Abstract
Recently, messenger RNAs in eukaryotes have shown to associate with antisense (AS) transcript partners that are often referred to as long noncoding RNAs (lncRNAs) whose function is largely unknown. Here, we have identified a natural AS transcript for tyrosine kinase containing immunoglobulin and epidermal growth factor homology domain-1 (tie-1), tie-1AS lncRNA in zebrafish, mouse, and humans. In embryonic zebrafish, tie-1AS lncRNA transcript is expressed temporally and spatially in vivo with its native target, the tie-1 coding transcript and in additional locations (ear and brain). The tie-1AS lncRNA selectively binds tie-1 mRNA in vivo and regulates tie-1 transcript levels, resulting in specific defects in endothelial cell contact junctions in vivo and in vitro. The ratio of tie-1 versus tie-1AS lncRNA is altered in human vascular anomaly samples. These results directly implicate noncoding RNA-mediated transcriptional regulation of gene expression as a fundamental control mechanism for physiologic processes, such as vascular development.
Introduction
Over the past few years, intensive unbiased analysis of transcriptome species has revealed that eukaryotic genomes contain a variety of RNA species. RNA molecules are essentially classified into 2 types, protein coding and nonprotein coding. The protein-coding transcripts or messenger RNA (mRNA) account for only approximately 2.3% of the human genome.1 The majority of transcription appears to be nonprotein coding or noncoding, and the function of these noncoding transcripts is largely unknown.2 Of the noncoding RNAs, the regulatory short noncoding RNAs, such as microRNAs, are well studied. The long noncoding RNAs (lncRNAs), which compose the largest portion of the mammalian noncoding transcriptome, are the least understood, especially its function.3,4 lncRNAs are oriented in sense or antisense (AS) direction with respect to a protein coding locus, and located in intronic or intergenic regions.5 In humans and mice, 61% to 72% of all transcribed regions possess lncRNAs in AS orientation,2,6 and AS lncRNA transcripts play important roles in pathogenesis. For instance, the BACE1-AS transcript was elevated in subjects with Alzheimer disease and in amyloid precursor protein transgenic mice.7 A growing body of evidence suggests that lncRNAs for most critical physiologic processes will be identified. Angiogenesis, the development of new vasculature from existing vasculature, is one of the fundamental developmental physiologic processes regulated in a developing vertebrate embryo.8 Here, we identify a natural AS transcript for tyrosine kinase containing immunoglobulin and epidermal growth factor homology domain-1 (tie-1), tie-1AS lncRNA in zebrafish, mouse, and humans. tie-1 is a cell-surface tyrosine kinase receptor for angiopoietin ligands that is known to play a role in vascular development in vertebrates.9-12 In embryonic zebrafish, tie-1AS lncRNA transcript is expressed temporally and spatially in vivo with its native target, the tie-1 coding transcript, and in additional locations (ear and brain). Its expression is controlled by a 3-kb genomic fragment in the 3′ region of tie-1, and the bioinformatic predicted hybrid structure between tie-1:tie-1AS was detected in vivo. Capped or uncapped tie-1AS lncRNA selectively binds tie-1 mRNA in vivo and regulates tie-1 transcript levels, resulting in specific defects in endothelial cell contact junctions in vivo and in vitro. Further, the ratio of tie-1 versus tie-1AS lncRNA is altered in human vascular anomaly samples, suggesting that the imbalance of tie-1 regulation by tie-1AS may be important in disease. This is the first report, to our knowledge, that identifies a long AS noncoding RNA in the tie-1 locus that regulates proper vessel formation in vivo.
Methods
Zebrafish and human studies
All zebrafish studies were performed according to Medical College of Wisconsin animal protocol guidelines under protocol no. 312-06-2. Research on human patient samples for this work was performed under the Medical College of Wisconsin–approved Institutional Review Board protocols. RNA from tissue sample was isolated by Trizol method.
Reagents
The transgenic Tg(flk:EGFP) line13 was obtained from M.A.'s laboratory in Basel, Switzerland. Probes used in this study, tie-1 and tie-1AS lncRNA, were generated by T3/T7 transcription on polymerase chain reaction (PCR) products of tie-1AS. T3 and T7 sequences were included into the PCR primers. Quantitative PCR for tie-1 was performed as described previously14 using gene-specific primer pairs for tie-1, cd-31, and zebrafish actin. All primer sequences are provided in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Molecular biology: cloning of tie-1AS lncRNA
Rapid amplification of cDNA ends (RACEs) was performed by the RLM-RACE Kit (Ambion) and SMART RACE cDNA Amplification Kit (Clontech). An embryo pool spanning 1 hour postfertilization (hpf) to 3 days old was used to prepare total RNA. The RNA was treated with DNase I for 30 minutes and recovered by RNeasy kit (QIAGEN). The RACE products were cloned into pCR4-TOPO vector (Invitrogen) and sequenced. RACE primers are listed in supplemental Table 1. The sequences have been deposited in GenBank ID number GU166385.
MO and microinjections
Morpholino (MO) injections were performed as described previously.15 Gene Tools Inc designed the tie-1 splice MO: CATGTCTACTTACAGATCCAGATTG. For tie-1 MO efficacy experiments, reverse-transcribed (RT)–PCR was performed using gene-specific primers in exons immediately flanking the targeted region on 24 hpf RNA isolated from uninjected and tie-1 MO-injected (5 and 10 ng) embryos. The mMESSAGE MACHINE kit (Ambion) was used for generating sense RNA for tie-1AS lncRNA. For quantitative PCR, RNA was extracted via the Trizol method and quantitative PCR was carried out using DyNAmo HS SYBR Green qPCR Kit (New England Biolabs). For RNA Pol inhibitor studies, 8 hpf zebrafish embryos were injected with α-amanitin and allowed to develop until 24 hpf. For tagetin, injections were performed at 1-cell stage. For tie-1AS lncRNA promoter cloning, the region 152648 to 149799 corresponding to clone CH211-286N19 in linkage group 7 (accession no. CR936976.15) was amplified using forward primer, TAAGGTACCGCGGGGACATCTAAAGACAC; and reverse primer, ATTGGATCCCTTGGTGGCCCAGAAAAC. The 3-kb PCR product was purified, digested with KpnI-BamHI, and cloned into the KpnI-BglII site of pGL3-Basic (Promega) vector and is referred to as pGL3-zfNcRNA-3kb construct. For microinjection, pGL3-hRobo4-3kb promoter construct16 and pGL3 vector were linearized with SalI, and pGL3-zfNcRNA-3kb promoter construct was linearized with BamHI; 100 pg of linearized DNA was injected into the cytoplasm of 1-cell stage zebrafish embryo, embryos were lysed at 26 hpf, and firefly luciferase activity was measured as per the instruction manual.
Endothelial cell culture and electroporation
Human umbilical vascular endothelial cells (HUVECs; passage 3-6) were cultured in M199 medium with 20% fetal bovine serum (HyClone) and endothelial cell growth supplement (BD Biosciences). Plasmid DNA transfection was performed using the MicroPorator from NanoEnTek. Briefly, 1 μg of plasmid DNA was mixed with 105 cells in 10 μL of resuspension buffer, and electroporation was performed at 1000 V and 30 ms. After electroporation, cells were seeded in regular HUVEC growth medium with transfection efficiency reaching 80%. The expression of Tie-1, Hsp90, and actin was examined by Western blot analysis at 72 hours after electroporation. The 3-dimensional tube formation assay was performed at 48 hours, and tube formation was observed at 72 hours after electroporation.
Results
Identification of tie-1AS in multiple species
Using the zebrafish information network database17,18 of in situ expression profile across embryonic developmental stages, we identified several novel expressed sequence tags (ESTs) that showed a vascular-specific expression profile. We focused on one EST (accession no. DU644726) that mapped to 73 bp downstream of the tie-1 coding locus. Because the 3′-untranslated region (UTR) of the zebrafish tie-1 gene was incomplete, we verified whether this EST represented the 3′-UTR of the tie-1 coding transcript or represented a new transcript. A set of primers derived from the EST was used to perform rapid amplification of cDNA ends (RACE) reaction (supplemental Figure 1A arrows). Interestingly, both the 5′ and 3′ RACE reactions in both directions generated end-extended products. The 3′ RACE reaction provided a product that extends the 3′UTR of the tie-1 gene to the polyA tail, whereas the 5′ RACE reaction generated a product in the opposite direction of tie-1 coding transcript, thus providing evidence for a novel transcript arising from the tie-1 genomic locus in the AS direction. The noncoding and novel transcripts associated with tie-1 have 456 bp of overlapped sequence with opposite transcriptional orientation. The 5′RACE transcript called “tie-1AS” was detected in the presence of RT from RNA isolated from an embryo pool of multiple stages (1-72 hpf; supplemental Figure 1B, +RT lane) and is 804 bp long. Sequencing of the tie-1AS revealed a 148-bp unique 5′ sequence, followed by a 456-bp region that is complementary to tie-1s' last exon, and a 200-bp 3′ region complementary to tie-1s' last intron-exon junction (supplemental Figure 1C). The single largest open reading frame predicted within this AS transcript, is 93 amino acids (data not shown). We performed additional RACE reactions and cDNA walking in the opposite orientation to multiple exons within the tie-1 locus and found that indeed multiple AS transcripts or long noncoding RNA are generated from the tie-1 locus (supplemental Figure 1D blue boxes). These data indicate that the tie-1AS represented by EST DU644726 is one of several noncoding RNAs transcribed from the tie-1 locus. We only focused on the extreme 3′-UTR tie-1AS for further studies and from hereon will be referring to it as tie-1AS lncRNA. Both mouse (supplemental Figure 1E) and human (supplemental Figure 1F blue boxes) RNA indeed have similar tie-1AS lncRNA as zebrafish and the nucleotide conservation across the 3 species is 28.5%. These results suggest that the tie-1 locus has evolutionarily conserved noncoding RNAs that may regulate tie-1 transcripts by similar mechanisms.
Expression profile of tie-1AS lncRNA and tie-1
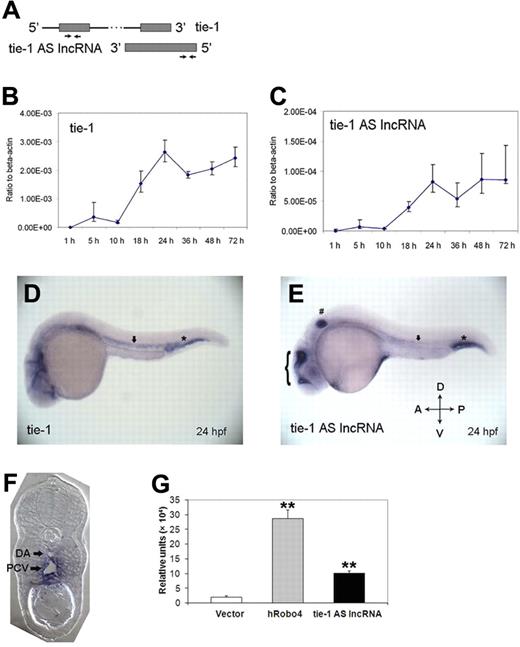
To investigate developmental expression pattern of tie-1AS lncRNA and tie-1, we performed quantitative real-time PCR for both transcripts using sequence-specific primers (Figure 1A arrows) that will amplify each transcript independently across multiple stages in embryonic zebrafish development. Starting at 1 hpf and until 10 hpf, the levels of both transcripts are low. After gastrulation, both transcripts show remarkable concordance at every stage until 72 hpf (compare Figure 1B tie-1 and Figure 1C tie-1AS lncRNA). The levels of tie-1 transcript are a log higher at each stage (compare ordinate axis in Figure 1B-C) compared with tie-1AS lncRNA, suggesting that the low amount of tie-1AS lncRNA might be sufficient to regulate endogenous tie-1 coding transcript levels.
Expression patterns of tie-1AS lncRNA. (A) The primer pair locations for quantitative PCR. Developmental expression profile of tie-1 (B) and tie-1AS (C) is shown. The total RNAs were extracted using Trizol followed by DNase I treatment. The RNAs were recovered by RNeasy kit (QIAGEN). (D) Tie-1 is expressed in the axial vessels ( ) and blood islands (*). (E) Tie-1AS lncRNA is expressed in the axial vessels (
) and blood islands (*). (E) Tie-1AS lncRNA is expressed in the axial vessels ( ), intermediate cell mass (*), otic vesicles (#), and brain (brace). (F) Transverse section of 24 hpf tie-1AS lncRNA embryo is shown.
), intermediate cell mass (*), otic vesicles (#), and brain (brace). (F) Transverse section of 24 hpf tie-1AS lncRNA embryo is shown.  indicates the location of DA and PCV. (G) Luciferase activity in vivo (26 hpf) for a 3-kb fragment upstream of zebrafish tie-1AS lncRNA. The hRobo4 promoter is used as a positive control. **P < .01.
indicates the location of DA and PCV. (G) Luciferase activity in vivo (26 hpf) for a 3-kb fragment upstream of zebrafish tie-1AS lncRNA. The hRobo4 promoter is used as a positive control. **P < .01.
Expression patterns of tie-1AS lncRNA. (A) The primer pair locations for quantitative PCR. Developmental expression profile of tie-1 (B) and tie-1AS (C) is shown. The total RNAs were extracted using Trizol followed by DNase I treatment. The RNAs were recovered by RNeasy kit (QIAGEN). (D) Tie-1 is expressed in the axial vessels ( ) and blood islands (*). (E) Tie-1AS lncRNA is expressed in the axial vessels (
) and blood islands (*). (E) Tie-1AS lncRNA is expressed in the axial vessels ( ), intermediate cell mass (*), otic vesicles (#), and brain (brace). (F) Transverse section of 24 hpf tie-1AS lncRNA embryo is shown.
), intermediate cell mass (*), otic vesicles (#), and brain (brace). (F) Transverse section of 24 hpf tie-1AS lncRNA embryo is shown.  indicates the location of DA and PCV. (G) Luciferase activity in vivo (26 hpf) for a 3-kb fragment upstream of zebrafish tie-1AS lncRNA. The hRobo4 promoter is used as a positive control. **P < .01.
indicates the location of DA and PCV. (G) Luciferase activity in vivo (26 hpf) for a 3-kb fragment upstream of zebrafish tie-1AS lncRNA. The hRobo4 promoter is used as a positive control. **P < .01.
Quantitative PCR analysis shows that the transcript levels of tie-1AS lncRNA and tie-1 are remarkably congruent in development but do not indicate the spatial location of the tie-1AS lncRNA transcript in relation to the tie-1 gene. To determine the expression pattern of tie-1AS lncRNA transcript, we performed in situ hybridization (ISH) for tie-1AS lncRNA using a digoxigenin-labeled tie-1AS lncRNA (or sense of tie-1) at 24 hpf and compared with tie-1 expression pattern previously demonstrated at the same stage.19 Tie-1 is expressed in the axial vessels as indicated by arrows in Figure 1D and in intermediate cell mass (Figure 1D *). Previous reports indicate tie-1 expression in intersomitic vessels (ISVs),19 which we have noticed in longer dark-stained embryos. Tie-1AS lncRNA is also expressed in the axial vessels, albeit weakly (Figure 1E arrow), but shows strong expression in the intermediate cell mass (Figure 1E *), otic vesicles (Figure 1E #), and brain (Figure 1E brace). We have been unable to detect strong tie-1AS lncRNA expression in ISVs, but it is worth mentioning that the axial vessel ISH results are in agreement with quantitative PCR results that showed lower amounts of tie-1AS lncRNA expression compared with tie-1 expression at any given stage, especially at 24 hpf. We have also performed ISH for tie-1AS lncRNA at other stages but did not notice any dramatic differences in spatial localization than at 24 hpf shown in Figure 1F. To fine-map the expression regions of tie-1AS lncRNA, we sectioned 24 hpf (Figure 1E) whole-mount stained embryos and mounted them as shown in Figure 1F. Tie-1AS lncRNA is expressed in both dorsal aorta (DA) and posterior cardinal vein (PCV) and at the junction of both structures (Figure 1F black arrows). The quantitative PCR and the ISH results together show that tie-1AS lncRNA and tie-1 transcripts are found at the same location and time in embryonic development, arguing for a regulatory role of the tie-1AS lncRNA on the tie-1 gene.
To investigate control of tie-1AS lncRNA transcription, we cloned a 3-kb genomic fragment downstream of AS to the tie-1 coding locus into a pGL-3 basic vector that contains luciferase reporter gene. This vector is specifically designed to determine promoter activity of DNA fragments. We also obtained a human Roundabout4 (Robo4) promoter fragment cloned in the same vector that was shown previously to be active in endothelial cells as a positive control.16 We injected wild-type zebrafish embryos with the empty pGL-3 vector, pRobo4promoter-luc, and ptie-1ASlncRNApromoter-luc at 1-cell stage. Injected embryos were assayed at 26 hpf when expression of Robo415 and tie-1AS lncRNA (Figure 1C) is high in the vasculature. Lysates from 26 hpf embryos were checked for luciferase activity. Compared with vector alone, the ptie-1ASlncRNApromoter-luc construct showed a 5-fold increase (Figure 1G black bar), whereas the pRobo4promoter-luc construct showed a 14-fold increase in luciferase activity (Figure 1G gray bar). This result suggests that a 3-kb DNA fragment downstream of tie-1 gene is sufficient to drive transcription of tie-1AS lncRNA in vivo.
Hybrid tie-1AS lncRNA:tie-1 RNA in vivo
We made use of the utilities RNAfold and RNAcofold from the Vienna RNA package20,21 to determine whether hybridization between tie-1AS lncRNA and tie-1 RNA was likely to occur. In our initial analysis, we set the temperature parameter to 28°C (representative of in vivo temperatures for zebrafish) and examined predicted RNA secondary structures and fold energies for tie-1AS lncRNA and tie-1 RNA singularly as well as in a duplex. Predicted secondary structures for the tie-1 RNA alone and in duplex with tie-1AS lncRNA are depicted in supplemental Figure 2A (the secondary structure for tie-1AS lncRNA is omitted). The hybridization energies for tie-1, tie-1AS lncRNA, and their duplex are −1418.83, −284.66, and −1885.96, respectively, implying that the tie-1 and tie-1AS lncRNA are anticipated to duplex under in vivo conditions.
To determine the specificity of this prediction, 2 additional studies were performed. The first examined whether tie-1AS lncRNA could be anticipated to have specific correspondence to tie-1 RNA, as opposed to other potential targets. We computed the ratio of the energy of the tie-1AS lncRNA/tie-1 mRNA structure and the sum of the individual energies of the tie-1AS lncRNA and tie-1 mRNA (1.107), and compared it with the analogous ratios for the tie-1AS lncRNA and each mRNA identified in the NCBI zebrafish reference sequence.22 The distribution of energy ratios is depicted in supplemental Figure 2B with a marker representing that of tie-1AS lncRNA and tie-1 mRNA. The ratio of tie-1AS lncRNA and tie-1 mRNA duplex to singular energies is greater than 99.97% of that of all other zebrafish mRNAs, implying that, compared with other zebrafish mRNAs, tie-1 mRNA represents a strong target for tie-1AS lncRNA.
Next, we studied whether the coding tie-1 mRNA could be anticipated to have other targeting interfering RNA with similar characteristics in terms of transcript length and base composition as tie-1AS lncRNA by computing a distribution of duplex energies between tie-1 mRNA and permuted tie-1AS lncRNA sequences, and then comparing the actual energy of the tie-1AS lncRNA and tie-1 mRNA with those observed (supplemental Figure 2C). The energy of the tie-1AS lncRNA and tie-1 mRNA duplex is substantially lower than that of any of the permuted tie-1AS lncRNAs, implying that, compared with other similar RNA molecules, tie-1AS lncRNA can be anticipated to particularly target tie-1 mRNA.
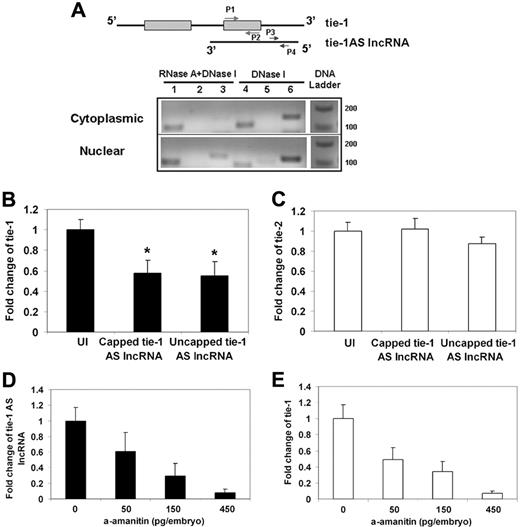
To confirm the bioinformatic prediction, we performed ribonuclease protection assay and RT-PCR to determine whether the tie-1AS lncRNA exists in vivo as a RNA-RNA duplex with tie-1 RNA (Figure 2A). RNA was isolated from 24 hpf wild-type zebrafish embryos, and the cytoplasmic (Figure 2A top panel) or nuclear (Figure 2A bottom panel) RNA fractions were isolated as described in supplemental Methods. Total RNA preparations were then digested with RNase A, which digests single-stranded RNA and not duplex RNA, plus DNaseI (Figure 2A lanes 1-3) and DNaseI alone (Figure 2A lanes 4-6). Using RT-PCR and primers specific to the overlapping region of tie-1 RNA (Figure 2A P1-P2), we detected a protected RNA fragment in both cytoplasmic and nuclear fractions of RNA, indicating that tie-1:tie-1AS lncRNA RNA-RNA duplex exists in vivo (Figure 2A lane 1). We did not observe a detectable signal (Figure 2A lane 2) using a second primer pair (Figure 2A P3-P4), specific to the nonoverlapping single-stranded region of tie-1AS lncRNA, although this sequence was amplified in nuclear DNase-treated extracts (Figure 2A lane 5 bottom panel), suggesting that indeed the lack of band in lane 2 and the presence of band in lane 1 is arising from a duplex species of tie-1 RNA and tie-1AS lncRNA. We have sequenced the amplicon in lane 1 and lane 4 and confirmed its identity as tie-1. To confirm complete digestion by RNase A, we checked β-actin mRNA, which was protected in nuclear fraction (Figure 2A lane 3 bottom gel) but not in cytoplasmic (Figure 2A lane 3 top gel), fraction. Assuming partial cleavage of RNA in nuclear fraction based on β-actin result and the complete lack of protected band for tie-1:tie-1AS lncRNA in nuclear fraction, an argument could be made that the protected tie-1:tie-1AS lncRNA hybrid species exists exclusively in the cytoplasmic fraction of embryo lysates. This result is consistent with regulation of endogenous tie-1 sense transcript by tie-1AS lncRNA at the posttranscriptional level.
Functional investigation of the tie-1AS lncRNA. (A) The RNA duplex was examined by ribonuclease protection assay. The zebrafish embryos at 24 hpf were digested with 0.25% trypsin into single cells, from which cytoplasmic and nuclear RNA was extracted. The RNAs were treated with DNase I and RNase A followed by RT-PCR. Primers used are as follows: lanes 1 and 4, P1 and P2, amplicon size 114 bp; lanes 2 and 5, P3 and P4, amplicon size 133 bp; and lanes 3 and 6, β-actin, amplicon size 147 bp. (B) Overexpression of tie-1AS lncRNA down-regulates the expression of tie-1. *P < .05. (C) No effect on the expression of tie-2. The zebrafish embryos were injected at the 1-cell stage with 150 pg of tie-1AS mRNA, and quantitative PCR was performed at 24 hpf. (D) Chemical treatment of zebrafish embryos with α-amanitin shows dose-dependent sensitivity to tie-1AS lncRNA and tie-1 (E) transcription.
Functional investigation of the tie-1AS lncRNA. (A) The RNA duplex was examined by ribonuclease protection assay. The zebrafish embryos at 24 hpf were digested with 0.25% trypsin into single cells, from which cytoplasmic and nuclear RNA was extracted. The RNAs were treated with DNase I and RNase A followed by RT-PCR. Primers used are as follows: lanes 1 and 4, P1 and P2, amplicon size 114 bp; lanes 2 and 5, P3 and P4, amplicon size 133 bp; and lanes 3 and 6, β-actin, amplicon size 147 bp. (B) Overexpression of tie-1AS lncRNA down-regulates the expression of tie-1. *P < .05. (C) No effect on the expression of tie-2. The zebrafish embryos were injected at the 1-cell stage with 150 pg of tie-1AS mRNA, and quantitative PCR was performed at 24 hpf. (D) Chemical treatment of zebrafish embryos with α-amanitin shows dose-dependent sensitivity to tie-1AS lncRNA and tie-1 (E) transcription.
Regulation of tie-1 by tie-1AS lncRNA
To investigate whether tie-1:tie-1AS lncRNA hybrid has a functional role in regulating tie-1 transcript levels in vivo, we injected the sense RNA of tie-1AS lncRNA in 1-cell zebrafish embryos and allowed embryos to develop to 24 hpf, at which point we harvested the RNA and performed quantitative PCR for tie-1 (Figure 2B) and tie-2 transcript levels (Figure 2C). We injected capped or uncapped sense RNA (100 pg) for tie-1AS lncRNA to determine whether tie-1 levels were modulated in these embryos. Interestingly, injection of either capped and uncapped tie-1AS lncRNA sense RNA reduced tie-1 expression at 24 hpf (Figure 2B, *P < .05) without affecting tie-2 transcript levels (Figure 2C), indicating that the hybridization of the tie-1AS lncRNA to tie-1 RNA in vivo results in down-regulation of the endogenous tie-1 transcript. Further, this effect is independent of the RNA capping process because both capped and uncapped sense RNA showed the same effect arguing for RNA based hybridization mediated mechanistic regulation of tie-1 transcript levels.
To investigate the RNA polymerase responsible for transcribing tie-1AS lncRNA in zebrafish, we performed the classic Pol II inhibitor (α-amanitin) treatment of zebrafish embryos and checked for transcript levels of tie-1 (Figure 2E) and tie-1AS lncRNA (Figure 2D) at 24 hpf. We observed a dose-dependent decrease in both tie-1 and tie-1AS lncRNA transcripts in α-amanitin–treated embryos starting at 50 pg, with robust inhibition observed at 450 pg. We also performed Pol III inhibitor (tagetin) treatment and did not observe any change in tie-1 or tie-1AS lncRNA transcript levels. These results indicate that RNA polymerase II is responsible for transcribing tie-1AS lncRNA in zebrafish embryo, which is in concordance with a recent report that suggests that the majority of noncoding RNA are transcribed by Pol II.23
Endothelial junction defects in tie-1AS GOF
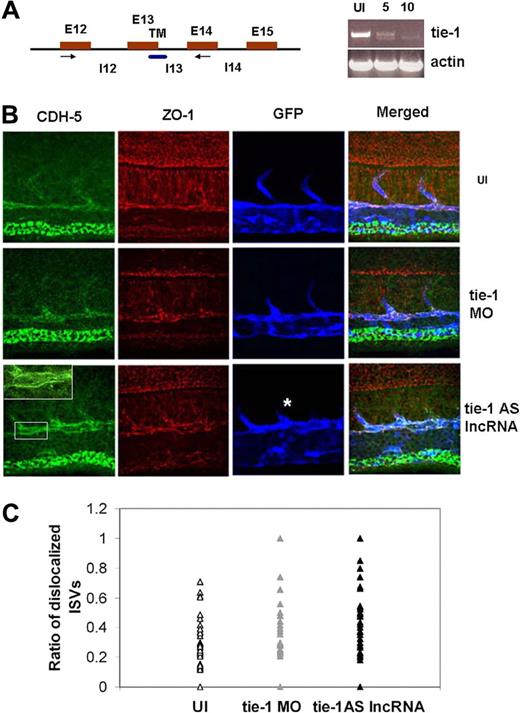
If tie-1 gene levels were reduced in embryos injected with tie-1AS lncRNA-injected embryos, we would expect to observe phenotype in injected embryos similar to conventional loss-of-function experiments. As a positive control for tie-1 knockdown, we chose the MO-mediated knockdown, which is routinely used in zebrafish research to determine the function of a given gene in development.24 We designed splice MO that targets the transmembrane region of the Tie-1 receptor kinase (Figure 3A blue box). To determine MO efficacy, we performed RT-PCR using specific primers in the exons spanning across the targeted exons of Tie-1 TM region. Compared with the uninjected sample (Figure 3A lane UI), embryos injected with 5 ng of the tie-1 MO (Figure 3A lane 5) show a second smaller transcript in addition to the native transcript. Indeed, increasing concentrations of tie-1 splice MOs (10 ng) resulted in knockdown of endogenous tie-1 transcript and replacement by an alternative transcript (Figure 3A, compare lanes 5 and 10), indicating that the MO affects endogenous tie-1 transcripts in a dose-dependent manner.
The phenotype of zebrafish tie-1AS lncRNA overexpression. (A) The MO targeting splice-site is complementary to the 13th exon-intron boundary. RT-PCR was performed to confirm MO-targeting effects. (B) Immunostaining of Tg(flk: EGFP) embryos injected with tie-1AS (150 pg) or tie-1 MO (5-10 ng) at the 1-cell stage and fixed at 24 hpf. Staining was performed using CDH5, ZO-1, and anti-GFP antibodies. Most tie-1AS lncRNA-injected embryos showed an asymmetric distribution of Cdh-5 staining on endothelial cell membrane in vivo as shown in the enlarged inset. ISVs (white asterisk) also show a truncated phenotype. Details of image capture are available in supplemental Methods. (C) Quantitation of the phenotype was performed as described in supplemental Methods, and the ratio of length of ISVs showing phenotype to an ISV with normal CDH-5 distribution is plotted. In general, the trend in tie-1AS lncRNA and tie-1 MO-injected embryos shows higher ratios indicating more ISVs with asymmetric CDH-5 distribution per embryo.
The phenotype of zebrafish tie-1AS lncRNA overexpression. (A) The MO targeting splice-site is complementary to the 13th exon-intron boundary. RT-PCR was performed to confirm MO-targeting effects. (B) Immunostaining of Tg(flk: EGFP) embryos injected with tie-1AS (150 pg) or tie-1 MO (5-10 ng) at the 1-cell stage and fixed at 24 hpf. Staining was performed using CDH5, ZO-1, and anti-GFP antibodies. Most tie-1AS lncRNA-injected embryos showed an asymmetric distribution of Cdh-5 staining on endothelial cell membrane in vivo as shown in the enlarged inset. ISVs (white asterisk) also show a truncated phenotype. Details of image capture are available in supplemental Methods. (C) Quantitation of the phenotype was performed as described in supplemental Methods, and the ratio of length of ISVs showing phenotype to an ISV with normal CDH-5 distribution is plotted. In general, the trend in tie-1AS lncRNA and tie-1 MO-injected embryos shows higher ratios indicating more ISVs with asymmetric CDH-5 distribution per embryo.
To investigate the possible phenotype of tie-1 LOF, we followed the well-established function of Tie-1 in maintaining the integrity of the mammalian vasculature.12 Tie-1–deficient mouse embryos show no perturbation of embryonic angiogenesis but loss of vessel integrity.9,10 We injected tie-1 MO or tie-1AS lncRNA into transgenic embryos where the vascular endothelial growth factor receptor-2 or flk promoter drives the expression of enhanced green fluorescent protein Tg(flk: EGFP)13 in the vasculature and investigated vessel integrity at 24 hpf. Injected embryos were immunostained for tight junction and adherens junction proteins using antibodies to zona occludens (ZO-1; Figure 3B ZO-1 panels) and Ve-cadherin (CDH-5; Figure 3B CDH-5 panels), respectively. The ZO-1 protein25 is present in all cell junctions, whereas the CDH-5 protein26 is localized exclusively in endothelial cell junctions. Both proteins have been shown previously in zebrafish to be in similar location13 as in mouse and humans.
We immunostained tie-1AS lncRNA injected, tie-1 MO injected, and uninjected embryos for ZO-1 and CDH-5 proteins at 24 hpf. Uninjected embryos (Figure 3B UI) show ISVs that were properly patterned with well-organized contact with endothelial cell of the DA and cardinal vein (CV) for both ZO-1 (Figure 3B UI, ZO-1) and CDH-5 (Figure 3B UI, CDH-5). In contrast, both the tie-1AS lncRNA-injected and the tie-1 MO-injected embryos showed aberrant ZO-1 staining (compare respective panels for tie-1 MO and tie-1AS lncRNA in Figure 3B). The ZO-1 staining in both tie-1 MO and tie-1AS lncRNA showed asymmetric distribution through the cell surface, whereas in uninjected embryos the ZO-1 staining was restricted to cell contact junctions. In terms of vascular-specific junctions, CDH-5 staining clearly shows asymmetric or uniform distribution of CDH-5 protein on all sides of the cell in tie-1AS lncRNA sample (Figure 3B), compared with the uninjected panel. The ISV structure in both tie-1AS lncRNA and the tie-1 MO-injected embryos appears thinner, and the ISV length from DA to dorsal surface is shorter. Quantification of the percentage CDH-5 mislocalization along the length of a single ISV in each sample group is shown in Figure 3C. In general, more ISV cells show CDH-5 mislocalization along the ISV length in tie-1 MO and tie-1AS lncRNA samples.
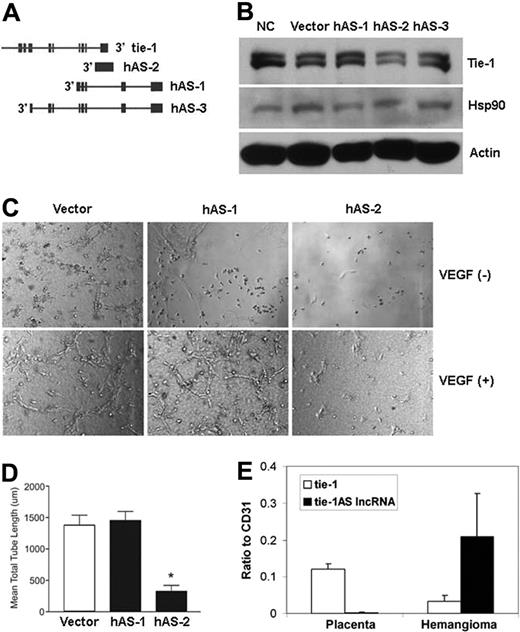
To investigate the effects of human tie-1AS lncRNA on endothelial tube formation in vitro, we electroporated HUVECs with AS transcripts encoding 3 different regions in the 3′-UTR of the human tie-1 gene locus as indicated in Figure 4A. The 3 human AS (hAS) transcripts are called hAS-1, hAS-2, and hAS-3. The hAS-1 and hAS-3 encodes identical regions with the hAS-3–encompassing region further upstream of hAS-1 (Figure 4A). The hAS-2 spans the last exon of the tie-1 gene and covers the last intron-exon junction. This AS contains both intron and exon sequences of tie-1 in the reverse orientation, which is similar to the zebrafish AS used in all the experiments here. First, we electroporated HUVECs with the 3 hASs and generated lysates to detect Tie-1 protein levels via Western blot. Among the different sample groups, only hAS-2–transfected lysate (Figure 4B lane hAS-2) showed appreciable difference in Tie-1 protein levels compared with hAS-1 (Figure 4B lane hAS-1), hAS-3 (Figure 4B lane hAS-3), untransfected (Figure 4B lane NC), or empty vector transfected (Figure 4B lane vector) cells. No appreciable differences were noticed for housekeeping proteins Hsp90 or actin (Figure 4B Hsp90/actin). This result indicates that hAS-2 like zebrafish tie-1AS lncRNA selectively targets tie-1 transcript, resulting in down-regulation of the Tie-1 protein.
Human lncRNA regulates Tie-1 expression in endothelial cells and is necessary for VEGF-induced tube formation and the disease connection. (A) A schematic of the 3 human lncRNA AS (h-AS) tie-1 transcripts is shown. (B) hAS-2 transfection in endothelial cells down-regulates Tie-1 protein levels compared with no-treatment control (NC), vector control (Vector), or hAS-1 or hAS-3 transfected lysates. (C) After transfection with empty vector and tie-1 noncoding RNAs h-AS1 or h-AS2, HUVECs were suspended in type I collagen gels and treated with vehicle or 100 ng/mL VEGF. After 24 hours of incubation, cells were photographed using QCapture Program (QImaging Inc). Representative images are shown for 2 independent experiments. Details of image capture are available in supplemental Methods. (D) Quantification of mean lengths of tubes formed with VEGF-treated transfected HUVECs. *P < .05. (E) Tissues from patients diagnosed with vascular anomalies have aberrant levels of tie-1AS lncRNA transcript compared with tie-1. Interestingly, RNA from normal placenta tissue shows an exact reverse pattern with high levels of tie-1 and low to nondetectable levels of tie-1AS lncRNA.
Human lncRNA regulates Tie-1 expression in endothelial cells and is necessary for VEGF-induced tube formation and the disease connection. (A) A schematic of the 3 human lncRNA AS (h-AS) tie-1 transcripts is shown. (B) hAS-2 transfection in endothelial cells down-regulates Tie-1 protein levels compared with no-treatment control (NC), vector control (Vector), or hAS-1 or hAS-3 transfected lysates. (C) After transfection with empty vector and tie-1 noncoding RNAs h-AS1 or h-AS2, HUVECs were suspended in type I collagen gels and treated with vehicle or 100 ng/mL VEGF. After 24 hours of incubation, cells were photographed using QCapture Program (QImaging Inc). Representative images are shown for 2 independent experiments. Details of image capture are available in supplemental Methods. (D) Quantification of mean lengths of tubes formed with VEGF-treated transfected HUVECs. *P < .05. (E) Tissues from patients diagnosed with vascular anomalies have aberrant levels of tie-1AS lncRNA transcript compared with tie-1. Interestingly, RNA from normal placenta tissue shows an exact reverse pattern with high levels of tie-1 and low to nondetectable levels of tie-1AS lncRNA.
We next performed an in vitro tubulogenesis assay in collagen gel in the presence and absence of vascular endothelial growth factor (VEGF). In both empty vector (Figure 4C vector) and hAS-1 (Figure 4C hAS-1) transfected endothelial cell panels, we notice tube formation in the presence of VEGF. However, in hAS-2–transfected endothelial cell (Figure 4C hAS-2), tube formation is absent in the presence of VEGF, indicating that down-regulation of Tie-1 protein results in disruption of endothelial tube formation in vitro. This result is in concordance with previous reports of Tie-1 function in modulating endothelial cell junctions and in turn stability of endothelial network.27,28 Quantitation of the VEGF-induced tube formation (Figure 4D) shows a statistically significant difference (*P < .05) between hAS-2 and vector or hAS-1–transfected cells.
Tie-1AS lncRNA disease connection
Mutations in Tie's have been previously implicated in patients with vascular malformations.29-33 To investigate whether the human tie-1AS lncRNA levels are altered in vascular malformation, we performed quantitative PCR for tie-1 and tie-1AS lncRNA on RNA isolated from vascular anomaly34 tissue from 12 patients previously diagnosed for various anomalies and compared them with RNA from normal placenta. In general, placenta share several markers with hemangioma tissue35 and therefore were used for comparisons. The quantitative PCR comparison was performed to the CD31 endothelial marker. We noticed that the tie-1AS lncRNA levels show a 5- to 10-fold increase (Figure 4E black bar, tie-1AS lncRNA) in vascular anomaly patient samples compared with normal placenta. Interestingly, normalized tie-1 levels (Figure 4E white bar, tie-1 panel) have not dramatically changed among the sample groups. Importantly, the ratio of tie-1 versus tie-1AS lncRNA is opposite in normal placenta tissue compared with vascular anomaly tissue. These results suggest that the balance between tie-1AS lncRNA and tie-1 is probably critical for regulation of tie-1 levels, and diseases where vasculature is implicated may observe a shift in this regulation, arguing for a specific function for tie-1AS lncRNA in vascular conditions.
Discussion
A growing body of evidence suggests that mammalian genomes encode many thousands of large intergenic transcripts. To date, the functional significance of these transcripts is poorly understood. In this study, we have identified a noncoding AS RNA in the 3′-UTR of the tie-1 gene locus that is expressed in the AS direction to the tie-1 gene. The salient features of this study include (1) a vascular-specific noncoding RNA that is expressed in both previously known and unknown regions of native tie-1 transcript expression in vivo, (2) selective targeting of tie-1 transcript by direct interaction of the noncoding RNA with its cognate RNA in vivo that results in down-regulation of the tie-1 transcript, (3) phenotypic consequences in vivo and in vitro of the noncoding RNA mediated down-regulation of tie-1 transcripts, and (4) potential imbalance in tie-1/noncoding tie-1AS ratios in vascular-related disease states.
The noncoding tie-1AS lncRNA is expressed in the temporal and, for the most part, spatial location that matches the endogenous tie-1 gene expression pattern. Interestingly, regions in the brain, such as the forebrain and hindbrain, and otic vesicles show strong expression of the noncoding species exclusively. This argues for a tie-1 gene role previously unrecognized in the brain and ear function, perhaps in the vasculature of these organs. Indeed, 2 recent studies implicate angiopoietins, ligands for Tie's, in lymphatic vessel development in ear.36,37 Whether the ear expression observed in the fish translates into a role for tie-1 in inner ear development or vasculature associated with ear development in mouse remains to be seen. Because the noncoding RNA for tie-1 is conserved in humans and mice, this paradigm of using noncoding RNA expression pattern to speculate putative novel gene function (simply based on expression alone) of well-studied genes is intriguing.
The presence of noncoding AS transcript at the same time and space as its native endogenous transcript suggests a regulatory paradigm; and based on our comprehensive zebrafish genome searches, it is clear that the energy of the tie-1AS lncRNA:tie-1 RNA complex is substantially lower than what is typically observed, which argues for selective duplex formation with one another. Indeed, experimental data confirm this prediction in vivo. Hybridization of the AS long noncoding RNA to its target is predicted to destabilize it and in turn result in loss of protein.4 This is probably the functional significance with the noncoding tie-1AS lncRNA both in vivo and in vitro. Both uncapped and capped noncoding tie-1AS lncRNA targets tie-1, suggesting that (1) the hybridization mediated down-regulation mechanism of target gene is independent of RNA capping and (2) this down-regulation is probably an RNA-mediated mechanism. However, definitive evidence is needed before reaching this conclusion.
Down-regulation of gene expression often leads to loss of function phenotypes, especially for haploinsufficient genes. Tie-1 loss of function is established in the late angiogenesis phase38 and is required cell autonomously for endothelial cell survival and vascular network extension during late embryogenesis.9 In tie-1AS lncRNA-injected embryos, we have similarly noticed that endothelial cell junctions typified by Ve-cadherin are mislocalized, therefore arguing for loss or aberrant cell contacts. This phenotype also corresponds well to the expression of tie-1AS at the junction between PCV and DA as shown by ISH. Importantly, the human tie-1AS lncRNA also prevents tube formation of endothelial cells in response to VEGF stimulus, arguing that the aberrant cell contact observed in vivo results in loss of cell-cell adhesion necessary for tube formation in vitro. Both humans and zebrafish share the same 3′-UTR tie-1AS lncRNA region that targets the tie-1 gene, which spans the exon-intron junction of the last tie-1 exon in the AS direction and in essence behaves mechanistically as a splice MO. Therefore, this argues that noncoding AS RNA-mediated down-regulation of target gene may mimic other AS technology.
At present, the function of noncoding RNA in disease is still unclear4 ; and to date, mutations in noncoding RNA have not been reported in disease. Mutations in Tie, especially Tie-2, have been implicated in vascular malformation, in particular congenital venous malformations.29 The imbalance in the ratio of tie-1 and noncoding lncRNA in patients diagnosed with vascular malformations suggests aberrant gene regulation in disease, but whether this regulation is critical for the pathogenesis of disease is unknown.
In conclusion, we have identified a novel long noncoding AS transcript that regulates tie-1 gene levels in vivo and in vitro, resulting in phenotypic consequences similar to mammals. To our knowledge, this is the first report on the identification of a long noncoding RNA that plays a functional regulatory role in vascular development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by start-up funds from the Children's Research Institute at the Medical College of Wisconsin and the National Institutes of Health (HL090712 grant; R.R.). K.L. and C.Z.C. are supported partly by funds from Advancing Healthier Wisconsin Grant (R.R.). G.V.S. is a recipient of the State of Wisconsin Breast Cancer Research postdoctoral fellowship.
National Institutes of Health
Authorship
Contribution: K.L., Y.B., A.V., Z.L., K.P., N.R.L., C.Z.C., G.V.S., B.Z., M.K.G., and M.A.H. performed experiments and analyzed data; K.L., Y.B., A.V., K.P., R.Q.M., G.A.W., and R.R. designed experiments; S.A.S. designed and performed bioinformatic analysis and analyzed data; P.E.N. provided human tissue samples; and G.A.W., M.A., and R.R. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.K.G. is Harvard Graduate School, Boston, MA.
Correspondence: Ramani Ramchandran, Department of Pediatrics, Medical College of Wisconsin, Children's Research Institute Developmental Vascular Biology Program, CRI C3420, 8701 Watertown Plank Rd, PO Box 26509, Milwaukee, WI 53226; e-mail: rramchan@mcw.edu.