Abstract
In this issue of Blood, Garzon and colleagues present evidence for a functional role of miR 29b in controlling DNMT levels in leukemia. Their work may provide insight into the mechanism of action of the azanucleotides and could potentially offer the first pharmacologically active miRNA.
MicroRNAs (miRNAs) are noncoding RNA sequences of 19 to 25 nucleotides in length, which bind through partial sequence homology to the 3′ or 5′ untranslated regions (3′ or 5′ UTRs) of target messenger RNAs (mRNAs). This results in either translational blockade or degradation of the mRNA.1 Recent work by a number of authors has demonstrated that miRNAs play key roles in tissue-specific and differentiation-specific expression of many proteins. Moreover, alterations in miRNA levels may play a role in carcinogenesis both in epithelial and hematapoietic malignancies.2
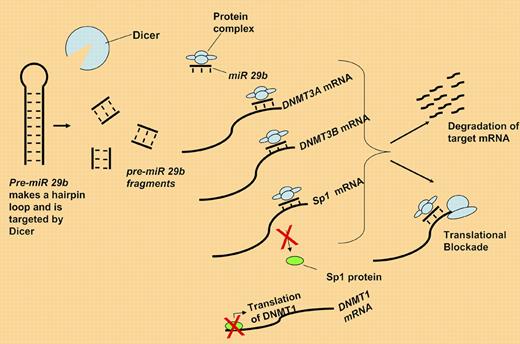
Pre-miR 29b is fragmented by dicer. The active miR 29b binds DNMT3A, 3B, and Sp1 mRNA either blocking translation or targeting them for degradation. Loss of Sp1 protein in turn down-regulates DNMT1 expression.
Pre-miR 29b is fragmented by dicer. The active miR 29b binds DNMT3A, 3B, and Sp1 mRNA either blocking translation or targeting them for degradation. Loss of Sp1 protein in turn down-regulates DNMT1 expression.
More than 1000 different miRNAs have now been identified, and work is ongoing to delineate their specific targets. Even in the absence of a complete understanding of their function, miRNA signatures have been shown to predict an aggressive phenotype in chronic lymphocytic leukemia (low miR 15a and 16-1) and non–small cell lung cancer (low miR 29 family expression).3,4 In addition, Marcucci and colleagues have recently demonstrated that miRNA signatures can discriminate outcome within the group of patients presenting with cytogenetically normal acute myeloid leukemia (AML) and a poor-risk molecular genotype (FLT3-ITD positive, NPM1 negative, or both).5 Further investigation of the functional significance of miRNA expression between cancer and normal tissues is now under way. What is clear is that miRNAs interact with both oncogenes and tumor suppressor genes. Furthermore, perturbations of miRNA expression by deletion, mutation, or hypermethylation contribute to the pathway dysregulation observed in many different malignancies.
Down-regulation of miR 29 family members in non–small cell lung cancer has been demonstrated to correlate with adverse prognosis. Recently, Fabbri and colleagues at The Ohio State University identified binding sites for miR 29 family members within the 3′ UTRs of DNA methyltransferases 3A and 3B (DNMT3A, 3B) and further demonstrated an inverse correlation between levels of miR 29 a, b, and c and the levels of DNMT3A and 3B in both lung cancer cell lines and primary non–small cell lung cancer samples.6 These investigators show that reintroduction of miR 29 family members (especially miR 29b) into lung cancer cell lines with low endogenous miR 29 expression in vitro resulted in both re-expression of methylated tumor suppressor genes and growth inhibition. The investigators postulated that synthetic miR 29s might be developed as a novel “epigenetic” therapy.
Here, Garzon and colleagues present evidence for a functional role of miR-29b in controlling levels of all 3 DNMTs within leukemia cells.7 As described in the figure, they demonstrate that miR 29b directly targets DNMT3A and 3B mRNA. Additionally, miR 29b interacts with the mRNA for Sp1, a known transcription factor for DNMT1, thus resulting in global DNA methyltransferase (DNMT) depletion. They further demonstrate that replacement of miR 29b oligonucleotides results in both re-expression of the methylated tumor suppressor genes p15INK4b and ESR1 (to an extent comparable to that using the DNMT inhibitor decitabine) and differentiation in AML cell lines.
The putative hypomethylating agents, 5-azacytidine and decitabine, are the first agents to have had a significant impact on prognosis for patients with high-risk myelodysplastic syndrome and have shown promising efficacy in elderly patients with AML.8 These agents are incorporated into DNA forming adducts with and inactivating DNMTs 1, 3A, and 3B which are responsible for maintenance and de novo methylation, respectively. Both aberrant DNA hypermethylation of tumor suppressor genes and overexpression of DNMT isoforms have been demonstrated in a variety of malignancies including AML. Although controversy remains with regard to the mechanism of action of these drugs, there is considerable circumstantial evidence that tumor suppressor gene re-expression plays an important role in their efficacy.9
The demonstration of a potentially pharmacologically active miRNA provides a unique opportunity for further insight into the activity of 5-azacytidine and decitabine. Debate continues over the mechanism of action of these so-called hypomethylating agents. However, no definitive evidence shows that methylation reversal is responsible for their clinical efficacy. In fact, if these agents exert their effects via hypomethylation of tumor suppressor genes (and resultant gene re-expression), their activity should be mimicked in in vitro and in vivo models by expression of exogenous miR 29b. While miRs could conceivably replace or augment the azanucleosides therapeutically, as with all genetically driven therapies, effective delivery to the tissue of interest will be a challenge. Ultimately, even if these agents do not prove to be useful pharmaceuticals, they should provide an opportunity to finally understand whether demethylation is central to the benefit seen with 5-azacytidine and decitabine.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■