Retro WT” are for gammaretroviral integration in CD34+ cells reported.25 (B) Integration frequency near annotated sequence features is quantified using the ROC area method.18 Increased integration near the indicated feature compared with random distribution is shown in red, decreased integration in blue. For many of the features, the strength of the trend was examined over several genomic length intervals. The interval lengths are shown to the right of the feature name (eg, for GC content, 1 kb indicates intervals of 1 kb around each integration site were used for analysis). Intervals marked “<” indicate measures of integration within the indicated distance of that feature. Intergenic width indicates the length of intervals between transcription units for those sites outside transcription units. The short intergenic regions (gene dense regions) indicated in blue were favored for integration. Effects of gene activity are captured in the expression intensity measure. Affymetrix expression data for lymphoid cells were used to annotate genes, then density of genes with different expression levels used to annotate integration sites as in the gene density analysis. For example, for the top 1/2 expression, the density of genes was analyzed at each integration site or random control, but only the most active 50% of genes was scored. For the top 1/16 expression, the most active 1/16th of genes was used. Because the datasets are large, in a few cases statistically significant differences were achieved for tiles where little color is evident. One anomalous dataset was excluded from the analysis as an extreme outlier (BstYI for patient no. 6).
Figure 2A presents an analysis of the distribution of SCID-X1 integration sites relative to epigenetic marks. For this analysis, we used data from genome-wide mapping of 10 forms of histone methylation, or chromatin-bound proteins in CD34+CD133+ cells.23 The significance of any departure from the matched random controls (supplemental Report 1) is indicated by asterisks in each tile of the heat map. Integration was favored near a collection of histone posttranslational modifications associated with promoters and active transcription units, including H3K4 me1, H3K9 me1, H3K27 me1, and bound RNA polymerase II. The histone variant H2AZ is associated with promoters, and it also was enriched near integration sites. A histone methylation mark associated with repression of transcription (H3K9 me3) was negatively associated with integration. The patterns did not show major differences in longitudinal analysis, indicating that they were mostly established during initial integration.
Vector integration was similarly quantified near genomic landmarks and compared with random distribution (Figure 2B).6,11,16 Integration was favored near gene boundaries, near features associated with promoters (CpG islands and DNase I hypersensitive sites), in gene-rich regions, and in G/C-rich regions (which are also gene-rich), as has been reported previously for integration by gammaretroviruses.6,24,25 These data generally parallel trends seen in previous studies, though for the first time allowing statistical comparisons over this many patients, time points, and genomic features. We note that another factor could also be involved, which is integration in active regions that might have promoted expression of the interleukin-2 receptor γ transgene and thereby promoted cell expansion.
Longitudinal abundance of cell clones
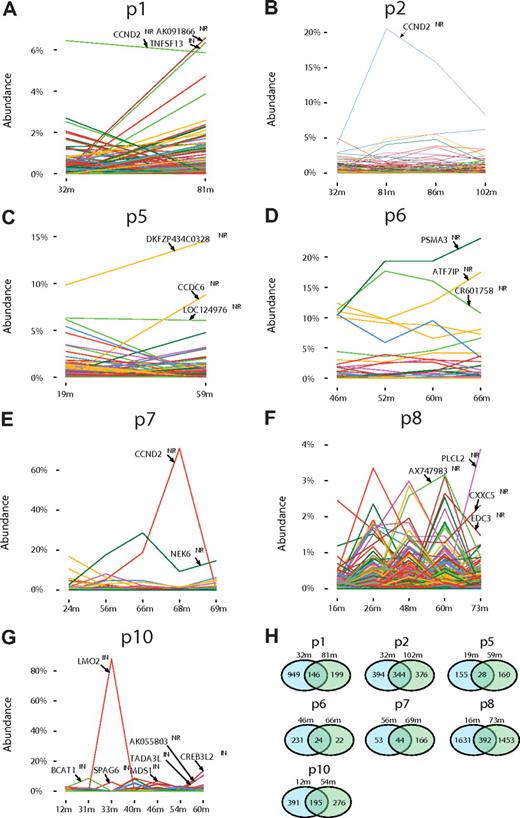
Figure 3 shows longitudinal analysis of the reconstructed clone abundance for 7 of the study subjects (for the eighth only one time point was available and so was not included). Notable changes in abundance of cell clones were seen over time in many of the patients, as has been suggested from less complete sampling in other gene therapy trials.26 For patients no. 7 and no. 10, expansions of clones harboring integration sites at CCND2 (patient no. 7) and LMO2 (patient no. 10) were associated with clinical adverse events (Figure 3E,G). Another site involved in the adverse event in patient no. 10, found within the SPAG6 locus and activating nearby BMI1, was recovered in lesser abundance because this site was difficult to isolate with the restriction enzymes used, and the correction procedure did not fully restore the missing counts.3
Longitudinal analysis of the relative abundance of gene-corrected cell clones. (A-G) The proportion of cells containing each integration site is shown on the y-axis; time after gene therapy in months (m) is on the x-axis. The proportion was calculated from the sequence counts as described in supplemental Reports 2, 3, and 4. The gene names for the most abundant clones are shown within each panel. “NR” indicates near the gene, “IN,” within the gene. The adverse events in the trial were as follows, designated by patient (p) number, genes involved, and time of event: p4, LMO2, 30m; p5, LMO2, 20m; p7, CCND2, 68m; p10, LMO2 and BMI1/SPAG6, 33m. (H) Comparison of unique integration site sequences at early versus late time points. Pairs of time points were chosen so that similar sets of restriction enzymes were used for analysis, because recovery using a greater number of restriction enzymes results in recovering a greater number of sites. Thus for some of the patients the last time point was not used in favor of earlier time points with more data. Restriction enzymes used were ApoI, AvrII/NheI/SpeI, BstYI, MseI, NlaIII, and Tsp509I (patients no. 1, no. 2, no. 6, and no. 8); ApoI, AvrII/NheI/SpeI, BstYI, and MseI (patient no. 10); and AvrII/NheI/SpeI and MseI (patients no. 5 and no. 7).
Longitudinal analysis of the relative abundance of gene-corrected cell clones. (A-G) The proportion of cells containing each integration site is shown on the y-axis; time after gene therapy in months (m) is on the x-axis. The proportion was calculated from the sequence counts as described in supplemental Reports 2, 3, and 4. The gene names for the most abundant clones are shown within each panel. “NR” indicates near the gene, “IN,” within the gene. The adverse events in the trial were as follows, designated by patient (p) number, genes involved, and time of event: p4, LMO2, 30m; p5, LMO2, 20m; p7, CCND2, 68m; p10, LMO2 and BMI1/SPAG6, 33m. (H) Comparison of unique integration site sequences at early versus late time points. Pairs of time points were chosen so that similar sets of restriction enzymes were used for analysis, because recovery using a greater number of restriction enzymes results in recovering a greater number of sites. Thus for some of the patients the last time point was not used in favor of earlier time points with more data. Restriction enzymes used were ApoI, AvrII/NheI/SpeI, BstYI, MseI, NlaIII, and Tsp509I (patients no. 1, no. 2, no. 6, and no. 8); ApoI, AvrII/NheI/SpeI, BstYI, and MseI (patient no. 10); and AvrII/NheI/SpeI and MseI (patients no. 5 and no. 7).
Unexpectedly, against the background of many distinct sites, the plot for both patients no. 1 and no. 2 showed high relative abundance of single sites in the CCND2 promoter in all the time points studied, and quantitative PCR analysis confirmed this observation (Figure 3A-B). Thus the pyrosequencing data document the presence of high-frequency cell clones harboring CCND2 sites in healthy patients, and also establish that clonal expansion associated with CCND2 insertion is not in itself sufficient for transformation over the time course studied. Previous studies have also reported that CCND2 and LMO2 are a common target for integration by gammaretroviruses.6,25,26 In patients no. 5, no. 6, and no. 8, high relative abundance sites were detected that were not associated with genes known to be involved in growth control (Figure 3C,D,F).
Another approach to quantifying the dynamics of transduced cell clones is to compare the unique sites seen at the first and last time points studied (Figure 3H). For patients no. 1 and no. 6, the number of unique sites recovered dropped between the first and last time points, and as many as half of the sites detected late were also detected early. This pattern suggests loss of long-term repopulating cell clones over time. These data also emphasize that we were unable to detect all of the gene-corrected cells present in a patient at early times after infusion because a substantial number of clones were detected late but not early. This is probably due either to the imperfect methods for integration site recovery or intermittent output from transduced cell clones.
We analyzed possible longitudinal changes in integration site frequency near cancer-related genes to determine whether selection for such sites was associated with favored cell growth. Analysis of the reconstructed abundance of cells harboring each integration site showed that most measures of proximity to cancer-related genes were not changing over time, though the number of integration sites within 50 kb of a cancer-related gene 5′ end were increasing slightly (supplemental Report 5, page 8).
Clustering of integration sites
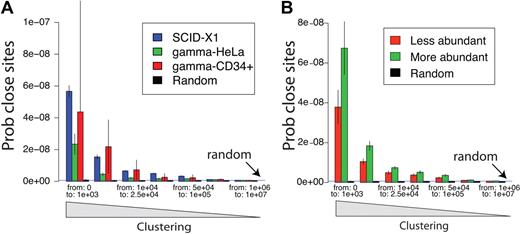
We next analyzed the clustering of integration acceptor sites on a genome-wide scale. Gammaretroviruses are known to favor integration near gene 5′ ends, so clustering is expected compared with random distributions.6,11,12,24,25 However, a key question is whether the SCID-X1 sites are more clustered than expected for gammaretroviral vector sites because increased clustering would be consistent with preferential outgrowth of clones with integrated vectors in genomic regions where insertional activation promotes cell growth. Figure 4A compares pooled SCID-X1 integration sites to gammaretroviral integration sites generated by infection of CD34+ cells25 or tissue culture cells.12,24,27 Clustering was quantified by measuring the number of bases between all adjacent pairs of integration sites, so that enrichment in short distances is indicative of clustering.17 We found that clustering in the SCID-X1 sample was indeed significantly greater than clustering for a control gammaretrovirus after tissue culture infections (P = .009; Figure 4A and as calculated in supplemental Report 5). SCID-X1 sites showed a trend toward more clustering than sites from infections of CD34+ cells, though this did not achieve significance, due in part to the comparatively small number of sites in CD34+ cells available for analysis.
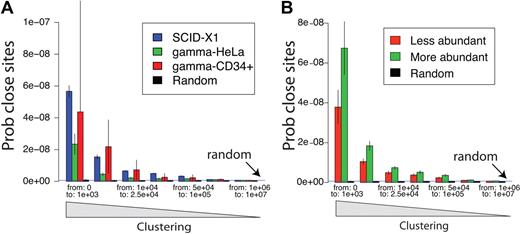
Clustering of integration sites. (A) Clustering in the SCID gene-corrected samples is greater than for a gammaretroviral vector in tissue culture. Clustering was analyzed by comparing the distribution of distances between integration sites (x-axis). That is, the lengths of chromosomal segments between integration sites is measured for all pairs and tabulated. Enrichment for short distances between pairs (left side of x-axis) indicates relatively greater clustering. The probability of encountering distances of the indicated lengths by chance (Prob close sites, y-axis) was normalized for the number of sites in each set. To obtain enough control gammaretroviral integration sites for comparison, sites from various studies were pooled.19,24,27 The dataset for gammaretroviral vector integration in CD34+ cells25 is smaller than the others, so the uncertainty is greater (larger error bars) because of the smaller sample size. The blue horizontal line (random) represents the probability expected for random control sites. The SCID sites were significantly more clustered than those of Moloney murine leukemia virus in HeLa cells. (B) Clustering is greater for frequently isolated SCID-X1 integration sites, reflecting selective expansion of cell clones with integration sites in clusters. The distance between integration sites is shown on the x-axis, and the probability of integration site distance is shown on the y-axis. The population of unique integration sites was annotated for the frequency of sequence reads for each, then the more abundant half (green) was compared with the less abundant half (red). The more abundant sites were significantly more clustered (P ≪ .05).
Clustering of integration sites. (A) Clustering in the SCID gene-corrected samples is greater than for a gammaretroviral vector in tissue culture. Clustering was analyzed by comparing the distribution of distances between integration sites (x-axis). That is, the lengths of chromosomal segments between integration sites is measured for all pairs and tabulated. Enrichment for short distances between pairs (left side of x-axis) indicates relatively greater clustering. The probability of encountering distances of the indicated lengths by chance (Prob close sites, y-axis) was normalized for the number of sites in each set. To obtain enough control gammaretroviral integration sites for comparison, sites from various studies were pooled.19,24,27 The dataset for gammaretroviral vector integration in CD34+ cells25 is smaller than the others, so the uncertainty is greater (larger error bars) because of the smaller sample size. The blue horizontal line (random) represents the probability expected for random control sites. The SCID sites were significantly more clustered than those of Moloney murine leukemia virus in HeLa cells. (B) Clustering is greater for frequently isolated SCID-X1 integration sites, reflecting selective expansion of cell clones with integration sites in clusters. The distance between integration sites is shown on the x-axis, and the probability of integration site distance is shown on the y-axis. The population of unique integration sites was annotated for the frequency of sequence reads for each, then the more abundant half (green) was compared with the less abundant half (red). The more abundant sites were significantly more clustered (P ≪ .05).
To investigate whether integration at sites marked by clusters could have promoted cell growth or persistence, we asked whether integration sites found in clusters were from relatively more abundant cell clones than integration sites outside of clusters. Figure 4B presents a test of this question, in which unique integration sites (pooled over all patients) were annotated by the number of sequence reads recovered for each site, then the pooled sites were divided into more abundant and less abundant halves. The extent of clustering was then compared between the 2 groups, again by quantifying the lengths of genomic intervals between sites. We found that the more clustered integration sites tended to be from more abundant clones (P = .019, as calculated in supplemental Report 5), consistent with the idea that cells harboring integration sites at loci marked by clustered sites grew out preferentially. A detectable but quite modest increase in clustering took place over time (P = .024, supplemental Report 5, page 6).
A variety of genomic features are significantly enriched near frequently isolated sites, including regions of high G/C content, DNase I cleavage sites, CpG islands, and cancer-related genes, though we note that the magnitude of these effects tended to be modest (supplemental Report 5).
Ontology of genes at integration sites
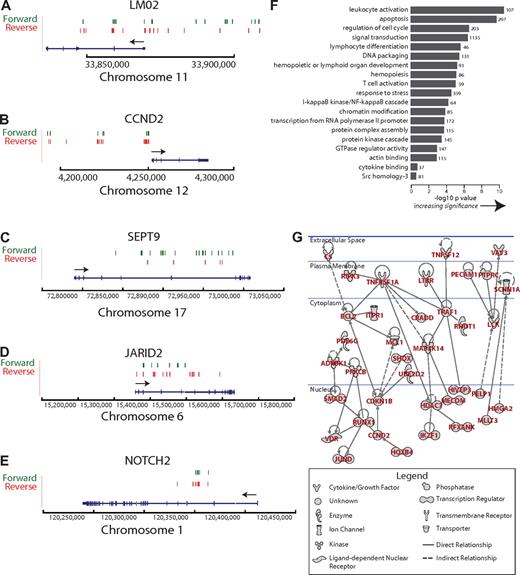
Clusters of integration sites mark genes that are either loci at which vector insertion promoted cell growth and persistence, or loci that are favored for initial integration. Previous smaller-scale studies have associated preferential gammaretroviral integration with genes associated with growth control.6,11,25,26 Integration sites associated with adverse events were near genes involved in growth control (LMO2, CCND2, and BMI1 genes3,6-8 ), and in the pooled integration site data, these genes were associated with large clusters of unique vector integration sites (LMO2, 38 sites; CCND2, 47 sites; and BMI1/SPAG6, 8 sites). Representative clusters are shown in Figure 5A to E. All clusters of integration sites can be viewed online together with user-configurable genomic annotation described in “Browsing integration sites on the human genome.”
Ontology and network analysis of genes at clustered integration sites. (A) Clustered integration sites at the LMO2 locus. The green and red lines indicate the position of vector integration sites. Forward indicates that the vector is oriented 5′ to 3′ relative to the chromosomal numbering system. Reverse indicates reverse orientation. The arrow indicates the direction of LMO2 transcription. Only selected splice variants are shown. Annotation similar in subsequent panels. (B) Clustered integration sites at the CCND2 locus. (C) Clustered integration sites at the SEPT9 locus. (D) Clustered integration sites at the JARID2 locus. (E) Clustered integration sites at the NOTCH2 locus. (F) Gene classes enriched near integration sites. The x-axis shows the statistical significance for enriched groups as the negative log of P after correction for multiple comparisons (Benjamini). The number at the end of each bar indicates the number of genes near integration sites in each category (categories were defined by the DAVID gene ontology). The raw ontology output was edited to remove uninformative high level classes or duplicative annotation. (G) A regulatory network defined by genes at clustered integration sites. The network was generated using Ingenuity, which uses published literature on interactions or affinity screens to link genes (solid line indicates direct relationships, dashed line indicates indirect relationships, arrow indicates directionality of the relationship). All networks are shown that involved more than 2 genes. No attempt was made to assess statistical significance of this network.
Ontology and network analysis of genes at clustered integration sites. (A) Clustered integration sites at the LMO2 locus. The green and red lines indicate the position of vector integration sites. Forward indicates that the vector is oriented 5′ to 3′ relative to the chromosomal numbering system. Reverse indicates reverse orientation. The arrow indicates the direction of LMO2 transcription. Only selected splice variants are shown. Annotation similar in subsequent panels. (B) Clustered integration sites at the CCND2 locus. (C) Clustered integration sites at the SEPT9 locus. (D) Clustered integration sites at the JARID2 locus. (E) Clustered integration sites at the NOTCH2 locus. (F) Gene classes enriched near integration sites. The x-axis shows the statistical significance for enriched groups as the negative log of P after correction for multiple comparisons (Benjamini). The number at the end of each bar indicates the number of genes near integration sites in each category (categories were defined by the DAVID gene ontology). The raw ontology output was edited to remove uninformative high level classes or duplicative annotation. (G) A regulatory network defined by genes at clustered integration sites. The network was generated using Ingenuity, which uses published literature on interactions or affinity screens to link genes (solid line indicates direct relationships, dashed line indicates indirect relationships, arrow indicates directionality of the relationship). All networks are shown that involved more than 2 genes. No attempt was made to assess statistical significance of this network.
We first investigated this gene set by quantifying the types of genes enriched at integration sites (pooled over all the SCID-X1 patients; Figure 5F). Strongly enriched categories included “leukocyte activation,” “lymphocyte differentiation,” and “apoptosis.” Thus either integration near some of these genes promoted cell growth and persistence or they were favored for initial integration.
We next extracted and analyzed genes near the largest clusters of integration sites, using several definitions of clustering (detailed analysis and gene lists are in supplemental Report 6). Several definitions of clustering were compared. Clustered integration sites from the SCID-X1 patients were significantly more commonly found near cancer-related genes than from the control groups (P < .001).
The genes at clustered integration sites were then analyzed using the Ingenuity pathway tool, which links genes based on interactions either from the peer-reviewed literature or by affinity-based screens. Graphs linking 2 or more genes specified candidate regulatory pathways potentially involved in progenitor cell proliferation or persistence (Figure 5G). A large network linked genes involved in phosphorylation (eg, ADRBK1, PRKCB, LCK, and MAP3K14), transcriptional control (RUNX1, JUND, and HOXB3), cell-cycle progression (CCND2 and CDKN1B), tumor necrosis factor (TNF) signaling (TNFRSF1A and TNFSF12), and negative control of apoptosis (BCL2). Thus the mapping of integration site clusters provides a detailed list of candidate pathways involved in hematopoietic progenitor cell growth and persistence.
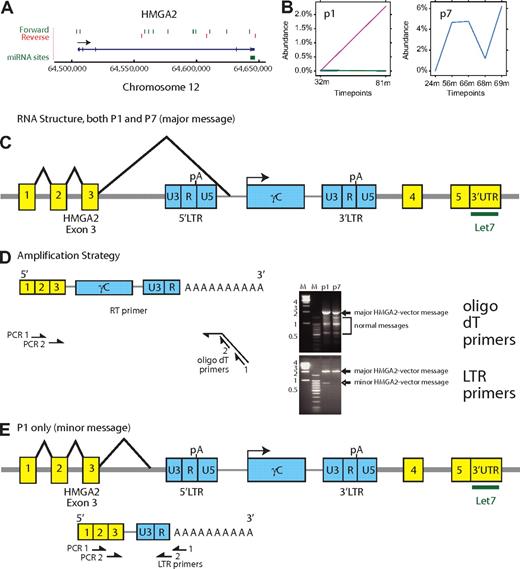
Inspection of the relationship of clustered integration sites and nearby genes showed that most clusters had distributions near gene 5′ ends, either upstream of the transcription start or in 5′ introns (eg, EVI1/MDS1 and HOXB2). If indeed these genes are subject to insertional activation, the inferred mechanism would be enhancer or promoter insertion. However, a minority of genes did not show this pattern, including HMGA2, NOTCH2, JARID2, and SEPT9 (Figures 5C-E, 6A). For these genes, integration events were clustered within the transcription units, and less common mechanisms of activation may potentially come in to play. The HMGA2 insertions are of special interest because of the association of this locus with a recently identified case of clonal skewing during stem cell–based lentiviral vector gene therapy (M.C.-C., E. Payen, O. Negre, G.P.W., K. Hehir, F. Fusil, J. Down, M. Denaro, T. Brady, R. Pawliuk, K. Westerman, R. Cavallesco, B. Gillet-Legrand, L. Caccavelli, F. Bernaudin, R. Girot, R. Dorazio, G.-J. Mulder, A. Polack, A. Bank, J. Soulier, J. Larghero, N. Kabbara, B. Dalle, B. Gourmel, G. Socié, S. Chrétien, N. Cartier, P. Aubourg, A.F., K. Cornetta, F. Galacteros, Y. Beuzard, E. Gluckman, F.D.B., S.H.-B.-A., P.L., manuscript submitted, January 27, 2010). Therefore, we studied these insertions in more detail.
Expansion of cell clones with integrated vectors in the HMGA2 third intron. (A) Map of integration sites detected in the HMGA2 locus, pooled over all the SCID-X1 patients. The green and red lines indicate the positions of vector integration sites. Forward indicates that the vector is oriented 5′ to 3′ relative to the chromosomal numbering system. Reverse indicates reverse orientation. (B) Longitudinal expansion of cell clones harboring integration events in HMGA2 in patient no.1 and patient no. 7. The x-axis shows the time after cell infusion, the y-axis shows the reconstructed percentage of all transduced cells contributed by cells harboring the HMGA2 integration site. Note the difference in the y-axis scale compared with Figure 3. (C) Structure of the major chimeric HMGA2-vector message. The major message (splice acceptor site at bp 1992 of the vector) was found in both patients no. 1 and no. 7. An alternative splice acceptor site at bp 2002 was found in patient no. 7. (D) Amplification strategy for determining the chimeric HMGA2-vector message structure using reverse-transcription PCR. The time points were 75 months (patient no. 1) and 56 months (patient no. 7). The bands marked “major” and “minor” HMGA2-vector message formed on the ethidium-stained gel were excised and subjected to Sanger DNA sequencing. Sequence analysis established that slower mobility bands corresponded exclusively to the chimeric HMGA2-vector forms. The mobility of the forms marked normal messages matched bands seen after amplification of control samples (data not shown). (E) Deduced structure of a minor form of the chimeric HMGA2-vector message found in lesser abundance in patient no. 1 only.
Expansion of cell clones with integrated vectors in the HMGA2 third intron. (A) Map of integration sites detected in the HMGA2 locus, pooled over all the SCID-X1 patients. The green and red lines indicate the positions of vector integration sites. Forward indicates that the vector is oriented 5′ to 3′ relative to the chromosomal numbering system. Reverse indicates reverse orientation. (B) Longitudinal expansion of cell clones harboring integration events in HMGA2 in patient no.1 and patient no. 7. The x-axis shows the time after cell infusion, the y-axis shows the reconstructed percentage of all transduced cells contributed by cells harboring the HMGA2 integration site. Note the difference in the y-axis scale compared with Figure 3. (C) Structure of the major chimeric HMGA2-vector message. The major message (splice acceptor site at bp 1992 of the vector) was found in both patients no. 1 and no. 7. An alternative splice acceptor site at bp 2002 was found in patient no. 7. (D) Amplification strategy for determining the chimeric HMGA2-vector message structure using reverse-transcription PCR. The time points were 75 months (patient no. 1) and 56 months (patient no. 7). The bands marked “major” and “minor” HMGA2-vector message formed on the ethidium-stained gel were excised and subjected to Sanger DNA sequencing. Sequence analysis established that slower mobility bands corresponded exclusively to the chimeric HMGA2-vector forms. The mobility of the forms marked normal messages matched bands seen after amplification of control samples (data not shown). (E) Deduced structure of a minor form of the chimeric HMGA2-vector message found in lesser abundance in patient no. 1 only.
Integration sites within the HMGA2 locus
In pooled SCID-X1 integration site data, 15 sites were found in the HMGA2 locus (Figure 6A), of which 12 are in the long third intron. Of these sites, 10 of 12 were in the sense orientation. This orientation is expected to disrupt HMGA2 mRNA synthesis because the vector splicing and polyA addition signals would be active. One hypothesis is that the HMGA2 gene may have been activated by removing 3′ untranslated region binding sites for the negatively acting microRNA let-7b, resulting in derepression of expression. This mechanism has been proposed to mediate activation via chromosomal translocations.28-31 Such a mechanism may be involved in clonal skewing observed during lentiviral vector-mediated gene correction of β-thalassemia (M.C.-C., E. Payen, O. Negre, G.P.W., K. Hehir, F. Fusil, J. Down, M. Denaro, T. Brady, R. Pawliuk, K. Westerman, R. Cavallesco, B. Gillet-Legrand, L. Caccavelli, F. Bernaudin, R. Girot, R. Dorazio, G.-J. Mulder, A. Polack, A. Bank, J. Soulier, J. Larghero, N. Kabbara, B. Dalle, B. Gourmel, G. Socié, S. Chrétien, N. Cartier, P. Aubourg, A.F., K. Cornetta, F. Galacteros, Y. Beuzard, E. Gluckman, F.D.B., S.H.-B.-A., P.L., manuscript submitted, January 27, 2010).
Analysis of the pyrosequence-based abundance data showed that in 2 of the 8 SCID-X1 patients, longitudinal expansions of cells harboring integrated vectors in HMGA2 could be detected (Figure 6B). These clones reached 2.3% of the population in patient no. 1 (127 sequence reads) and 6% of the population in patient no. 7 (629 sequence reads). Both of these vectors were integrated in the sense orientation in the HMGA2 third intron.
Because activation by 3′ truncation would require formation of a new hybrid message, we used reverse-transcription PCR to ask whether a chimeric HMGA2-vector message could be detected and if so whether it was overexpressed (Figure 6C-E). The most abundant truncated form, identified in both patient no. 1 and patient no. 7, involved splicing of the HMGA2 third intron splice donor to a site upstream of the transgene within the vector (Figure 6C-D). Analysis using an amplicon extending from the polyA tail to the HMGA2 third intron showed that the chimeric message was over-represented compared with the normal forms, even though cells containing this integration site comprised a minority of the population analyzed (Figure 6D, ethidium bromide gel for oligo dT primers). In this chimeric message, the HMGA2 reading frame was terminated after 7 additional nonsense amino acids derived from vector sequences. A minor form, seen only in patient no. 1, involved splicing into sequences upstream of the 5′ vector LTR, followed by termination at the vector polyA site at the R/U5 boundary (Figure 6E). This chimeric message encoded 5 C-terminal nonsense amino acids. Additional minor splice forms were also detected (data not shown). These data thus establish that integration with the HMGA2 locus resulted in formation of chimeric HMGA2-vector messages, which were selectively overexpressed and may have contributed to the observed expansion of cell clones.
Assessing the outcome of chemotherapy during adverse events
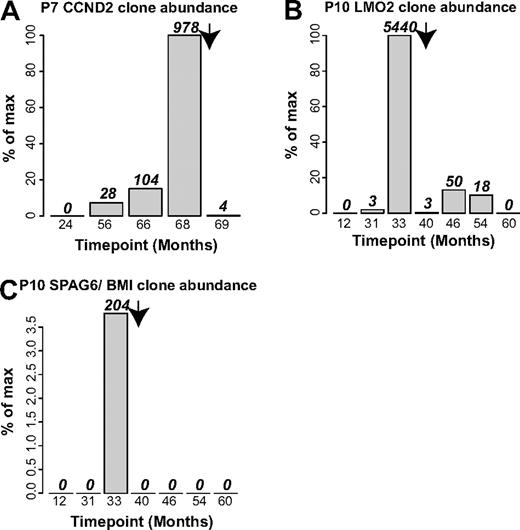
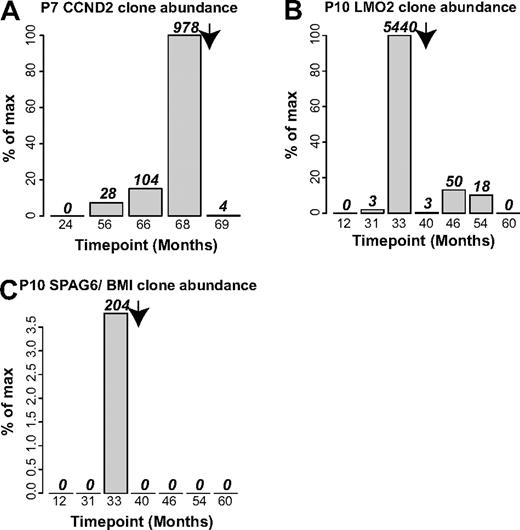
Lastly, we used integration site data to investigate the durability of chemotherapy to treat adverse events. For patients no. 7 and no. 10, longitudinal samples were available for time points after chemotherapy, allowing quantification of the abundance of cells involved in adverse events by tracking integration sites unique to these cells (Figure 7). For patient no. 7, the number of sequence reads for the CCND2 blast cell site increased to 978 at the time of the adverse event (68 months after infusion), but after chemotherapy at 69 months, only 4 CCND2 sequence reads were detected (Figure 7A). For patient no. 10, 2 integration sites were involved, LMO2 and SPAG6/BMI1, which could be analyzed independently. The LMO2 site reached 5440 sequence reads at the time of the adverse event (33 months after infusion), then trailed off in the 4 subsequent time points, so that by 60 months after infusion, no LMO2 sites were detected (Figure 7B). For the SPAG6/BMI1 sites, 204 sites were detected at the time of the adverse event, then none at subsequent time points (Figure 7C). Thus the integration site data documented a durable reversal of the clonal expansions associated with the adverse events.
Integration site sequence data document durable control of blast cell expansions by chemotherapy in patient no. 7 and patient no. 10. (A) Control of blast cells containing the CCND2 integration site in patient no. 7. (B) Control of blast cells containing the LMO2 site in patient no. 10. (C) Control of blast cells containing the SPAG6/BMI1 site in patient no. 10. Arrow indicates the time of initiation of chemotherapy.
Integration site sequence data document durable control of blast cell expansions by chemotherapy in patient no. 7 and patient no. 10. (A) Control of blast cells containing the CCND2 integration site in patient no. 7. (B) Control of blast cells containing the LMO2 site in patient no. 10. (C) Control of blast cells containing the SPAG6/BMI1 site in patient no. 10. Arrow indicates the time of initiation of chemotherapy.
Discussion
In this study, we carried out longitudinal pyrosequencing analysis of vector integration site distributions from 8 gene-corrected SCID-X1 patients. These data support the following conclusions: (1) The number of gene-corrected cells initially infused correlated with the number and diversity of integration sites subsequently detected. (2) Integration sites accumulated near sites of histone methylation and acetylation associated with gene 5′ ends and active transcription units. (3) Relatively abundant cell clones were detected in many patients, including 2 abundant clones in healthy patients that harbored integration sites near CCND2, establishing that clonal expansion associated with integration near these cancer-associated genes is not invariably associated with a rapid onset of leukemia. (4) Genes marked by clustered integration sites specified candidate genes and pathways potentially involved in regulating growth and persistence of progenitor cells. (5) The HMGA2 locus harbored a cluster of integration sites, and expansions of clones in 2 patients harboring these sites were documented. The HMGA2 messages were fused to vector sequences, and these were overexpressed compared with the normal messages. (6) Tracking of clones involved in adverse events after chemotherapy documented durable control of leukemia.
A question in analyzing the clusters of integration sites seen here and in previous studies centers on whether the clusters arose by (1) proliferation of cell clones attributable to a growth advantage conferred by insertional activation of nearby genes, or (2) favoring of particular chromosomal regions as integration targets during initial transduction of progenitor cells. In favor of the first possibility, integration sites at LMO2 and CCND2 were associated with cell proliferation during adverse events, and each of these genes were sites of large clusters of integration sites, suggesting that in nontransformed cells vector integration at these locations might have promoted cellular outgrowth. Clustering was more pronounced in patient samples than in integration sites from tissue culture cells, and clustering was increasing slowly over time, suggestive of a growth advantage, though the effect was quite modest (supplemental Report 5). Cancer-associated genes were strongly enriched near clustered integration sites (supplemental Report 6). However, in favor of the initial targeting model, some of the expanded cell clones detected in patients were not obviously involved in cell growth or persistence, consistent with the idea that these cells expanded for reasons that were not related to vector integration. Furthermore, the majority of clustered integration sites were not near known cancer-related genes. Thus for most clusters of integration sites and most expanded cell clones, it is unclear whether they arose because of favored initial integration or subsequent selection.
The longitudinal data establish that key features of the corrected cell populations were established by the first time point analyzed. This included clone number and diversity, proximity to genomic features such as sites of histone modification, integration site clustering, and expansion of some (though not all) specific cell clones. Some cell clones appeared and disappeared over time in all patients, possibly reflecting T-cell activation in response to antigen or intermittent stem cell activity, though others were abundant from the earliest time point measured. Taken together, these findings focus attention on events during the early steps of the gene-correction protocol, where further optimization may have the greatest chance of maximizing therapeutic benefit and minimize adverse events.
Although insertional activation has been associated with adverse events in 5 SCID-X1 patients, chemotherapy successfully controlled the leukemia in 4 of the patients. Here we show how pyrosequence data could be used to document continued suppression of the leukemic clone in 2 of the treated patients (patient no. 7 and patient no. 10). These patients continue to benefit from the therapeutic gene transfer.
Lastly, our data also raise questions regarding mechanisms of insertional activation. Did the integrated vectors in the HMGA2 third intron seen here and in lentivirus-based gene therapy for β-thalassemia (M.C.-C., E. Payen, O. Negre, G.P.W., K. Hehir, F. Fusil, J. Down, M. Denaro, T. Brady, R. Pawliuk, K. Westerman, R. Cavallesco, B. Gillet-Legrand, L. Caccavelli, F. Bernaudin, R. Girot, R. Dorazio, G.-J. Mulder, A. Polack, A. Bank, J. Soulier, J. Larghero, N. Kabbara, B. Dalle, B. Gourmel, G. Socié, S. Chrétien, N. Cartier, P. Aubourg, A.F., K. Cornetta, F. Galacteros, Y. Beuzard, E. Gluckman, F.D.B., S.H.-B.-A., P.L., manuscript submitted, January 27, 2010) contribute to clonal outgrowth, or were they merely adventitiously associated with expanded cell clones? Extensive studies of HMGA2 in other contexts support the idea that the part of the protein encoded by the 3 5′ exons is sufficient for transformation, and in model studies truncation in the long third intron, removing the binding sites for the let7b microRNA, are sufficient for overexpression and transformation.28-31 In a transgenic mouse model, overexpression of a truncated HMGA2 protein including the first 3 A/T hooks resulted in lymphomas.32 The HMGA2 gene has also been implicated in preventing senescence of stem cells.29 These findings are consistent with the idea that vector integration in HMGA2 during therapeutic gene correction promoted limited clonal expansion, though further studies will be needed to test this idea rigorously. Clustered integration sites within the transcription units of the cancer-associated genes NOTCH2, JARID2, and SEPT9 are also suggestive of mechanisms of activation that may differ from simply increasing the rate of transcription initiation, and would similarly be interesting topics for further study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH AI52845 and AI66290; F.D.B.). G.P.W. was supported by the NIH National Institute of Allergy and Infectious Diseases Training Grant in Infectious Diseases (T32 AI07634) and by the University of Pennsylvania School of Medicine Department of Medicine Measey Basic Science Fellowship Award.
National Institutes of Health
Authorship
Contribution: G.P.W., N.M., and F.D.B. provided sequence determination; G.P.W., C.C.B., N.M., and F.D.B. performed analysis; G.P.W., C.C.B., N.M., P.L., A.F., S.H.-B.-A., M.C.-C., and F.D.B. provided interpretation; and P.L., A.F., S.H.-B.-A., and M.C.-C. contributed samples.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for G.P.W. is Department of Medicine, University of Florida College of Medicine, Gainesville, FL.
Correspondence: F. D. Bushman, Department of Microbiology, University of Pennsylvania School of Medicine, 3610 Hamilton Walk, Philadelphia, PA 19104-6076; e-mail: bushman@mail.med.upenn.edu.